Abstract
The polymerization domain of ϕ29 DNA polymerase acquires a toroidal shape by means of an arch-like structure formed by the specific insertion TPR2 (Terminal Protein Region 2) and the thumb subdomain. TPR2 is connected to the fingers and palm subdomains through flexible regions, suggesting that it can undergo conformational changes. To examine whether such changes take place, we have constructed a ϕ29 DNA polymerase mutant able to form a disulfide bond between the apexes of TPR2 and thumb to limit the mobility of TPR2. Biochemical analysis of the mutant led us to conclude that TPR2 moves away from the thumb to allow the DNA polymerase to replicate circular ssDNA. Despite the fact that no TPR2 motion is needed to allow the polymerase to use the terminal protein (TP) as primer during the initiation of ϕ29 TP–DNA replication, the disulfide bond prevents the DNA polymerase from entering the elongation phase, suggesting that TPR2 movements are necessary to allow the TP priming domain to move out from the polymerase during transition from initiation to elongation. Furthermore, the TPR2-thumb bond does not affect the equilibrium between the polymerization and exonuclease activities, leading us to propose a primer-terminus transference model between both active sites.
INTRODUCTION
To accomplish efficient and fast genome replication, replicative DNA polymerases (also called replicases) rely on their functional association to other replicative proteins as helicases, that unwind the double-stranded DNA (dsDNA) allowing progression of the replication fork, and to processivity factors that held the DNA polymerase bound to the template strand (1,2). In addition, most of the replicases depend also on the presence of a short RNA or DNA molecule to act as primer, as they are unable to start de novo DNA synthesis.
Bacteriophage ϕ29 DNA polymerase is a proofreading family B DNA-dependent DNA polymerase (3) endowed with three distinctive features that allow it to overcome the replication issues mentioned above. It initiates DNA replication at the origins placed at both ends of the linear genome, which possesses a terminal protein (TP) covalently linked at its 5′ ends (TP–DNA), by catalyzing the addition of the initial dAMP onto the hydroxyl group of Ser232 of a primer TP (4–7). After a transition stage during which a sequential switch from TP priming to DNA priming takes place, the same polymerase replicates the entire TP–DNA molecule processively, without dissociating from the DNA (8), coupling processive DNA polymerization to strand displacement (8).
Resolution of the ϕ29 DNA polymerase structure has given insights into its strand displacement and processivity capacities (9). This replicase is formed by a N-terminal exonuclease domain and a C-terminal polymerization one, the relative orientation of them being similar to that described in other family B DNA polymerases (10–12). The polymerization domain of ϕ29 DNA polymerase contains the three universal thumb, palm and fingers subdomains that form an U-shaped groove adapted to bind dsDNA. In addition, a long β-hairpin, corresponding to the protein insertion named TPR2 (from Terminal Protein Region 2) and specifically present in the protein-primed DNA polymerases (13,14), emerges between the fingers and palm subdomains and opposite to the thumb, closing the polymerization domain. The resulting tunnel-like structure formed by TPR2, thumb, fingers and palm subdomains encircles the upstream dsDNA in a manner reminiscent of clamp proteins [(9,15), see Figure 1], conferring the high binding stability that accounts for the processivity displayed by ϕ29 DNA polymerase (16). Additionally, the TPR2 insertion, together with the fingers and palm subdomains, and residues from the exonuclease domain, forms another tunnel through which the single-stranded 5′ template overhang is threaded (9,15). The narrow dimensions of this tunnel compels to the separation of the two DNA strands before the template could reach the polymerization active site, the TPR2 subdomain acting as a wedge and providing the structural basis of the strand displacement capacity of ϕ29 DNA polymerase (9,16).
Figure 1.
Ribbon representation of ϕ29 DNA polymerase binary complex. Crystallographic data are from Protein Data Bank ID code 2PZS (15). DNA polymerase subdomains are coloured as follows: exonuclease domain in red, and palm, fingers, thumb, TPR1 and TPR2 subdomains in magenta, dark blue, green, orange and cyan, respectively. Primer and template strands are coloured in grey and yellow, respectively. The upstream dsDNA and downstream template tunnels are indicated with arrows. G410 and P562 residues, placed at the apexes of the TPR2 and thumb subdomains of ϕ29 DNA polymerase, respectively, were changed into cysteines and depicted as spheres.
Prior to a start of the ϕ29 TP–DNA replication, ϕ29 DNA polymerase forms a heterodimer with a primer TP to catalyse the formation of the TP–dAMP initiation product, as described above. The recently reported structure of the ϕ29 DNA polymerase/TP heterodimer shows that the TP forms an elongated three-domain structure. It contains a N-terminal domain, that does not interact with the DNA polymerase, an intermediate domain, that makes extensive contacts with DNA polymerase TPR1 (from Terminal Protein Region 1) subdomain, a protein insertion specifically present in protein-primed DNA polymerases, and a C-terminal priming domain that mimics duplex product DNA in its electrostatic profile and binding site in the polymerase (17). The structural fitting between the DNA polymerase upstream DNA tunnel and the TP priming domain allows TP Ser232 residue to be placed in a competent orientation to prime the initiation of ϕ29 DNA replication (17).
Comparison of the ϕ29 DNA polymerase apoenzyme, the binary complex and the DNA polymerase/TP heterodimer structures shows that the general conformation of the polymerization domain does not change upon binding the substrates, the downstream template and upstream DNA tunnels remaining closed (9,17,15). However, the ability displayed by the enzyme to polymerize on circular single-stranded DNA (ssDNA) implies that the arch-like structure formed by the TPR2 and thumb subdomains should not be a static one. Thus, ϕ29 DNA polymerase must be able to open the downstream template tunnel to allow the circular DNA to be properly bound at the polymerization domain (9). A similar opening should take place during the termination of ϕ29 genome replication, to allow the DNA polymerase to dissociate from the DNA to start a new replication round (9).
In this work, we study the role of ϕ29 DNA polymerase TPR2 subdomain movement by analyzing the consequences that the blockage of such motion has in the activities of the enzyme.
MATERIALS AND METHODS
Nucleotides and DNAs
[α-32P]dATP (3000 Ci/mmol) and [γ-32P]ATP (3000 Ci/mmol) were obtained from Amersham International Plc. Unlabelled nucleotides were purchased from Amersham Pharmacia Biotech Inc. Oligonucleotide sp1 (5′GATCACAGTGAGTAC) was 5′-labelled with [γ-32P]ATP and phage T4 polynucleotide kinase and purified electrophoretically on 8 M urea-20% polyacrylamide gels. Labelled sp1 was hybridized to oligonucleotide sp1c+6 (5′ TCTATTGTACTCACTGTGATC) in the presence of 0.2 M NaCl and 50 mM Tris–HCl, pH 7.5, resulting in a primer/template molecule that can be used in the coupled DNA polymerization/exonuclease and in the gel retardation assays. M13mp18 ssDNA was hybridized to the universal primer in the presence of 0.2 M NaCl and 60 mM Tris–HCl, pH 7.5, and the resulting molecule was used as a primer/template to analyse processive DNA polymerization coupled to strand displacement by ϕ29 DNA polymerase. Terminal protein-containing ϕ29 DNA (ϕ29 TP–DNA) was obtained as described (18).
