Abstract
The products of the recF, recO, and recR genes are thought to interact and assist RecA in the utilization of single-stranded DNA precomplexed with single-stranded DNA binding protein (Ssb) during synapsis. Using immunoprecipitation, size-exclusion chromatography, and Ssb protein affinity chromatography in the absence of any nucleotide cofactors, we have obtained the following results: (i) RecF interacts with RecO, (ii) RecF interacts with RecR in the presence of RecO to form a complex consisting of RecF, RecO, and RecR (RecF–RecO–RecR); (iii) RecF interacts with Ssb protein in the presence of RecO. These data suggested that RecO mediates the interactions of RecF protein with RecR and with Ssb proteins. Incubation of RecF, RecO, RecR, and Ssb proteins resulted in the formation of RecF–RecO–Ssb complexes; i.e., RecR was excluded. Preincubation of RecF, RecO, and RecR proteins prior to addition of Ssb protein resulted in the formation of complexes consisting of RecF, RecO, RecR, and Ssb proteins. These data suggest that one role of RecF is to stabilize the interaction of RecR with RecO in the presence of Ssb protein. Finally, we found that interactions of RecF with RecO are lost in the presence of ATP. We discuss these results to explain how the RecF–RecO–RecR complex functions as an anti-Ssb factor.
Keywords: RecF pathway, multiprotein complexes, protein affinity chromatography, immunoprecipitation
Mutations in the recF gene have a wide variety of phenotypes: they increase UV sensitivity (1), delay the induction of the genes involved in DNA repair processes, reduce plasmid recombination, and decrease mutagenesis of single stranded (ss) DNA phage (2–6). Several genetic studies have suggested that the products of the recO and recR genes are involved and presumably interact with RecF in many of these processes. (i) The recF, recO, and recR genes belong to the same epistasis group (7). (ii) Certain phenotypes associated with recF, recO, and recR are suppressed by recA803 (8–10), recA441, recA730, and other recA mutations (11). (iii) The recombination of bacteriophage λ red, ninB mutant phage in recB recC sbcB sbcC require Escherichia coli host recF, recO, and recR gene products (12). (iv) Overproduction of recO and recR partially suppress the mutant phenotypes associated with recF mutations (13).
The single-stranded DNA binding protein (Ssb), which is crucial for DNA replication, also plays an important role in genetic recombination presumably affecting RecA protein catalyzed recombination and repair reactions (14, 15). The Ssb protein when added prior to RecA interferes with RecA binding to ssDNA thereby inhibiting joint-molecule formation and strand-exchange processes whereas, when added after RecA, Ssb protein stimulates RecA binding to ssDNA resulting in stimulation of the formation of the joint molecule and of strand-exchange processes (for review, see ref. 15). The interference by Ssb protein in RecA protein catalyzed recombination reactions appears to be important in vivo because elevated intracellular levels of Ssb protein cause defects in the RecF pathway of recombination (16, 17) and because a mutant recA, recA803 is a partial phenotype suppressor for recF, recO, and recR mutations (8, 9, 18). These studies have shown that the products of recF, recO, and recR assist RecA protein to negate the interference due to single-stranded DNA binding (Ssb) protein in recombination and repair reactions.
Umezu and Kolodner (19) have shown strong physical interactions between RecO and RecR and between RecO and Ssb proteins and weak interactions between RecO and RecF proteins, all in the absence of ATP and DNA. These authors have also identified complexes consisting of RecO and Ssb when either the preformed complexes of RecO–RecR were challenged with Ssb or when RecO and RecR were preincubated along with Ssb (19). Webb et al. (20) have shown that RecR protein interacts with RecF in the presence of double-stranded (ds) DNA and ATP. In vitro, RecO and RecR, independent of RecF, assist RecA in the utilization of ssDNA precomplexed with Ssb protein in joint molecule formation (21). These studies neither defined a role of RecF in DNA repair and recombination nor provided any evidence that RecF, RecO, and RecR proteins interact to form a complex. In this study, we have evaluated the interactions of RecF with RecO, RecR, and Ssb proteins individually and together. Since RecF binds ATP (22), the effect of ATP on these interactions has also been examined. Our results indicate that RecF protein physically interacts with RecO and with RecO–RecR complex. While RecF, like RecR, did not interact with Ssb protein, a complex of RecF and Ssb protein was obtained in the presence of RecO. Furthermore the RecF–RecO–RecR complex interacted with Ssb protein to form a RecF–RecO–RecR–Ssb complex. The interaction of RecF with RecO was lost in the presence of ATP. Since RecF protein has been shown to exhibit ATPase activity (20), we discuss how the RecF–RecO–RecR complex might function in DNA repair and recombination.
MATERIALS AND METHODS
Reagents.
PBE-94 resin, Superdex S-75 FPLC column, Phenyl-Sepharose CL-6B fast flow, and Superose-12 (FPLC) column were obtained from Pharmacia. Cibachrome blue 3GA-agarose 100, spermidine, ssDNA-cellulose resin, bovine serum albumin, and ATP were from Sigma. Phosphocellulose P-11 was from Whatman. Affi-Gel 10 and protein A-agarose beads were from Bio-Rad laboratories. Restriction endonuclease was from New England BioLabs, and Taq DNA polymerase was from Hoffman–La Roche. Anti-RecF and anti-Ssb antibodies were obtained by immunizing rabbits with SDS/polyacrylamide gel-purified RecF and Ssb proteins, respectively.
