Abstract
The increased protein proportion of archaeal and eukaryal ribonuclease (RNase) P holoenzymes parallels a vast decrease in the catalytic activity of their RNA subunits (P RNAs) alone. We show that a few mutations toward the bacterial P RNA consensus substantially activate the catalytic (C-) domain of archaeal P RNA from Methanothermobacter, in the absence and presence of the bacterial RNase P protein. Large increases in ribozyme activity required the cooperative effect of at least two structural alterations. The P1 helix of P RNA from Methanothermobacter was found to be extended, which increases ribozyme activity (ca 200-fold) and stabilizes the tertiary structure. Activity increases of mutated archaeal C-domain variants were more pronounced in the context of chimeric P RNAs carrying the bacterial specificity (S-) domain of Escherichia coli instead of the archaeal S-domain. This could be explained by the loss of the archaeal S-domain's capacity to support tight and productive substrate binding in the absence of protein cofactors. Our results demonstrate that the catalytic capacity of archaeal P RNAs is close to that of their bacterial counterparts, but is masked by minor changes in the C-domain and, particularly, by poor function of the archaeal S-domain in the absence of archaeal protein cofactors.
INTRODUCTION
The ribonucleoprotein enzyme ribonuclease P (RNase P) throughout phylogeny removes 5′ leader sequences of precursor tRNAs (ptRNAs) to generate mature tRNA 5′ ends. The enzyme harbors a single RNA subunit (P RNA) of common ancestry, but only bacterial P RNAs are efficient catalysts in vitro in the absence of the single bacterial protein cofactor (1,2). Very low RNA-alone activity of archaeal, eukaryal and organellar P RNAs (3–5) correlates with the presence of multiple protein subunits (usually 4 in Archaea and 9–10 in Eukarya).
Bacterial P RNAs are composed of two independent folding domains, the catalytic (C-) domain and the specificity (S-) domain (6,7; Figure 1), the latter playing a crucial role in substrate binding via its interaction with the T stem-loop region of ptRNAs (for review, see 8). The single bacterial RNase P protein (P protein), dispensable in vitro but essential in vivo (9,10), binds to the P2–J2/3–J3/4–P4–J18/2 region in the C-domain of P RNA (11–14). The protein increases substrate affinity through interaction with the 5′-leader (14,15), which entails tighter binding of key metal ions involved in substrate positioning and catalysis (16).
Figure 1.
Secondary structure presentation according to Massire et al. (20) of M. thermoautotrophicus ΔH RNase P RNA (MM), E. coli P RNA (EE), and chimeras with the C-domain from M. thermoautotrophicus and the S-domain from E. coli (ME) and vice versa (EM). The 13 nucleotide exchanges and the single 1-nt insertion in P RNA of strain M. thermoautotrophicus strain Marburg (MM-Marburg in the tables) relative to M. thermoautotrophicus ΔH are indicated along the sequence in the secondary structure of MM RNA. The C-domain is depicted in green and the S-domain is depicted in blue. Grey nucleotides at the 5′- and 3′-ends indicate non-natural additions present in the T7 in vitro transcripts.
Activity of archaeal P RNA in the absence of the archaeal RNase P protein cofactors Pop5, Rpp21, Rpp29 and Rpp30 was observed solely for one of the two types of archaeal P RNAs, type A, but not type M (3). Archaeal type A RNAs are structurally related to bacterial P RNAs, but lack the P13/14 stem and helix-P18 among several other deviations from the bacterial consensus (17; Figure 1). Type A archaeal P RNA-alone activity was observed to be low and limited to conditions of very high ionic strength for optimal activity, such as 3 M NH4+ and 0.3 M Mg2+ in the case of P RNA from the euryarchaeon Methanothermobacter thermoautotrophicus (3). Addition of the P protein from Bacillus subtilis activated the archaeal P RNA to some extent at lower ionic strength (3).
For bacterial type A RNA, three loop-helix interdomain contacts (L8–P4, L9–P1 and L18–P8) have been identified (18–21) and are thought to orient the C- and S-domains toward each other (Figure 1). Only one of these contacts, L8–P4, can form in the M. thermoautotrophicus P RNA structure, since the P18 element is missing and the reported 5′- and 3′-ends of the RNA (3) only permit formation of a P1 helix too short to serve as a receptor site for L9 (Figure 1).