Proteins
Phage T4 polynucleotide kinase was obtained from New England Biolabs. Wild-type ϕ29 DNA polymerase was purified from E. coli BL21(DE3) cells harbouring plasmid pJLPM (a derivative of pT7-4w2), as described (19). ϕ29 TP was purified as described (20).
Site-directed mutagenesis of ϕ29 DNA polymerase
The ϕ29 DNA polymerase G410C/P562C double mutant was obtained by using the QuikChange site-directed mutagenesis kit provided by Stratagene. Plasmid pJLPM, containing the ϕ29 DNA polymerase gene, was used as template of the mutagenesis reaction. Primers are complementary and designed to hybridize to opposite strands of the plasmid just where the mutation is located. Following temperature cycling using PfuTurbo DNA polymerase, and after treatment with DpnI endonuclease, the synthesized DNA was transformed into XL1-Blue supercompetent cells. The presence of the desired mutations and the absence of other mutations were confirmed by sequencing the entire gene.
Design and production of the mutants
Sites for insertion of the disulfide bridge were selected using the program SS-BOND (21). This program uses the backbone coordinates from the three-dimensional structure to select pairs of residues based on the calculated Cβ–Cβ distances. Subsequently, Sγ positions with ideal or nearly ideal geometries were generated for the selected pairs. An energy minimization procedure was used to select acceptable conformations. Taking into account these considerations, the residues G410 and P562 of the ϕ29 DNA polymerase, located in the TPR2 and thumb subdomains, respectively, were mutated to Cys. The ϕ29 DNA polymerase G410C/P562C double mutant was purified from E. coli BL21(DE3) cells harbouring the corresponding recombinant plasmid, essentially as described for the wild-type enzyme (19), except that β-mercaptoethanol was omitted during the final stage of purification. Mutant DNA polymerase was estimated to be >95% pure by 10% SDS–PAGE and further staining with Coomassie Brilliant Blue.
The samples were further diluted with a storage buffer containing 50 mM Tris–HCl, pH 8.0, 1 mM EDTA, 0.5 M NaCl, 50% (v/v) glycerol and 20 mM DTT to prevent the disulfide bond formation (reducing buffer), or 2 mM oxidized glutathione to favour disulfide bond formation (oxidative buffer), as described (22). After overnight incubation at 4°C, both fractions (reduced and oxidized) of each protein were incubated with a loading buffer lacking reducing agents and further analysed by 10% non-denaturing SDS-PAGE (without β-mercaptoethanol). The proteins were visualized by staining with Coomassie Brilliant Blue.
Polymerase/exonuclease coupled assay
The primer/template molecule sp1/sp1c+6 (15/21mer) contains a six-nucleotide long 5′-protruding end, and therefore, the primer strand can be used as substrate for both the 3′–5′ exonuclease activity and DNA-dependent DNA polymerization. The incubation mixture contained, in 12.5 μl, 50 mM Tris–HCl, pH 8.0, 10 mM MgCl2, 4% (v/v) glycerol, 0.1 mg/ml bovine serum albumin (BSA), 1.3 nM of 5′-labelled sp1/sp1c+6, 121 nM of wild-type or mutant ϕ29 DNA polymerase, respectively, and the indicated concentration of the four dNTPs. After incubation for 5 min at 25°C, the reactions were stopped by adding EDTA up to 10 mM. Samples were analysed by 8 M urea-20% PAGE and autoradiography. Polymerization or 3′–5′ exonuclease is detected as an increase or decrease in the size (15mer) of the 5′-labelled primer, respectively.
DNA gel retardation assay
The interaction of the wild-type and the ϕ29 DNA polymerase mutant with the primer/template structure was assayed using the 5′-labelled sp1/sp1c+6 (15/21mer) DNA. The incubation mixture contained, in a final volume of 20 µl, 12 mM Tris-HCl, pH 8.0, 1 mM EDTA, 20 mM ammonium sulphate, 0.1 mg/ml BSA, 10 mM MgCl2, 1 nM of sp1/sp1c+6 and the indicated increasing amounts of wild-type or mutant ϕ29 DNA polymerase. After incubation for 5 min at 4°C, the samples were subjected to electrophoresis in 4% (w/v) polyacrylamide gels (80:1, monomer:bis), containing 12 mM Tris-acetate, pH 7.5 and 1 mM EDTA, and run at 4°C in the same buffer at 8 V/cm, essentially as described (23). After autoradiography, ϕ29 DNA polymerase-dsDNA complexes were detected as a mobility shift (retardation) in the migrating position of the labelled DNA, and quantified by densitometry of the autoradiograms.
Replication of primed M13 DNA
The incubation mixture contained, in 25 µl, 50 mM Tris–HCl, pH 8.0, 4% (v/v) glycerol, 0.1 mg/ml BSA, 20 µM each dCTP, dGTP, dTTP and [α-32P]dATP (1 µCi), 8.5 nM of primed M13mp8 ssDNA, and 60 nM of wild-type or mutant ϕ29 DNA polymerase. To break the disulfide bond, proteins were previously incubated with 20 mM DTT for the indicated times. In all cases, reactions were started by adding 10 mM MgCl2. After incubation for 40 min at 30°C, the reactions were stopped by adding 10 mM EDTA and 0.1% SDS, and the samples were filtered through Sephadex G-50 spin columns. Relative activity was calculated from the Cerenkov radiation corresponding to the excluded volume. For size analysis, the labelled DNA was denatured by treatment with 0.7 M NaOH and subjected to electrophoresis in alkaline 0.7% agarose gels, as described (24). After electrophoresis, gels were dried and autoradiographed.
TP–dAMP formation (protein-primed initiation assay)
The incubation mixture contained, in 25 µl, 50 mM Tris–HCl, pH 8.0, 10 mM MgCl2, 20 mM ammonium sulfate, 4% (v/v) glycerol, 0.1 mg/ml BSA, 0.2 µM dATP, [α-32P]dATP (2 µCi), 1.6 nM of ϕ29 TP–DNA, 60 nM of purified TP and 60 nM of wild-type or mutant ϕ29 DNA polymerase, and incubated for the indicated times at 30°C. The reactions were stopped by adding 10 mM EDTA and 0.1% SDS, filtered through Sephadex G-50 spin columns, and further analysed by SDS-PAGE, as described (18). Quantitation was done by densitometric analysis of the labelled band corresponding to the TP–dAMP complex, detected by autoradiography.