Cloning and Overexpression of recR.
The E. coli strain containing the plasmid pSJS1027 expressing recR+ under the control of the bacteriophage pL promoter (13) was provided for our use by Steven Sandler. A pET vector expressing a mutant recR gene under the T7 promoter was obtained from T. V. Wang (Chang Gung Medical College, Taiwan). Nucleotide sequencing data revealed that the region between the initiating codon and the HpaI site at codon 12 (23) of recR cloned under T7 promoter had a wild-type sequence. To eliminate the mutant sequence downstream of HpaI site, we removed the recR sequence from the HpaI site to the C-terminal end of recR from the pET vector and replaced with the corresponding sequence obtained from pSJS1027 to obtain pMQ6527. The integrity of the entire recR gene in this construct was verified by direct sequencing in Pharmacia automated laser fluorescence DNA sequencer using the autocycle sequencing kit and protocol.
Proteins.
The RecF (24), RecR (21), and Ssb (25) proteins were purified essentially as described. RecO was purified from E. coli RDK2582 containing the recO-overexpressing plasmid pRDK205 (26) on PBE-94 followed by Ssb protein affinity chromatography. Conditions used to overexpress RecO, subsequent cell lysis, ammonium sulfate precipitation, and chromatography of the PBE-94 column were essentially as described by Luisi-DeLuca and Kolodner (26). The flow through obtained from the PBE-94 column was directly applied to a Ssb protein affinity column preequilibrated in running buffer R (20 mM Tris·HCl, pH 7.5/0. 1 mM EDTA/0.1 mM DTT/100 mM NaCl), washed, and eluted with a linear gradient salt to 1 M NaCl in buffer R. Peak fractions containing RecO were eluted at approximately 650 mM NaCl were pooled, dialyzed against buffer containing 20 mM Tris·HCl (pH. 7.5), 1 mM DTT, 0.1 mM EDTA, and 60% glycerol. In some cases, a second chromatography step on Ssb affinity column was performed. The RecO protein purified from the Ssb protein affinity column was homogenous on silver-stained polyacrylamide gels.
Immunoprecipitation Experiments.
Polyclonal anti-RecF and anti-Ssb antibodies were used to precipitate RecF and Ssb proteins, respectively, along with any other proteins that interact with them. Specificity testing showed that antibodies were specific to the antigen used to raise antibodies. Reactions were carried out in a final volume of 10 μl of buffer I (35 mM Tris·HCl, pH 7.5/10 mM MgCl2/1 mM DTT/100 mM NaCl) containing RecF, RecO, RecR, and Ssb proteins, as indicated, each at a final concentration of 6 μM. Reaction mixes were incubated on ice for 30 min. In some experiments, buffer I also contained 1 mM ATP. At the end of the incubation period, 2 μl of a 1:100 dilution of either anti-RecF antisera or stock (5 mg/ml) of affinity-purified anti-Ssb antibodies was added and the reactions were continued for an additional 30 min at 37°C. At the end of the incubation, 2 μl of protein A-agarose beads (Bio-Rad) was added and the samples were incubated on ice with gentle rocking for a further 10 min. Protein A-agarose bound proteins were then separated from free proteins by centrifugation in an Eppendorf refrigerated microcentrifuge at 12,000 × g for 30 min followed by washing three times with buffer I. Samples were finally resuspended in Laemmli solubilization buffer, boiled, and electrophoresed (27). The Pharmacia Fast Gel system was used to separate proteins that were then visualized by Coomassie blue staining. When equimolar amounts of RecF, RecO, and RecR proteins were stained for 15 min using the Pharmacia Fast Gel system, we found that the RecO protein stained poorly. When stained manually for longer periods of time, the RecO retained more dye, but the intensity of dye binding was always less than that observed with other proteins. Comparisons of the known amount of pure proteins with the proteins present in the complexes precipitated by anti-RecF antibodies revealed that all input RecO appeared to coimmunoprecipitate with RecF. In some experiments, proteins in various lanes along with the known amounts of standards were quantified by densitometry in Millipore Bioimage densitometer. The measured intensity of each band was divided by the molecular weight of the respective protein and compared with each other to estimate molar ratios.
Ssb Protein Affinity Chromatography.
Purified Ssb protein (6 mg/ml) was coupled to an activated Affiprep-10 matrix (Bio-Rad) essentially as described by Formosa and Albert (28). Typically 3–4 mg of Ssb protein per ml of resin was bound under standard coupling conditions. After coupling, the resin was washed, packed in a Pharmacia HR 5/5 column (5 × 50 mm), and equilibrated with 10 bed volumes of buffer I prior to use. Proteins in various combinations, as indicated, were loaded, the column was washed to elute unbound proteins, and then bound proteins were eluted with a linear gradient of salt to 1 M NaCl using a Biologic chromatography system (Bio-Rad). Peak fractions by UV absorption at 280 nm were pooled and examined by SDS/PAGE in Pharmacia Fast Gel System.