Previous attempts to increase the RNA-alone activity of archaeal or organellar P RNAs by insertion of structural elements or by in vitro selection have failed to identify variants with improved RNA-alone activity relative to the corresponding wild-type RNA (5,17,22). We have reinvestigated this issue in a more systematic manner by constructing a series of archaeal C-domain variants with confined alterations towards the bacterial P RNA consensus. The aim of this approach was to identify specific deficits of archaeal P RNAs that limit their RNA-alone activity and make them depend so much more on protein cofactors than their bacterial homologs. Combining the archaeal C-domain from M. thermoautotrophicus P RNA with the S-domain of Escherichia coli P RNA we were able to increase activity—up to three orders of magnitude—by simultaneously introducing a few minor alterations into the archaeal C-domain. All variants were screened at two Mg2+ concentrations, 100 mM in the RNA-alone reaction and 4.5 mM in the holoenzyme reaction. Observed activity gains may well include contributions from improved Mg2+ binding, which in turn could have affected folding, substrate binding, catalysis or binding of the RNase P protein. Thus, activity differences between tested P RNA variants might be mitigated to some extent at Mg2+ concentrations higher than those applied here (see Discussion section). We further provide evidence that the genuine 5′-and 3′-ends of P RNA from Methanothermobacter are longer than previously determined (3). This longer P1, which resulted in 100–200-fold higher RNA-alone activity, may exert its effect primarily through formation of the L9–P1 contact. Finally, analysis of ternary complex formation (P RNA, P protein, ptRNA) revealed that the archaeal S-domain has lost the capacity of bacterial S-domains to confer affine and productive interaction with the substrate, thereby masking the catalytic capacity of archaeal C-domains.
MATERIAL AND METHODS
Construction of plasmids for complementation and in vitro transcription
Plasmids were constructed by standard cloning and PCR methods as described in (5). For specific details of plasmid construction and the oligonucleotides used, see Supplementary Information.
In vitro transcription and 5′ endlabelling of P RNAs and substrate ptRNA
Run-off transcription with bacteriophage T7 RNA polymerase and 5′ and 3′ endlabelling with T4 polynucleotide kinase and T4 RNA ligase were performed essentially as described (23,24). Templates used were pSBpt3′HH linearized with BamHI for the Thermus thermophilus ptRNAGly substrate (23), pDW98 linearized with BsaAI for E. coli P RNA (24) and pUC119_T7_M.th._rnpB linearized with BsaI for M. thermoautotrophicus wild-type P RNA (3). Mutants of the latter were transcribed from the pUC19 constructs described in Supplementary Information; all pUC19 derivatives were linearized with EcoRI.
Expression and purification of recombinant E. coli RNase P protein
E. coli RNase P protein (tagged with an N-terminal hexahistidyl peptide leader MRGSHHHHHHGS, encoded in plasmid pQE-30) was recombinantly expressed in E. coli JM109 and purifed by Ni-NTA affinity chromatography as described (25).
RNase P processing assays
The ptRNAGly substrate (23) was preincubated in assay buffer for 5 min at 55°C and 25 min at 37°C, P RNAs (separately) for 5 min at 55°C and 35 min at 37°C. For holoenzyme reactions, E. coli P protein was added to the P RNA 5 min before combining enzyme and substrate. Conditions of RNA-alone reactions were single turnover with trace amounts (<1 nM) of 5′-32P-labelled substrate, 300 nM or 10 µM P RNA, 100 mM Mg(OAc)2, 100 mM NH4OAc, 50 mM MES and 2 mM EDTA, pH 6.0 and 37°C. Holoenzyme reactions were performed as multiple turnover reactions at concentrations of 10 nM P RNA, 50 nM P protein, 100 nM ptRNAGly, and buffer KN (20 mM HEPES-KOH pH 7.4, 4.5 mM Mg[OAc]2, 150 mM NH4OAc, 2 mM spermidine, 0.05 mM spermine and 4 mM β-mercaptoethanol) at 37°C. Analysis of reactions by denaturing PAGE and evaluation of data was done as described (23); all quantifications were based on at least three independent experiments.
Folding analysis by native PAGE
3′-End-labelled P RNAs were preincubated at a concentration of 10 nM in buffer KN for 5 min at 55°C and then 50 min at 37°C. After addition of 1 volume of gel loading buffer (10% [v/v] glycerol, 4.5 mM MgCl2, xylene cyanol and bromophenol blue) at room temperature, the samples were immediately loaded onto non-denaturing polyacrylamide gels (11.25% [v/v] polyacrylamide, 66 mM HEPES, 33 mM Tris [pH 7.4], 0.1 mM EDTA, 100 mM NH4OAc and 4.5 mM MgCl2). Gels were run and the RNAs visualized as described (25).
UV melting profiles
UV melting profiles were recorded at 260 nm on a CARY 100 Bio UV-Visible spectrophotometer, using the Cary Win UV software version 3.00.
Bacterial strains
Total RNAs for RACE experiments were prepared from cells of M. thermoautotrophicus strain Marburg (DSM 2133, syn. M. marburgensis), whereas the Methanothermobacter RNase P RNA used throughout this study is that of M. thermoautotrophicus strain ΔH (DSM 1053).
Determination of equilibrium dissociation constants
Equilibrium dissociation constants (Kd) of complex formation between RNase P holoenzymes and ptRNA substrate were determined by spin column assays essentially as described (26,27) in a binding buffer containing 50 mM Tris-acetate, pH 7.1, 200 mM NH4OAc, 0.05% Nonidet P-40, and 15 mM Ca(OAc)2. Ca2+ was used instead of Mg2+ to prevent substrate cleavage during binding and the concentration of 15 mM was chosen to increase sensitivity of the assay, because the Kd's measured at this Ca2+ concentration are substantially lower than those obtained at lower concentrations (e.g. 5 mM Ca2+). Before the assay, holoenzymes were reconstituted from P RNA variants and E. coli P protein by incubating 1 µM P RNA in binding buffer for 1 h at 37°C and then adding E. coli P protein to 5 µM, followed by another 15 min at 37°C. Holoenzymes were then diluted in binding buffer to desired concentrations, incubated for 5 min at 37°C, mixed with trace amounts (<1 nM) of 5′-32P-labelled substrate (preincubated for 30 min at 37°C in binding buffer) and incubated at 37°C for 5 min.