Replication assay (protein-primed initiation plus elongation) using ϕ29 TP–DNA as template
The incubation mixture contained, in 25 µl, 50 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 20 mM ammonium sulfate, 4% (v/v) glycerol, 0.1 mg/ml BSA, 20 µM each dCTP, dGTP, dTTP and [α-32P]dATP (1 µCi), 1.6 nM of ϕ29 TP–DNA, 60 nM of purified TP and 60 nM of wild-type or mutant ϕ29 DNA polymerase. After incubation for the indicated times at 30°C, the reactions were stopped by adding 10 mM EDTA-0.1% SDS, and the samples were filtered through Sephadex G-50 spin columns. Relative activity was calculated from the Cerenkov radiation corresponding to the excluded volume. For size analysis, the labelled DNA was denatured by treatment with 0.7 M NaOH and subjected to electrophoresis in alkaline 0.7% agarose gels, as described (24). After electrophoresis, gels were dried and autoradiographed. For the analysis of the transition products, 150 nM of purified TP and 60 nM of wild-type or mutant ϕ29 DNA polymerase were used with the concentration of dNTP indicated in each case. After incubation for 6 min at 30°C, the samples were processed and subjected to 12% SDS-PAGE (360 × 280 × 0.5 mm3) electrophoresis to obtain enough resolution to distinguish TP bound to the first elongation products.
RESULTS
Formation of a disulfide bond between the TPR2 and thumb subdomains in ϕ29 DNA polymerase
The spatial nearness between residues G410 and P562, located at the very tip of the TPR2 and thumb subdomains, respectively, as well as the spatial orientation of their side chains (21), led us to replace those residues by cysteines (see Materials and Methods section) to analyse the effect that the introduction of a disulfide bond between both subdomains has in the ϕ29 DNA polymerase activities, as it would prevent potential TPR2 movement (Figure 1).
ϕ29 DNA polymerase double mutant G410C/P562C was overexpressed and purified, as described under Materials and Methods section. The wild-type and mutant ϕ29 DNA polymerases were stored in both, reducing and oxidative buffers, to prevent and allow disulfide bond formation, respectively, in the G410C/P562C mutant (Materials and methods section). The presence of the expected disulfide bond was analysed by non-reducing SDS-PAGE (in the absence of either DTT or ß-mercaptoethanol; Figure 2) comparing the mobility of the proteins under the reduced and oxidized state. As expected, there were no differences in the electrophoretic mobility between both states of the wild-type enzyme. On the contrary, whereas the reduced form of the G410C/P562C mutant migrated as the wild-type enzyme, the oxidized form migrated slightly faster (Figure 2), as it occurs in other proteins containing disulfide bonds (25,26). Further incubation of the oxidized form of mutant G410C/P562C in a buffer containing 20 mM DTT caused the recovery of the wild-type mobility in essentially 100% of the mutant protein (Figure 2B). This result indicates that the expected disulfide bond was formed between the two cysteines introduced in the ϕ29 DNA polymerase mutant. ϕ29 DNA polymerase contains seven cysteine residues that are not solvent accesible and far from the engineered ones. The absence of multimers of the ϕ29 DNA polymerase in the non-reducing SDS-PAGE indicates that no intermolecular disulfide bonds were formed.
Figure 2.
Effect of disulfide bond formation on the electrophoretic mobility of ϕ29 DNA polymerase G410C/P562C mutant. (A) Both, wild-type and G410C/P562C mutant polymerases were subjected to non-reducing 10% polyacrylamide-SDS gel electrophoresis, as described in Materials and methods section. Lanes a and c show the migration of the wild-type and mutant protein, respectively, stored in the reducing buffer (red) containing 20 mM DTT. Lanes b and d show the electrophoretic mobility of wild-type and mutant DNA polymerases, respectively, previously stored under oxidative (oxi) conditions, in the presence of 2 mM oxidized glutathione. Previous to the electrophoresis, the samples were incubated with a loading buffer lacking DTT and β-mercaptoethanol. The mobility displayed by the wild-type and G410C/P562C double mutant under the reducing and oxidative conditions are indicated at the right. (B) Both, wild-type and G410C/P562C mutant polymerases were stored overnight under oxidative conditions, in the presence of 2 mM oxidized glutathione (lanes e and g, respectively) and further incubated for 40 min in the presence of 20 mM DTT (lanes f and h). Previous to the electrophoresis, the samples were incubated with a non-reducing loading buffer. The change in the mobility displayed by the G410C/P562C double mutant from the oxidized to the reduced state is indicated at the right. Parts A and B come from two independent experiments.
The covalent linkage between the TPR2 and thumb subdomains of ϕ29 DNA polymerase does not affect the equilibrium between the polymerization and the 3′–5′ exonuclease activities on linear DNA
ϕ29 DNA polymerase is endowed with a 3′–5′ exonuclease activity that proofreads the polymerization errors (27,28). Despite the location of the polymerization and exonuclease active sites in two structurally independent domains, both activities work in concert to ensure a productive and faithful DNA synthesis, preventing the accumulation of errors in the newly synthesized strand while allowing a proper elongation rate (28). It has been shown that the wild-type equilibrium between synthesis and degradation in ϕ29 DNA polymerase depends on their relative catalytic rates, since these could be modulated in vitro as a function of the dNTP concentration (28), or by partial or total inactivation of specific residues belonging to each active site (29,30).
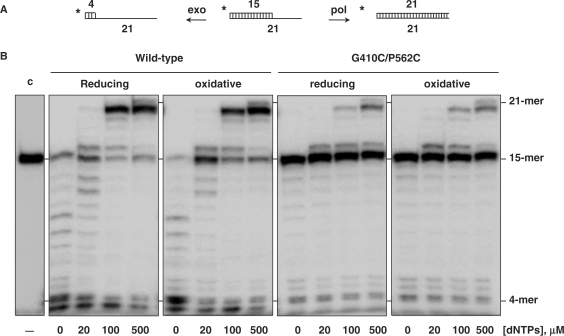
To analyse whether the introduced disulfide bond had an effect on replication of linear dsDNA by ϕ29 DNA polymerase, as well as on the equilibrium between its 3′-5′ exonuclease and 5′–3′ polymerization activities, the functional coupling between synthesis and degradation on the primer/template molecule sp1/sp1c+6 was analysed as a function of dNTP concentration (see Materials and methods section and scheme in Figure 3A). Thus, in the absence of nucleotides, the lack of competition by the polymerization activity caused the exonucleolytic degradation of most of the primer molecules by the wild-type enzyme (Figure 3B). By adding increasing amounts of dNTPs, exonucleolysis was progressively challenged, 100 nM dNTP being required to get a net polymerization balance. As it can be also noticed, the behaviour of the wild-type enzyme was independent of its oxidation state. This was also the case for the G410C/P562C mutant polymerase, although 80% of the DNA remained intact at the 15mer position. Interestingly, the formation of the disulfide bond (oxidative conditions) did not change the polymerization/exonucleolysis balance of the DNA polymerase mutant with respect to the wild-type enzyme.