Size Exclusion Chromatography.
Superose 12 (Mr range, 3000–300,000) column was used to detect protein complexes. Proteins were loaded in a final volume of 100 μl in buffer I at indicated concentrations. In some experiments protein complexes were prepared by incubating respective proteins in buffer I on ice for 30 min prior to loading onto the column. The peak fractions containing proteins were detected by UV absorption at 280 nm, collected, and analyzed by SDS/PAGE.
RESULTS
Interactions of RecF with RecO and RecR Proteins: RecF Interactions with RecR Are Mediated by RecO.
Immunoprecipitation experiments with anti-RecF antibodies indicated that RecF antibodies precipitated RecF but neither RecO nor RecR (compare Fig. 1, lane 1 with lanes 2 and 3). Immunoprecipitation of RecO by anti-RecF antibodies required only RecF (compare Fig. 1, lane 2 with lane 4), whereas precipitation of RecR by anti-RecF antibodies required both RecO and RecF (compare Fig. 1, lane 6 with lane 5). Since neither RecO nor RecR was individually precipitated by RecF antibodies (lanes 2 and 3) and precipitation of these proteins in various combinations by anti-RecF antibodies required RecF protein, we interpreted this to indicate that RecF physically interacted with RecO and with the RecO–RecR complex. Quantification of proteins present in lanes 4 and 6 revealed that the RecF and RecO (lane 4) and RecF, RecO, and RecR (lane 6) were present in 1:1 and 1:1:1 molar ratios, respectively. Similar ratios were also obtained when anti-Ssb antibodies were used in place of anti-RecF antibodies (see below). RecO was also found to be precipitated along with RecF by anti-RecF antibodies even in the presence of 150 mM NaCl but not in the presence of 300 mM NaCl. Under these conditions RecF protein was precipitated by anti-RecF antibodies (data not shown).
Figure 1.
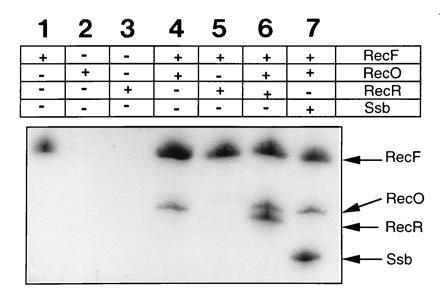
Immunoprecipitation of RecF, RecO, and RecR proteins by RecF antibodies. RecF, RecO, and RecR proteins (each at 6 μM) in various combinations were mixed in buffer I containing 35 mM Tris·HCl (pH 7.5), 10 mM MgCl2, 1 mM DTT, and 100 mM NaCl. RecF antibodies were added, incubated for 30 min on ice followed by the addition of protein A-agarose beads. Samples were then incubated at 4°C with gentle rocking for 10 min. Protein A-agarose-bound antigen–antibody complexes were separated from free proteins by centrifugation at 12,000 × g for 10 min followed by washing three times in buffer I. Samples were resuspended in Laemmli buffer, and proteins were separated on SDS/polyacrylamide gels and visualized by staining with Coomassie blue. Protein combinations used in these experiments are indicated.
Size-Exclusion Chromatography.
To further confirm the interactions among RecF, RecO, and RecR proteins, we analyzed the various complexes by size-exclusion chromatography on Superose-12 gel filtration columns (Mr cut-off, 3000–300,000). The elution profile of RecF protein compared with standards such as Ferritin (400 kDa; elution time, 16.32 min), catalase (230 kDa; elution time, 16.48 min), and aldolase (130 kDa; elution time, 17.23 min) suggested that native RecF protein exists in a range of multimeric forms (Fig. 2A) and was eluted predominantly in the void volume of the column. These results are consistent with our earlier published results (9) indicating that purification of monomeric RecF protein required the use of buffers containing high salt concentrations (1 M NaCl). The trailing of RecF protein probably reflects a heterogeneity in self association state of the molecule. Consistent with the data of Umezu and Kolodner (19) elutions of native RecO (26 kDa) (Fig. 2B; elution time, 28.4 min) and RecR (22 kDa) (Fig. 2C; elution time, 25 min) correspond to the positions of monomer and dimer respectively. When mixed RecO and RecR eluted as a complex (Fig. 2D; elution time, 24 min). When RecF protein was mixed either with RecO (Fig. 2E) or with both RecO and RecR (Fig. 2F) only one single peak eluted at the column void volume. (Note the disappearance of peaks corresponding to RecO and RecO–RecR and the appearance of single peak corresponding to RecF position.) SDS/PAGE analysis confirmed the presence of the various proteins in each peak (data not shown). Consistent with the data of Webb et al. (23), no interactions between RecF and RecR were detected (data not shown) nor could we identify complexes consisting of RecF and Ssb when pure proteins were mixed in buffer I and incubated prior to applying on to column (data not shown). Quantification of protein bands present in the sharp peak corresponding to Fig. 2F and comparisons with the known amounts of pure proteins revealed that the RecF, RecO, and RecR proteins were present in a 1:1:1 molar ratio. Elution profiles of the RecF–RecO–RecR complex indicate that it consist of at least three molecules of RecF (41 kDa), three of RecO (27 kDa), and three of RecR (22 kDa) proteins, which corresponds to 276 kDa.
Figure 2.
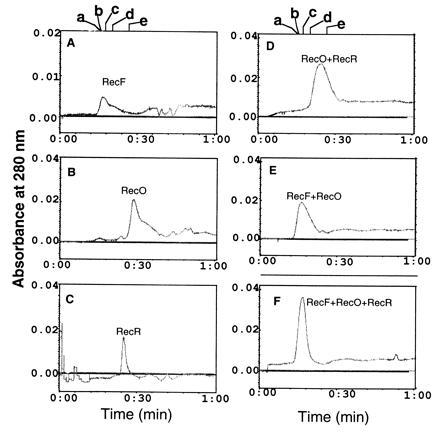
Elution profiles of RecF, RecO, and RecR proteins on Superose-12 sizing columns. RecF (1.5 μM) or RecO (6 μM) or RecR (6 μM) in a final volume of 100 μl of buffer I was loaded on Superose-12 HR 10/30 (Pharmacia) and eluted at a flow rate of 0.5 ml/min in the same buffer. Elution time is shown on the x axis and eluted proteins were collected by absorption at 280 nm. For detecting complexes, RecF, RecO, and RecR (each at 6 μM) in the indicated combinations were mixed in buffer I and incubated for 30 min on ice prior to loading on to the column. A, RecF; B, RecO; C, RecR; D, RecO–RecR complex; E, RecF–RecO complex; F, RecF–RecO–RecR complex. The trailing shoulder peak of RecO (B) indicate some interaction of RecO with the resin. The position of molecular mass standards labeled a–e is shown above A and D. a, Ferritin, 400 kDa; b, catalase, 230 k:da; c, aldolase, 130 kDa; d, ovalbumin, 45 kDa; e, chymotrypsin, 25 kDa.
Interactions of RecF with Ssb Protein Are Mediated by RecO.
The Ssb protein has been shown to disrupt RecO–RecR complexes to form RecO–Ssb complexes (19). To test whether Ssb protein disrupts RecF–RecO complexes to form RecO–Ssb complexes, immunoprecipitation with anti-RecF and anti-Ssb antibodies was carried out. The anti-RecF antibodies failed to precipitate Ssb protein in the presence of RecF, indicating the lack of any direct interaction between these two proteins (data not shown). However, when RecO was present along with RecF and Ssb proteins, anti-RecF antibodies did precipitate a RecF–RecO–Ssb complex (Fig. 1, lane 7). These data indicated that the interaction of RecF with RecO was not affected by the presence of Ssb protein. Similar results were obtained when anti-Ssb antibodies were used in place of anti-RecF antibodies (data not shown). Only RecO in the presence of Ssb and RecF when added along with RecO and Ssb were precipitated by anti-Ssb antibodies. Quantification of RecF and RecO proteins present in the complex precipitated by anti-Ssb antibodies revealed that RecF and RecO were present in a molar ratio of 1:1 (data not shown). As expected when mixed with Ssb protein, RecF and RecR individually or together were not precipitated by anti-Ssb antibodies. To further substantiate these results complexes were analyzed by Ssb protein affinity chromatography. Results were consistent with immunoprecipitation experiments. RecO (Fig. 3B) but not RecF (Fig. 3A) was retained on the Ssb column. Approximately 660 mM NaCl was found to be required to disrupt RecO–Ssb interactions. RecF protein was only bound to Ssb affinity column when in complex with RecO, indicating that its interactions with Ssb were mediated by RecO (Fig. 3D). Neither RecR alone (Fig. 3C) nor a mixture containing RecF and RecR (Fig. 3E) was retained on the Ssb affinity column. SDS/PAGE analysis confirmed the presence of the various proteins in each peak (Fig. 3 F, peaks for A–E).
Figure 3.
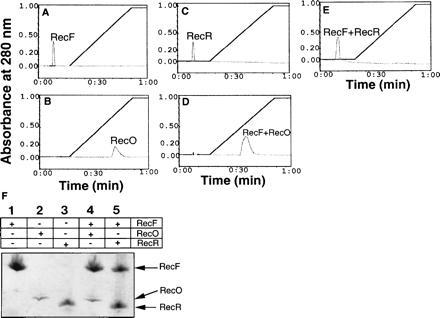
Elution profiles of proteins bound to the Ssb affinity column: RecF (38 μM), RecO (56 μM), and RecR (68 μM) proteins in reaction buffer I were applied to an Ssb protein affinity column preequilibrated with the same buffer I. The column was washed and bound proteins were eluted in a linear gradient of salt to 1 M NaCl. To form complexes proteins as indicated were mixed in the buffer and incubated on ice for 30 min prior to applying on to the column. Total time taken to complete a run is given on the x axis. A, RecF; B, RecO; C, RecR; D, RecF–RecO; E, RecF–RecR. 3F: Peak fractions were analyzed by SDS/PAGE and proteins were visualized by Coomassie blue staining. Fractions corresponding to each peak are indicated. The RecO protein appears to stain less under these conditions.
Interactions of the RecF–RecO–RecR Complex with Ssb.