Figure 3.
ϕ29 DNA polymerase G410C/P562C double mutant shows polymerization activity on a linear DNA molecule under both reducing and oxidative conditions. (A) Scheme of polymerization and 3′-5′ exonuclease activities on a primer/template molecule 15/21mer. (B) Polymerization and exonucleolysis competition. The assay was carried out as described in Materials and methods section using 32P-labelled molecule 15/21mer as primer/template DNA and the indicated concentration of dNTPs. Polymerization or 3′–5′exonuclease activities are detected as an increase or decrease, respectively, in the size (15mer) of the 5′-labelled primer. The 4mer position (last nucleotide before distributive exonucleolysis), the 15mer position (non-elongated primer), and the 21mer position (completely elongated primer) are indicated.
The results described above suggest that: (i) the placement of linear dsDNA in ϕ29 DNA polymerase would not require any movement of the TPR2 subdomain; and (ii) neither the G410 and P562 residues nor the TPR2 motion seem to be involved in the coordination between polymerization and exonucleolysis, as these activities were similarly affected in the mutant polymerase. On the other hand, the poor use of the substrate by the G410C/P562C mutant in its reduced and oxidized states is likely because of a deficient interaction with the dsDNA as a consequence of the mutations introduced.
To determine the DNA-binding efficiency of the G410C/P562C mutant under both the reducing and oxidative conditions, gel-shift assays were carried out using as substrate the same sp1/sp1c+6 molecule described above (see Materials and methods section). As shown in Figure 4, the wild-type enzyme rendered a single shifted band that most likely corresponds to a stable protein–DNA binding in which the primer-terminus is stabilized at the polymerization active site (31), and whose intensity depends on the amount of enzyme added. As it can be also observed in Figure 4, the DNA-binding capacity shown by the wild-type DNA polymerase was not dependent on its oxidation state. As expected, the G410C/P562C mutant was very deficient in the formation of a stable complex with the DNA in comparison with the wild-type enzyme, as much higher amounts of protein were required to give rise to a retardation band similar to that obtained with the wild-type enzyme, under both reducing and oxidative conditions (Figure 4 and Table 1).
Figure 4.
ϕ29 DNA polymerase G410C/P562C double mutant shows a reduced DNA binding capacity. The assay was carried out as described in Materials and methods section, using as substrate the synthetic 5′-labelled hybrid molecule sp1/sp1c+6, in the presence of the indicated concentrations of wild-type or mutant ϕ29 DNA polymerase under reducing and oxidative states. After gel electrophoresis, the mobility of free DNA (spl/splc+6) and that of the polymerase–DNA complex was detected by autoradiography.
Table 1.
Enzymatic activities of ϕ29 DNA polymerase G410C/P562C mutant
| Activity assay | Substrate(s) | G410C/P562C |
|
|---|---|---|---|
| reduced | oxidized | ||
| Stable DNA binding | sp1/sp1c+6 | 3 | 2 |
| M13 DNA replication | Primed M13 DNA, dNTP | 15 | 2 |
| ϕ29 TP–DNA replication | ϕ29 TP–DNA, dNTP, TP | 21 | <1 |
| ϕ29 TP–DNA initiation | ϕ29 TP–DNA, dATP, TP | 40 | 20 |
Numbers indicate the average percentage of activity relative to the wild-type enzyme (100%) obtained from several experiments.
These results suggest, on the one hand, that the double mutation introduced in the ϕ29 DNA polymerase affected the stable binding of the enzyme with dsDNA, explaining the impairment in the use of the DNA shown by the G410C/P562C mutant, and, on the other hand, that the formation of the disulfide bond does not entail an additional hindrance to bind linear DNA.
The formation of a disulfide bridge between the TPR2 and thumb subdomains prevents the ϕ29 DNA polymerase mutant G410C/P562C to polymerize on circular DNA
A circular template that lacks ends would be the best substrate to evaluate the TPR2 movement capacity, since an opening of the downstream template tunnel is required to place this molecule at the polymerization active site of ϕ29 DNA polymerase. Thus, we tested the ability of the mutant DNA polymerase to perform primed M13 DNA rolling circle replication (see Materials and methods section). In this assay, ϕ29 DNA polymerase starts polymerization from the 3′-OH group of a short DNA primer, and once the 5′ terminus is reached, strand displacement is required for ongoing polymerization (8). As shown in Figure 5A, the oxidation state of the wild-type polymerase did not influence its capacity to carry out efficiently M13 DNA replication, the length and amount of the replicated products being the same under the reducing and oxidative conditions. As expected, the low amount of replication products obtained by the G410C/P562C mutant was likely reflecting a poor binding efficiency to the dsDNA portion of the primed M13, in good agreement with the gel shift data presented above (Table 1). Nevertheless, the length of such products indicated that the double mutation introduced did not affect either the polymerization rate or the strand displacement ability of the ϕ29 DNA polymerase. On the contrary, the formation of the disulfide bond in mutant G410C/P562C (oxidative conditions) prevented the use of the circular M13 ssDNA as template, since no replication products were detected (Figure 5A and Table 1). As a control, progressive breakage of the disulfide bond, by incubating the oxidized form of the G410C/P562C mutant with 20 mM DTT, caused the gradual recovery of the polymerization activity, reaching the level initially displayed by the reduced form (Figure 5B).
Figure 5.
(A) The formation of a disulfide bond between the TPR2 and thumb subdomains prevents ϕ29 DNA polymerase G410C/P562C mutant to replicate a circular DNA molecule. Replication of primed M13 DNA was carried out as described in Materials and methods section using 60 nM of either wild-type or mutant DNA polymerase under reducing and oxidative states, and 40 µM dNTPs. After incubation for 40 min at 30°C, the length of the synthesized DNA was analysed by alkaline agarose gel electrophoresis. The migration position of unit-length M13 DNA is shown at the right. (B) Gradual disruption of the disulfide bond allows ϕ29 DNA polymerase G410C/P562C mutant to recover the ability to polymerize on circular ssDNA templates. The assay was carried out as described in Materials and methods section. The mutant ϕ29 DNA polymerase in the oxidative condition was incubated with 20 mM DTT for the indicated times, and the reaction was started by adding 10 mM MgCl2. After incubation for 40 min at 30°C, the length of the synthesized DNA was analysed by alkaline agarose gel electrophoresis. The migration position of unit-length M13 DNA is shown at the right. The separate panels come from the same experiment.
These results show the ability of ϕ29 DNA polymerase to accommodate a circular template at its polymerization active site by opening the arch-like structure formed by TPR2 and thumb subdomains.