Since the interaction of RecF with RecR is mediated by RecO (Fig. 1) and since RecF, RecO, and RecR proteins function as complex in repair and recombination processes, we reasoned that one role of RecF protein might be to stabilize the interactions of RecO–RecR in the presence of Ssb and to prevent RecR dissociation from the RecO–Ssb complex. To test this hypothesis, RecF, RecO, and RecR proteins were preincubated to form RecF–RecO–RecR complex. The resulting complex was then applied to the Ssb affinity column and eluted. As a control complexes containing RecO and RecR proteins were also applied to Ssb column and eluted under the same conditions. When preformed complexes of RecO and RecR were applied to the Ssb affinity column, a fraction of the total protein did not bind (Fig. 4), while elution of bound protein with a linear gradient of salt resulted in two protein peaks—a minor peak was eluted at 200 mM NaCl and a major peak was eluted at 650 mM NaCl (Fig. 4A). SDS/PAGE analysis identified the unbound protein as RecR and the bound proteins that eluted at 200 mM salt as RecO and RecR and that eluted at 650 mM salt as RecO alone (see Fig. 4A). This result is consistent with that of Umezu and Kolodner (19) indicating that Ssb displaces RecR when it binds to RecO, but in our experiments we do see a small amount of RecO–RecR bound to Ssb column. As expected when a RecF–RecO–RecR complex was applied, most of the proteins in the complex were retained and eluted as a single major peak at approximately 200 mM NaCl. The peak could also consist of the complexes of both RecF–RecO–RecR and RecO–RecR. SDS/PAGE analysis confirmed the presence of RecF, RecO, and RecR proteins in this complex (see Fig. 4C).
Figure 4.
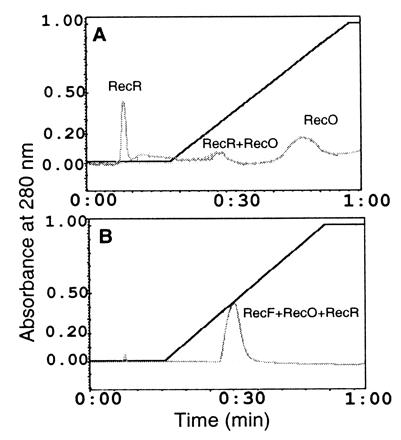
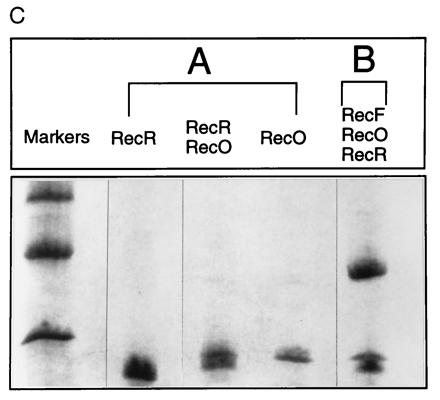
RecO at 150 μg and RecR at 150 μg, which correspond to 56 μM and 68 μM, respectively, or RecF at 150 μg, RecO at 150 μg, and RecR at 150 μg, which correspond to 38 μM, 56 μM, and 68 μM, respectively, were mixed in buffer I, and incubated on ice for 30 min prior to applying on Ssb affinity column. Bound proteins were eluted in linear gradient of salt. (A) RecO–RecR complex on Ssb affinity column. (B) RecF–RecO–RecR complex on Ssb affinity column. (C) Peak fractions were analyzed by SDS/PAGE and proteins were visualized by Coomassie blue staining. (A) Peak fractions obtained when RecO–RecR was applied to the Ssb column. (B) The peak fraction obtained when the RecF–RecO–RecR complex was applied. Each lane represent proteins present in corresponding peak fractions.
Further, to confirm these results we carried out immunoprecipitation experiments (Fig. 5). In these experiments, either all proteins were mixed together or RecF, RecO, and RecR were mixed first to form the RecF–RecO–RecR complex and then Ssb protein was added prior to treating with RecF antibodies. Additionally, RecF, RecO, and Ssb were mixed to form the RecF–RecO–Ssb complex and then challenged with RecR prior to adding RecF antibodies. RecR was precipitated by RecF antibodies only when a preformed RecF–RecO–RecR complex interacted with Ssb protein but not when all proteins were incubated together or when RecF–RecO–Ssb complex was formed first and then challenged with RecR (Fig. 5). Consistent results were obtained by Ssb antibodies (data not shown). Quantification of proteins present in various complexes and comparisons with the standards revealed that RecF/RecO and RecF/RecO/RecR were present in a molar ratios of 1:1 and 1:1:1, respectively (data not shown).
Figure 5.