The formation of a disulfide bridge between the TPR2 and thumb subdomains hinders transition from TP-primed initiation to DNA-primed elongation during ϕ29 TP–DNA replication
Replication of bacteriophage ϕ29 TP–DNA involves the TP-primed initiation at both origins of the linear TP–DNA molecule. Thus, ϕ29 DNA polymerase catalyses the template-directed formation of a covalent complex between the viral TP residue Ser232 (which acts as primer) and 5′-dAMP (initiation step). The TP priming domain (containing Ser232 residue) moves out of the active site as polymerase elongates the initiation product, the TP remaining bound to the DNA polymerase through a tight interaction between the polymerase TPR1 subdomain and the TP intermediate domain (transition step) (17,32,33). After incorporation of the tenth nucleotide, the TP dissociates from the DNA polymerase, entering in the DNA-primed elongation step to produce full-length ϕ29 DNA (19 285 bp) (4,6,32). To analyse the effect of the disulfide bond formation between the TPR2 and thumb subdomains of ϕ29 DNA polymerase in the protein-primed DNA replication process, we used a minimal ϕ29 TP–DNA replication system containing only primer TP and DNA polymerase (8). As it can be observed in Figure 6A, in its reduced state, the G410C/P562C mutant was able to synthesize full-length ϕ29 TP–DNA at a similar rate as the wild-type enzyme, although with a fivefold lower efficiency (Figure 6A and Table 1), most likely reflecting a deficient interaction with the substrates (TP and/or TP–DNA), similar to that displayed with DNA (see above). Interestingly, the formation of the disulfide bond in the G410C/P562C mutant (oxidative conditions) prevented TP–DNA replication, despite the linear shape of this substrate (Figure 6A and Table 1).
Figure 6.
(A) The formation of a disulfide bridge between the TPR2 and thumb subdomains prevents TP–DNA replication by ϕ29 DNA polymerase G410C/P562C mutant. The assay was carried out as described in Materials and methods section in the presence of 60 nM of wild-type or mutant ϕ29 DNA polymerase and 60 nM of TP. After incubation for 10 min at 30°C, the length of the synthesized DNA was analysed by alkaline agarose gel electrophoresis. The migration position of unit length ϕ29 TP–DNA is indicated at the right. (B) Both reducing and oxidative conditions allow ϕ29 DNA polymerase G410C/P562C double mutant to catalyse the formation of the TP–dAMP initiation product. The assay was performed as described under Materials and methods section in the presence of 10 mM MgCl2, 60 nM of ϕ29 DNA polymerase, 60 nM of TP and 1.6 nM of ϕ29 TP–DNA. After incubation at 30°C for the indicated times, samples were analysed by SDS–PAGE and autoradiography. The position of the TP–dAMP initiation complex is indicated.
To analyse whether the impairment shown by the oxidized form of the G410C/P562C mutant polymerase to replicate ϕ29 TP–DNA was due to a defect in its initiation capacity, we tested its ability to catalyse the template-directed addition of the initiating dAMP onto the TP (initiation reaction, see Materials and methods section). As shown in Figure 6B, both, the reduced and oxidized form of the G410C/P562C mutant synthesized TP–dAMP to a similar extent, showing a 2.5-fold (reduced) and 5-fold (oxidized) lower activity than the wild-type enzyme (Table 1). Thus, whereas such initiation level would explain the low TP–DNA replication activity displayed by the reduced form of the polymerase mutant, it would not account for the lack of replication products observed with the oxidized form.
To study the ability of the G410C/P562C mutant to accomplish the transition step, truncated elongation assays were carried out (see Materials and methods section). As shown in Figure 7, by providing dATP as the only nucleotide (initiation reaction), the wild-type enzyme yielded TP–dAMP and TP–(dAMP)2. In the presence of dATP, dGTP and dTTP, initiation products were elongated mainly up to TP–(dNMP)8 and TP–(dNMP)11, the expected sizes taking into account the sequence at the left and right origin, respectively (Figure 7). The pattern of products synthesized by the G410C/P562C mutant in its reduced state, as well as the ratio of elongation to initiation products were similar to those obtained with the wild-type DNA polymerase. By the contrary, in its oxidized state, the G410C/P562C mutant did not yield elongation products (Figure 7). Thus, the introduction of the disulfide bond between the TPR2 and thumb subdomains appears to impair the transition from initiation to elongation during ϕ29 DNA replication, most likely being the cause of the inability of the oxidized form of the G410C/P562C mutant to fulfil ϕ29 TP–DNA replication.
Figure 7.
A disulfide bond between the TPR2 and thumb subdomains blocks the transition stage. The assay was performed as described in Materials and methods section, using 10 mM MgCl2, 1.6 nM of ϕ29 TP–DNA, 150 nM of TP, 60 nM of wild-type or mutant ϕ29 DNA polymerase and 40 µM of the indicated dNTPs. After incubation for six minutes at 30°C, the different transition products were detected and analysed by high-resolution SDS-PAGE. Brightness and contrast of the image were adjusted with the aim to detect elongation products with G410C/P562C mutant in the presence of dATP, dGTP and dTTP under oxidative conditions. The position of the different transition products is indicated at the left. Every panel presented corresponds to the same experiment.
DISCUSSION
ϕ29 DNA polymerase is an outstanding enzyme endowed with high processivity and strand displacement capacities that allow it to replicate the ϕ29 double-stranded linear genome without unwinding proteins and processivity factors, unlike most replicases (8). Such intrinsic abilities are conferred by the presence of the TPR2 insertion between the fingers and palm subdomains, as it closes the universally conserved U-shaped dsDNA groove in the polymerization domain by forming an arch-like structure with the thumb subdomain. This fact entails the formation of an upstream DNA tunnel that encircles the DNA product, conferring processivity, and a downstream template tunnel that forces the separation of the DNA strands as replication fork progresses. Here, we have studied whether the arch-like structure needs to be open with certain DNA templates. To this end, the TPR2 and thumb subdomains have been covalently linked through the formation of a disulfide bond between cysteines at positions 410 (TPR2) and 562 (thumb) of the ϕ29 DNA polymerase by constructing the G410C/P562C double mutant.
The TPR2 movement is required to polymerize on circular DNA
DNA binding depends mostly on specific ligands located at the thumb and TPR2 subdomains (34,15). In the Results section, it has been shown that the change of residues G410 and P562 into cysteine in the G410C/P562C mutant impairs the ability of the DNA polymerase to bind the linear primer/template DNA, even when the disulfide bond formation is not allowed (reducing conditions). Despite their nearness, residues G410 (TPR2 subdomain) and P562 (thumb subdomain) do not contact each other in any of the reported structures of ϕ29 DNA polymerase (9,15,17). However, modelling of the TPR2 and thumb subdomains structure in the DNA polymerase G410C/P562C mutant predicts a steric hindrance between the cysteines introduced at those positions (Figure 1). This fact could prevent the proper placement of TPR2 and thumb subdomains, affecting negatively the binding of the DNA at the polymerization domain. Interestingly, under oxidative conditions, the presence of the disulfide bond in mutant G410C/P562C DNA polymerase did not prevent the replication of linear primer/template structures. Thus, it can be inferred that the TPR2 subdomain does not need to move to allow the binding and subsequent elongation of this kind of linear DNA molecules which could diffuse through the upstream DNA tunnel, the protruding template strand passing through the downstream template tunnel (Figure 8A, left panel).
Figure 8.
(A) Binding of linear and circular DNA substrates by ϕ29 DNA polymerase. ϕ29 DNA polymerase subdomains are coloured as in Figure 1. The binding of linear primer/template (left panel) would involve the simple diffusion of the 5′-overhanged end of the template strand through the downstream template tunnel (green arrow indicates the diffusion direction), without requiring the previous opening of the TPR2-thumb arch-shaped structure, the dsDNA portion being encircled and stabilized by the downstream DNA tunnel. Conversely, binding of circular ssDNA (right panel) requires the TPR2 subdomain to peel away (indicated with a green arrow) to allow this substrate to be properly positioned at the downstream tunnel. Once bound, the dsDNA portion, by means of the contacts established with residues of the TPR2 and thumb subdomains, will crosslink such subdomains, enclosing the downstream DNA tunnel by a backwards motion of TPR2. (B) Close-up view of the primer-terminus at the polymerization and exonuclease active sites. The figure was done by superposition of the ϕ29 DNA polymerase binary complex [PDB code 2PZS (15)] with the structure of the apoenzyme with a 5 nt ssDNA bound at its 3′–5′ exonuclease active site [PDB 1XHZ; (9)]. DNA polymerase is space-filling represented. Template strand is coloured in yellow, whereas primer strand is coloured in dark blue and green in the polymerization and editing modes, respectively.
Conversely, and as expected, the covalent linkage of the TPR2 and thumb subdomains prevented the DNA polymerase to replicate circular ssDNA templates. The gradual recovery of the activity on such DNA as the disulfide bond breaks indicates that the TPR2 subdomain moves away from the thumb subdomain to allow positioning of the template at the downstream template tunnel (Figure 8A, right panel). In contrast, the channel-shaped polymerization domain of the rest of the reported DNA polymerases would not pose any structural restriction to bind circular DNAs as they do with linear DNAs (35–37).
It has been demonstrated that ϕ29 DNA polymerase replicates circular templates processively due to a tight DNA binding (8). This feature, together with the presence of DNA ligands in the TPR2 and thumb subdomains of ϕ29 DNA polymerase, lead us to hypothesize that once the DNA is bound at the polymerase after the opening of the TPR2-thumb arch-shaped structure, it would act as an electrostatic crosslink between both subdomains, preventing motions that, otherwise, could cause premature dissociation. In other replicative DNA polymerases, the channel-shaped structure of their polymerization domains causes high-rate DNA dissociation (38). However, these DNA polymerases remain attached to their corresponding processivity factors to guarantee efficient and fast DNA replication, as it has been demonstrated for T4 DNA polymerase (39).
Primer-terminus switching between polymerization and exonuclease active sites: a passive diffusion transference model
Comparison of the structures of many apo DNA polymerases with their corresponding binary complexes showed that the major conformational changes occur mainly in their thumb subdomains, composed of two microdomains with a clear helicoidal character, linked by a flexible region (40–45). Conversely, the ϕ29 DNA polymerase thumb subdomain has an unusual structure since it is small and has very little helical character, being mainly constituted by a long β-hairpin structure without identifiable microdomains (9). Moreover, comparison of the apo enzyme with the binary complexes shows that the thumb subdomain does not rotate upon DNA binding (15). In the present work, it has been shown that the prevention of a potential thumb tip movement by introducing the disulfide bond in the ϕ29 DNA polymerase G410C/P562C mutant, did not affect the partitioning of the primer-terminus between the polymerization and editing active sites. From this result it could be concluded: (i) in ϕ29 DNA polymerase the primer-terminus switching between both active sites is not guided by a rotation of the thumb tip; (ii) the impeded motion of the TPR2 subdomain, together with the narrow dimensions of the downstream template tunnel (∼10 Å) suggest that rotation of the DNA is not required to transfer the primer-terminus between the polymerization and editing active sites in ϕ29 DNA polymerase, most likely as there is not any structural barrier in between (Figure 8B). An intriguing question which remains to be solved is how the frayed terminus travels to the exonuclease active site. Considering the ϕ29 DNA polymerase thumb subdomain as a nearly static structure, the primer switching would be accomplished by a passive diffusion of the frayed primer-terminus (Figure 8B). The energetically unfavourable gradual melting of three to four base pairs should be progressively offset by new and specific interactions established with DNA ligands of the thumb subdomain, as previously suggested (34). Such interactions would also channel the primer-terminus in the appropriate orientation, and also with ssDNA ligands of the exonuclease domain responsible for the stabilization of the primer-terminus at the editing active site (9,34,46,47).
TPR2 motion is required during transition from initiation to elongation in ϕ29 TP–DNA replication
As described above, ϕ29 DNA polymerase is an unique enzyme endowed with the ability to use a protein (TP) as primer to initiate ϕ29 genome replication (4–6). The structure of the ϕ29 DNA polymerase/TP heterodimer has been recently reported, the conformation of the polymerase in the complex being largely unchanged from that of the apo polymerase (17). TP forms an extended structure with a C-terminal priming domain comprised of a four-helix bundle containing the Ser232 residue, which provides the priming OH-group for DNA synthesis and lies in a loop at the end of the domain, close to the active site of the polymerase. The overall dimensions as well as the negative charge of the priming domain of TP mimics DNA in its interactions with the polymerization domain (17). Thus, the similar TP-primed initiation activity displayed by mutant G410C/P562C in both, its reduced and oxidative states, shows that a TPR2 motion is not required to allow the priming domain of TP to prime TP–dAMP formation, in good agreement with the results obtained with linear primer/template molecules. In addition, as initiation of ϕ29 DNA replication is a template-directed reaction, it is worth noting that the ability of mutant G410C/P562C to catalyse the templated formation of the TP–dAMP product also implies that the 3′ end of the template strand of ϕ29 TP–DNA diffuses through the downstream template tunnel to reach the polymerization active site without requiring any TPR2 movement. It should be noticed that here the diffusion direction through the template tunnel is opposite to that described to occur with the linear primer/template DNA (see above).