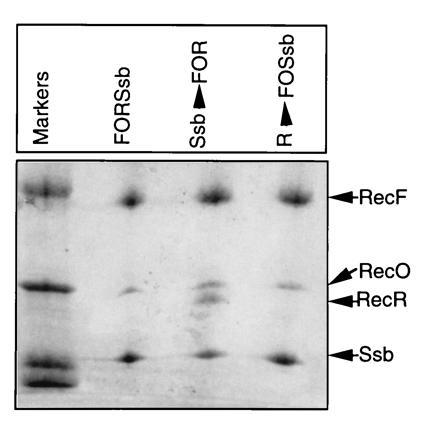
Immunoprecipitation of RecF, RecO, RecR, and Ssb protein complexes: RecF, RecO, RecR, and Ssb proteins (each at 6 μM) in indicated combinations in buffer I were incubated on ice for 30 min prior to treating with RecF antibodies. Antigen–antibody mixtures were processed essentially as described in Fig. 1. Order of addition of protein components is indicated. FORSsb indicates that all proteins were incubated together. Ssb before arrow indicates that Ssb protein was added to a preformed RecF–RecO–RecR complex. Similarly R before arrow indicates that RecR protein was added to a complex containing RecF–RecO–Ssb.
Interactions of RecF with RecO Are Lost in the Presence of ATP.
The RecF protein binds ATP, and binding of RecF to ATP is essential for its binding to dsDNA (22) and for it to exhibit preferential binding to gapped (g) DNA (24). Hence the effect of ATP on the interaction of RecF with RecO was examined using the immunoprecipitation techniques. RecF antibodies failed to precipitate RecO in the presence of ATP and RecF. Since interaction of RecF with RecR and Ssb are mediated by RecO, no other protein was precipitated under these conditions (data not shown). The lack of interaction of RecF with RecO in the presence of ATP was further confirmed on Ssb affinity columns where only RecO but not RecF was retained when a sample containing RecF, RecO, and ATP were loaded to Ssb affinity column that was preequilibrated with buffer containing 1 mM ATP (Fig. 6).
Figure 6.
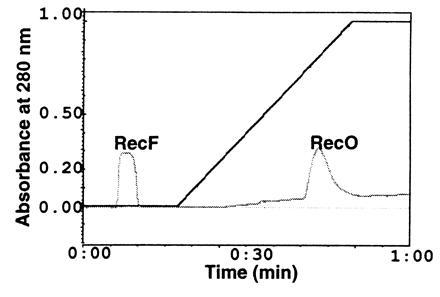
RecF interaction with RecO on Ssb affinity column in the presence of ATP: RecF (38 μM) and RecO (56 μM) were mixed in buffer I containing 1 mM ATP and incubated on ice. At the end of the incubation, the sample was applied to Ssb affinity column preequilibrated in buffer I containing 1 mM ATP. Bound proteins were eluted in a linear gradient of salt as described in Fig. 5. The identity of proteins present in the peak fractions was determined by SDS/PAGE and the proteins were visualized by Coomassie blue staining (data not shown).
DISCUSSION
The results presented in this paper show that RecF, RecO and RecR proteins form a complex in a 1:1:1 molar ratio. The elution profile of the complex in sizing columns (Fig. 2F) indicates that it appears to be at least heterotrimeric (276 kDa). Further experiments are required to define the precise composition of the complex.
Identification of complexes consisting of RecF, RecO, and RecR proteins is consistent with genetic data that indicate that recF, recO, and recR genes act at the same presynaptic step in recombination and repair (7, 8, 11, 18). It, however, presents a problem, which is the function of RecF protein in this complex. On one hand, genetic and biochemical evidence from a recA mutation that partially suppresses recF, recO, and recR mutations suggests that the function of the complex is to assist RecA protein in removing Ssb protein bound to ssDNA (8, 9). On the other hand, in vitro experiments show (19, 21) that RecO and RecR proteins can accomplish this assistance without RecF protein and in vivo experiments show (13) that overexpression of recR can partially suppress recF mutations in a recO+-dependent way. If RecF is at least partially dispensable, what then might its function be? We suggest that RecF targets the anti-Ssb activity of RecO and RecR proteins to gDNA substrates.
A targeting function for RecF protein is consistent with its preferential binding, in the presence of adenosine 5′-[γ-thio]triphosphate but not in the absence of nucleotide, to junctions between dsDNA and ssDNA at the boundaries of a gap (24). Such targeting could serve to restrict the anti-Ssb activity of RecO and RecR proteins to ssDNA substrates, one or both of whose junctions with dsDNA would not be occupied by polymerase or nuclease molecules. As suggested by Clark and Sandler (29), such a gDNA substrate could be one that is not under active metabolism by replication and, therefore, would require recombination to acquire a complementary strand. Another feature of the targeting hypothesis is that it could permit an orderly removal of Ssb and its replacement by RecA protein, while keeping the double-strand ends of the gap protected from other enzyme action.
A cartoon depicting our hypothesis is shown in Fig. 7. One feature of the cartoon is that contact is made between the RecF(ATP)–gDNA complex and RecO–RecR (step 2). A contact between RecR and RecF in the complex is in accord with the biochemical and electron microscopic results of Webb et al. (20). The contact with RecO seems to contradict our finding here that ATP causes the association of RecF and RecO to be lost. However, because DNA allows RecR to interact with RecF in the presence of ATP, we assume that the physiological DNA substrate, by hypothesis gDNA, will stabilize the association of RecO–RecR with RecF in the presence of ATP. RecF might make contacts with RecO directly as shown in step 3 or indirectly through RecR. Although systematic experiments on the effects of DNA binding are required, preliminary experiments indicate that anti-RecF antibodies precipitate RecF, RecO, and RecR proteins when they are mixed together in the presence of dsDNA and ATP (M.V.V.S.M. and S. P. Hegde, unpublished data), thus favoring the stabilization assumption. We are also identifying conditions under which the contacts of RecF with RecO and RecO–RecR complex in the presence of ATP can be stabilized. We found that the preformed complexes of RecF–RecO and RecF–RecO–RecR in the absence of DNA are not disrupted by ATP (M.V.V.S.M. and S. P. Hegde, unpublished data), thus supporting the existence of the RecF–RecO and RecF–RecO–RecR complexes in the presence of ATP under some conditions.