Interestingly, the disulfide bond prevented the transition from the initiation to the elongation phase of ϕ29 DNA replication. During this stage, as synthesis proceeds, the duplex product occupies the upstream DNA tunnel, causing the progressive displacement of the priming domain of TP (17). Since this TP domain is simultaneously bound by TPR2 and thumb residues, a dynamic formation/breakage of interactions between both proteins, DNA polymerase and TP, would be necessary. Our results suggest that TPR2 movement is required to accomplish the continuous accommodation of the DNA polymerase subdomains to the changing dimensions of the TP priming domain as it leaves the DNA polymerase following a predicted helicoidal fashion (17). Although such TPR2 motions could potentially favour dissociation of the DNA polymerase from the DNA, in most of the replicating complexes the dissociation risk by the TPR2 movement could be counteracted by the stable interaction between the DNA polymerase TPR1 subdomain and the intermediate domain of TP (17,33), both proteins remaining bound to each other until the 10th nucleotide is added (32). From this point, the newly synthesized DNA would be held firmly by the TPR2 and thumb subdomains to guarantee processive replication, the DNA acting as a crosslinker between both protein regions. Once ϕ29 DNA replication is finished, TPR2 should move out to allow DNA polymerase to dissociate from the DNA, as the last templating nucleotide is covalently linked to a TP molecule whose dimensions impede the passage through the template tunnel. In this replication stage, such a movement could be facilitated by the absence of contacts between the template strand and the TPR2 subdomain that would release its electrostatic crosslinking to the thumb subdomain. Additionally, the covalently linked TP molecule to the 5′ end of the template strand would help the TPR2 to move away from the thumb, acting as a ‘wedge’ to allow the 5′ templating nucleotide to reach the polymerization active site to direct the last nucleotide insertion event.
FUNDING
Spanish Ministry of Education and Science (grant BFU 2005-00733 to M.S.); Comunidad Autónoma de Madrid (grant S-0505/MAT-0283 to M.S.); Institutional grant from Fundación Ramón Areces to the Centro de Biología Molecular ‘Severo Ochoa’. Funding for open access charge: Spanish Ministry of Education and Science (grant BFU 2005-00733).
Conflict of interest statement. None declared.
REFERENCES
- 1.Kornberg A, Baker T. DNA Replication. 2nd. New York: W.H. Freeman; 1992. [Google Scholar]
- 2.Watson J, Baker T, Bell S, Gann A, Levine M, Losick R. Molecular Biology of the Gene. 5th. Plainview, New York: Cold Spring Harbor Lab Press; 2004. [Google Scholar]
- 3.Bernad A, Zaballos A, Salas M, Blanco L. Structural and functional relationships between prokaryotic and eukaryotic DNA polymerases. EMBO J. 1987;6:4219–4225. doi: 10.1002/j.1460-2075.1987.tb02770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salas M. Protein-priming of DNA replication. Annu. Rev. Biochem. 1991;60:39–71. doi: 10.1146/annurev.bi.60.070191.000351. [DOI] [PubMed] [Google Scholar]
- 5.Salas M, Miller J, Leis J, DePamphilis M. Mechanisms for Priming DNA Synthesis. New York: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 6.Salas M. Mechanisms of initiation of linear DNA replication in prokaryotes. Genet. Eng. 1999;21:159–171. doi: 10.1007/978-1-4615-4707-5_8. [DOI] [PubMed] [Google Scholar]
- 7.Salas M, de Vega M. In: Bacteriophage Protein-Primed DNA Replication. Hefferon KL, editor. Trivandrum, Kerala (India): Ithaca, Research Signpost; 2006. [Google Scholar]
- 8.Blanco L, Bernad A, Lázaro JM, Martín G, Garmendia C, Salas M. Highly efficient DNA synthesis by the phage ϕ29 DNA polymerase. Symmetrical mode of DNA replication. J. Biol. Chem. 1989;264:8935–8940. [PubMed] [Google Scholar]
- 9.Kamtekar S, Berman AJ, Wang J, Lázaro JM, de Vega M, Blanco L, Salas M, Steitz TA. Insights into strand displacement and processivity from the crystal structure of the protein-primed DNA polymerase of bacteriophage ϕ29. Mol. Cell. 2004;16:609–618. doi: 10.1016/j.molcel.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 10.Wang J, Sattar AK, Wang CC, Karam JD, Konigsberg WH, Steitz TA. Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69. Cell. 1997;89:1087–1099. doi: 10.1016/s0092-8674(00)80296-2. [DOI] [PubMed] [Google Scholar]
- 11.Hopfner KP, Eichinger A, Engh RA, Laue F, Ankenbauer W, Huber R, Angerer B. Crystal structure of a thermostable type B DNA polymerase from Thermococcus gorgonarius. Proc. Natl Acad. Sci. USA. 1999;96:3600–3605. doi: 10.1073/pnas.96.7.3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hashimoto H, Nishioka M, Fujiwara S, Takagi M, Imanaka T, Inoue T, Kai Y. Crystal structure of DNA polymerase from hyperthermophilic archaeon Pyrococcus kodakaraensis KOD1. J. Mol. Biol. 2001;306:469–477. doi: 10.1006/jmbi.2000.4403. [DOI] [PubMed] [Google Scholar]
- 13.Blasco MA, Blanco L, Parés E, Salas M, Bernad A. Structural and functional analysis of temperature-sensitive mutants of the phage ϕ29 DNA polymerase. Nucleic Acids Res. 1990;18:4763–4770. [PMC free article] [PubMed] [Google Scholar]
- 14.Dufour E, Méndez J, Lázaro JM, de Vega M, Blanco L, Salas M. An aspartic acid residue in TPR-1, a specific region of protein-priming DNA polymerases, is required for the functional interaction with primer terminal protein. J. Mol. Biol. 2000;304:289–300. doi: 10.1006/jmbi.2000.4216. [DOI] [PubMed] [Google Scholar]
- 15.Berman AJ, Kamtekar S, Goodman JL, Lázaro JM, de Vega M, Blanco L, Salas M, Steitz TA. Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases. EMBO J. 2007;26:3494–3505. doi: 10.1038/sj.emboj.7601780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodríguez I, Lázaro JM, Blanco L, Kamtekar S, Berman AJ, Wang J, Steitz TA, Salas M, de Vega M. A specific subdomain in ϕ29 DNA polymerase confers both processivity and strand-displacement capacity. Proc. Natl Acad. Sci. USA. 2005;102:6407–6412. doi: 10.1073/pnas.0500597102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamtekar S, Berman AJ, Wang J, Lázaro JM, de Vega M, Blanco L, Salas M, Steitz TA. The phi29 DNA polymerase:protein-primer structure suggests a model for the initiation to elongation transition. EMBO J. 2006;25:1335–1343. doi: 10.1038/sj.emboj.7601027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peñalva MA, Salas M. Initiation of phage ϕ29 DNA replication in vitro: formation of a covalent complex between the terminal protein, p3, and 5′-dAMP. Proc. Natl Acad. Sci. USA. 1982;79:5522–5526. doi: 10.1073/pnas.79.18.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lázaro JM, Blanco L, Salas M. Purification of bacteriophage ϕ29 DNA polymerase. Methods Enzymol. 1995;262:42–49. doi: 10.1016/0076-6879(95)62007-9. [DOI] [PubMed] [Google Scholar]