Figure 7.
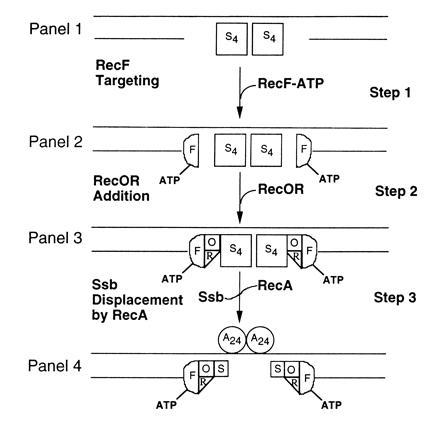
Cartoon for the RecF–RecO–RecR action in gap repair. For the sake of simplicity only presynaptic events are shown. The first letter of each protein is used to represent the respective protein: S, Ssb; F, RecF; O, RecO; R, RecR; A, 24. (Step 1) gDNA bound by Ssb tetramers (S4). (Step 2) RecF–ATP complex bound at gap junctions of gDNA. It is assumed that RecF protein, in the presence of ATP (step 3) recognizes and subsequently binds at gap junctions. (Step 3) Multiprotein–DNA complexes consisting of RecF–ATP, RecO, RecR, and Ssb. The RecO–RecR are targeted to RecF–ATP–gDNA complexes (step 3) and RecO makes direct contacts with Ssb. (Step 4) Presynaptic filaments consisting of RecF–RecO–RecR–Ssb and RecA proteins. Ssb tetramer displacement is initiated by RecA protein in the presence of RecF–RecO–RecR complex (step 3). Salient features of the cartoon are described in the text.
Another feature of the step 2 of the cartoon agrees with our findings. We have shown that RecF stabilizes association of RecR with a RecO–Ssb complex. Stabilization could occur through interaction of RecR with RecF or alteration of the strength of binding of RecR with RecO or with Ssb. Tests of the stability of the RecF–RecO–RecR complex in the presence of gDNA and Ssb need to be performed.
Other features of the cartoon require experimental test. For example, the cartoon assumes that the single-strand portion of the gDNA is bound by Ssb tetramers and that the RecF–ATP–RecO–RecR complex interacts with tetramers of Ssb protein from both sides of the gDNA substrate (step 3). The first assumption mandates the replacement of four Ssb molecules by 24 RecA molecules, since under some conditions each Ssb tetramer binds about 72 nt (14, 15) and each RecA monomer binds about 3 nt (for review, see ref. 15). The cartoon also shows in step 4 an Ssb monomer bound to the RecF–ATP–RecO–RecR complex after replacement of the Ssb by RecA protein. This remaining Ssb monomer is suggested by the RecO–RecR and Ssb proteins retained by the RecA–ssDNA presynaptic complex after RecO–RecR assists RecA in displacing Ssb in vitro (19). If the hypothetical contact between Ssb and RecO in the four-protein complex is stable in the presence of gDNA and ATP, it may limit the number of Ssb tetramers a RecF–ATP–RecO–RecR complex is able to assist RecA in removing without detaching from the gDNA. The weak ATPase activity of RecF protein, stimulated by RecR (20), may facilitate this turnover. Alternatively the multimeric character of the RecF–RecO–RecR complex found here may be retained by the hypothetical RecF–ATP–RecO–RecR–gDNA complex allowing a stoichiometric number of Ssb tetramers to be displaced.
Our results shown in Figs. 4 and 6 provide clues to how anti-Ssb activity (step 3) is initiated at the gap junctions. For example, binding of RecF to ATP could decrease the affinity of RecO toward RecF without actually affecting its affinity toward Ssb (Fig. 6), whereas removal of ATP presumably due to hydrolysis by the complex could increase affinity of RecO toward RecF and RecR with concomitantly decreasing its affinity toward Ssb (Fig. 4). These events could lead to modulation of Ssb–ssDNA complexes at gap junctions with concomitant nucleation of RecA.
The presence of a RecF–RecO–RecR–Ssb complex at the gap junctions of the presynaptic complex shown in the final panel of Fig. 6 suggests additional hypotheses concerning the functioning of RecF–RecO–RecR complex in recombination. Full details of the model we have in mind for will appear elsewhere (A.J.C., M.V.V.S.M., and S. J. Sandler, unpublished results).