- 20.Zaballos A, Lázaro JM, Méndez E, Mellado RP, Salas M. Effects of internal deletions on the priming activity of the phage ϕ29 terminal protein. Gene. 1989;83:187–195. doi: 10.1016/0378-1119(89)90104-2. [DOI] [PubMed] [Google Scholar]
- 21.Hazes B, Dijkstra BW. Model building of disulfide bonds in proteins with known three-dimensional structure. Protein Eng. 1988;2:119–125. doi: 10.1093/protein/2.2.119. [DOI] [PubMed] [Google Scholar]
- 22.Mukherjee S, Brieba LG, Sousa R. Discontinuous movement and conformational change during pausing and termination by T7 RNA polymerase. EMBO J. 2003;22:6483–6493. doi: 10.1093/emboj/cdg618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carthew RW, Chodosh LA, Sharp PA. An RNA polymerase II transcription factor binds to an upstream element in the adenovirus major late promoter. Cell. 1985;43:439–448. doi: 10.1016/0092-8674(85)90174-6. [DOI] [PubMed] [Google Scholar]
- 24.McDonell MW, Simon MN, Studier FW. Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J. Mol. Biol. 1977;110:119–146. doi: 10.1016/s0022-2836(77)80102-2. [DOI] [PubMed] [Google Scholar]
- 25.Schlegel RA, Thomas CA., Jr Some special structural features of intracellular bacteriophage T7 concatemers. J. Mol. Biol. 1972;68:319–345. doi: 10.1016/0022-2836(72)90216-1. [DOI] [PubMed] [Google Scholar]
- 26.Mansfeld J, Vriend G, Dijkstra BW, Veltman OR, Van den Burg B, Venema G, Ulbrich-Hofmann R, Eijsink VG. Extreme stabilization of a thermolysin-like protease by an engineered disulfide bond. J. Biol. Chem. 1997;272:11152–11156. doi: 10.1074/jbc.272.17.11152. [DOI] [PubMed] [Google Scholar]
- 27.Blanco L, Salas M. Characterization of a 3′-5′ exonuclease activity in the phage ϕ29-encoded DNA polymerase. Nucleic Acids Res. 1985;13:1239–1249. doi: 10.1093/nar/13.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garmendia C, Bernad A, Esteban JA, Blanco L, Salas M. The bacteriophage ϕ29 DNA polymerase, a proofreading enzyme. J. Biol. Chem. 1992;267:2594–2599. [PubMed] [Google Scholar]
- 29.Blanco L, Salas M. Mutational analysis of bacteriophage ϕ29 DNA polymerase. Methods Enzymol. 1995;262:283–294. doi: 10.1016/0076-6879(95)62024-9. [DOI] [PubMed] [Google Scholar]
- 30.Blanco L, Salas M. Relating structure to function in ϕ29 DNA polymerase. J. Biol. Chem. 1996;271:8509–8512. doi: 10.1074/jbc.271.15.8509. [DOI] [PubMed] [Google Scholar]
- 31.Méndez J, Blanco L, Lázaro JM, Salas M. Primer-terminus stabilization at the ϕ29 DNA polymerase active site. Mutational analysis of conserved motif TX2GR. J. Biol. Chem. 1994;269:30030–30038. [PubMed] [Google Scholar]
- 32.Méndez J, Blanco L, Salas M. Protein-primed DNA replication: a transition between two modes of priming by a unique DNA polymerase. EMBO J. 1997;16:2519–2527. doi: 10.1093/emboj/16.9.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pérez-Arnaiz P, Longás E, Villar L, Lázaro JM, Salas M, de Vega M. Involvement of phage ϕ29 DNA polymerase and terminal protein subdomains in conferring specificity during initiation of protein-primed DNA replication. Nucleic Acids Res. 2007;35:7061–7073. doi: 10.1093/nar/gkm749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pérez-Arnaiz P, Lázaro JM, Salas M, de Vega M. Involvement of ϕ29 DNA polymerase thumb subdomain in the proper coordination of synthesis and degradation during DNA replication. Nucleic Acids Res. 2006;34:3107–3115. doi: 10.1093/nar/gkl402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hübscher U, Maga G, Spadari S. Eukaryotic DNA polymerases. Annu. Rev. Biochem. 2002;71:133–163. doi: 10.1146/annurev.biochem.71.090501.150041. [DOI] [PubMed] [Google Scholar]
- 36.Bebenek K, Kunkel TA. Functions of DNA polymerases. Adv. Protein Chem. 2004;69:137–165. doi: 10.1016/S0065-3233(04)69005-X. [DOI] [PubMed] [Google Scholar]
- 37.Rothwell PJ, Waksman G. Structure and mechanism of DNA polymerases. Adv. Protein Chem. 2005;71:401–440. doi: 10.1016/S0065-3233(04)71011-6. [DOI] [PubMed] [Google Scholar]
- 38.Capson TL, Peliska JA, Kaboord BF, Frey MW, Lively C, Dahlberg M, Benkovic SJ. Kinetic characterization of the polymerase and exonuclease activities of the gene 43 protein of bacteriophage T4. Biochemistry. 1992;31:10984–10994. doi: 10.1021/bi00160a007. [DOI] [PubMed] [Google Scholar]
- 39.Yang J, Zhuang Z, Roccasecca RM, Trakselis MA, Benkovic SJ. The dynamic processivity of the T4 DNA polymerase during replication. Proc. Natl Acad. Sci. USA. 2004;101:8289–8294. doi: 10.1073/pnas.0402625101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beese LS, Derbyshire V, Steitz TA. Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science. 1993;260:352–355. doi: 10.1126/science.8469987. [DOI] [PubMed] [Google Scholar]
- 41.Eom SH, Wang J, Steitz TA. Structure of Taq polymerase with DNA at the polymerase active site. Nature. 1996;382:278–281. doi: 10.1038/382278a0. [DOI] [PubMed] [Google Scholar]
- 42.Doubliè S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 43.Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shamoo Y, Steitz TA. Building a replisome from interacting pieces: sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex. Cell. 1999;99:155–166. doi: 10.1016/s0092-8674(00)81647-5. [DOI] [PubMed] [Google Scholar]
- 45.Franklin MC, Wang J, Steitz TA. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 46.de Vega M, Lázaro JM, Salas M, Blanco L. Primer-terminus stabilization at the 3′-5′ exonuclease active site of ϕ29 DNA polymerase. Involvement of two amino acid residues highly conserved in proofreading DNA polymerases. EMBO J. 1996;15:1182–1192. [PMC free article] [PubMed] [Google Scholar]
- 47.de Vega M, Lázaro JM, Salas M, Blanco L. Mutational analysis of ϕ29 DNA polymerase residues acting as ssDNA ligands for 3′-5′ exonucleolysis. J. Mol. Biol. 1998;279:807–822. doi: 10.1006/jmbi.1998.1805. [DOI] [PubMed] [Google Scholar]