Acknowledgments
We thank Drs. Steven J. Sandler and R. D. Kolodner for the recF, recO, and recR overexpression plasmids. We also acknowledge with gratitude receiving the RecO protein purification protocol prior to publication from Dr. Richard D. Kolodner. We thank Dr. Charles M. Radding for his interest and helpful comments on this manuscript, Dr. S. J. Sandler and Ivan Sandler for help in preparing the model figure, and Dr. V. M. Rao for helpful suggestion on immunoprecipitation experiments. This work is supported by Grant NP-847 from the American Cancer Society to M.V.V.S.M. and Grant AI-05371 from the National Institutes of Health to A.J.C.
Footnotes
Abbreviations: ssDNA, single-stranded DNA; dsDNA, double-stranded DNA; gDNA, gapped DNA; Ssb, single-stranded DNA binding protein.
References
- 1.Horii Z I, Clark A J. J Mol Biol. 1973;80:327–344. doi: 10.1016/0022-2836(73)90176-9. [DOI] [PubMed] [Google Scholar]
- 2.Volkert M R. J Bacteriol. 1989;171:99–103. doi: 10.1128/jb.171.1.99-103.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thoms B, Wackernagel W. J Bacteriol. 1987;169:1731–1736. doi: 10.1128/jb.169.4.1731-1736.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sassanfar M, Roberts J M. J Bacteriol. 1991;173:5869–5875. doi: 10.1128/jb.173.18.5869-5875.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark A J, Low K B. In: The Recombination of Genetic Material. Low K, editor. San Diego: Academic; 1988. pp. 155–215. [Google Scholar]
- 6.Ciesla Z, O’Brien P, Clark A J. Mol Gen Genet. 1987;207:1–8. doi: 10.1007/BF00331483. [DOI] [PubMed] [Google Scholar]
- 7.Lloyd R G, Buckman C. Biochimie (Paris) 1991;73:313–320. doi: 10.1016/0300-9084(91)90218-p. [DOI] [PubMed] [Google Scholar]
- 8.Madiraju M V V S, Templin A, Clark A J. Proc Natl Acad Sci USA. 1988;85:6592–6596. doi: 10.1073/pnas.85.18.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madiraju M V V S, Clark A J. Nucleic Acids Res. 1991;19:6295–6300. doi: 10.1093/nar/19.22.6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Volkert M R, Hartke M A. J Bacteriol. 1984;157:498–506. doi: 10.1128/jb.157.2.498-506.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang T C V, Chang H Y, Hung J L. Mutat Res. 1993;294:157–166. doi: 10.1016/0921-8777(93)90024-b. [DOI] [PubMed] [Google Scholar]
- 12.Sawitzke J A, Stahl F W. Genetics. 1992;130:7–16. doi: 10.1093/genetics/130.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sandler S S, Clark A J. J Bacteriol. 1994;176:3661–3672. doi: 10.1128/jb.176.12.3661-3672.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer R, Laine P S. Microbiol Rev. 1990;54:342–380. doi: 10.1128/mr.54.4.342-380.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kowalczykowski S C, Eggleston A K. Annu Rev Biochem. 1994;63:991–1043. doi: 10.1146/annurev.bi.63.070194.005015. [DOI] [PubMed] [Google Scholar]
- 16.Moreau P L. J Bacteriol. 1988;170:2493–2500. doi: 10.1128/jb.170.6.2493-2500.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moreau P L. J Mol Biol. 1987;194:621–643. doi: 10.1016/0022-2836(87)90239-7. [DOI] [PubMed] [Google Scholar]
- 18.Clark A J. Biochimie (Paris) 1991;73:523–532. doi: 10.1016/0300-9084(91)90124-j. [DOI] [PubMed] [Google Scholar]
- 19.Umezu K, Kolodner R D. J Biol Chem. 1994;269:30005–30013. [PubMed] [Google Scholar]
- 20.Webb B L, Cox M M, Inman R B. J Biol Chem. 1995;270:31397–31401. doi: 10.1074/jbc.270.52.31397. [DOI] [PubMed] [Google Scholar]
- 21.Umezu K, Chi N W, Kolodner R D. Proc Natl Acad Sci USA. 1993;90:3875–3879. doi: 10.1073/pnas.90.9.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madiraju M V V S, Clark A J. J Bacteriol. 1992;174:7705–7710. doi: 10.1128/jb.174.23.7705-7710.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahdi A A, Lloyd R G. Mol Gen Genet. 1989;216:503–510. doi: 10.1007/BF00334397. [DOI] [PubMed] [Google Scholar]
- 24.Hegde S, Rajagopalan M, Madiraju M V V S. J Bacteriol. 1996;178:184–190. doi: 10.1128/jb.178.1.184-190.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lohman T M, Green J M, Beyer R S. Biochemistry. 1986;25:21–25. doi: 10.1021/bi00349a004. [DOI] [PubMed] [Google Scholar]
- 26.Luisi-DeLuca C, Kolodner R. J Mol Biol. 1994;236:124–138. doi: 10.1006/jmbi.1994.1123. [DOI] [PubMed] [Google Scholar]
- 27.Laemmli E K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Formosa T, Burke R L, Alberts B M. Proc Natl Acad Sci USA. 1983;80:2442–2446. doi: 10.1073/pnas.80.9.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark A J, Sandler S J. CRC Rev Microbiol. 1994;20:125–142. doi: 10.3109/10408419409113552. [DOI] [PubMed] [Google Scholar]