RESULTS
Analysis of E. coli and M. themoautotrophicus wild-type and chimeric P RNAs
Starting from M. thermoautotrophicus wild-type P RNA (henceforth called MM RNA) and E. coli wild-type P RNA (termed EE RNA; Figure 1), we constructed P RNA chimeras by combining the S-domain of E. coli P RNA with the C-domain of M. thermoautotrophicus (termed ME RNA) or the E. coli C-domain with the M. thermoautotrophicus S-domain (termed EM RNA; Figure 1). The chimeric P RNAs allowed us to address if the heterologous domains are able to cooperate and if there is a major asymmetry in the contribution of the archaeal C- and S-domains to the low RNA-alone activity of the archaeal P RNA.
RNA-alone single turnover reactions (E>>S) at pH 6.0 in the presence of 300 nM P RNA, trace amounts (<1 nM) ptRNAGly and 0.1 M each Mg2+ and NH4+ revealed only very weak cleavage by EM RNA (2 × 10−4 min−1) and no activity at all with ME and MM P RNA (Table 1). The activity of EM RNA increased linearly with concentration (tested from 300 nM to 10 µM, data not shown), indicating low substrate affinity. At 10 µM P RNA concentration, activities were detectable for all P RNA variants, with the chimeras EM and ME being 34–44-fold more active than the archaeal MM RNA (Table 1). The finding of activity at 10 µM but not at 0.3 µM for the MM and ME variants suggested that weak substrate affinity is also a major cause for the poor performance of these two P RNAs. The lower RNA-alone activity of MM versus EM and ME RNA (Table 1) showed that deficits of MM RNA are located in its S- as well as its C-domain.
Table 1.
ptRNA processing activity of E. coli/M. thermoautotrophicus P RNA domain swop chimeras in RNA-alone and holoenzyme assays
| P RNA alone |
Holoenzyme |
||
|---|---|---|---|
| P RNA | kobs [min–1 × 10−4] | P RNA | kobs [min–1 × 10−4] |
| EE (0.3 μM) | 51 000 ± 4000 | EE (10 nM) | 82 000 ± 2000 |
| EM (0.3 μM) | 2 ± 1 | EM | Not analyzed |
| ME (0.3 μM) | Not detectable | ME | Not analyzed |
| MM (0.3 μM) | Not detectable | MM | Not analyzed |
| EM (10 μM) | 88 ± 19 | EM (10 nM) | 70 ± 10 |
| ME (10 μM) | 67 ± 17 | ME (10 nM) | 2 ± 1 |
| MM (10 μM) | 2 ± 0.1 | MM (10 nM) | 2 ± 1 |
| Ecat (10 μM) | Not detectable | Ecat (10 nM) | 77 ± 6 |
Assay conditions for the RNA-alone reaction were 50 mM MES pH 6.0 at 37°C, 2 mM EDTA, 100 mM NH4OAc, 100 mM Mg(OAc)2, trace amounts of ptRNA and 10 µM P RNA unless otherwise indicated. Holoenzyme activity assays were performed at 20 mM HEPES pH 7.4 at 37°C, 2 mM spermidine, 0.05 mM spermine, 4 mM β-mercaptoethanol, 150 mM NH4OAc, 4.5 mM Mg(OAc)2, 100 nM ptRNA, 10 nM P RNA and 50 nM recombinant E. coli P protein with N-terminal His tag. Errors are standard deviations; all values are based on at least three independent experiments. For further experimental details see Materials and methods section. Values for EE and Ecat RNA are taken from ref. (5).
As in the RNA-alone reactions, RNase P holoenzymes reconstituted with the E. coli P protein and tested for multiple turnover under near-physiological salt conditions (buffer KN, 4.5 mM Mg2+, 10 nM P RNA, 50 nM P protein, 100 nM ptRNAGly) yielded very low cleavage rates for MM RNA and the ME chimera (Table 1). The EM variant was 35-fold more active than MM and ME, though still a factor of >1000 slower than EE RNA. Higher activity of EM RNA, harbouring the E. coli C-domain, was anticipated because the primary binding site of the bacterial P protein is in the C-domain (11–14). The fact that EM RNA activity equaled that of the isolated C-domain of E. coli P RNA (Ecat; Table 1) indicates that the archaeal S-domain made no functional contribution under the applied conditions of the holoenzyme assay. Not unexpected, alleles encoding MM, EM and ME RNAs were unable to rescue the mutant phenotype of a bacterial RNase P mutant strain (Table S3).
ME RNA variants with specific structural alterations in the archaeal C-domain
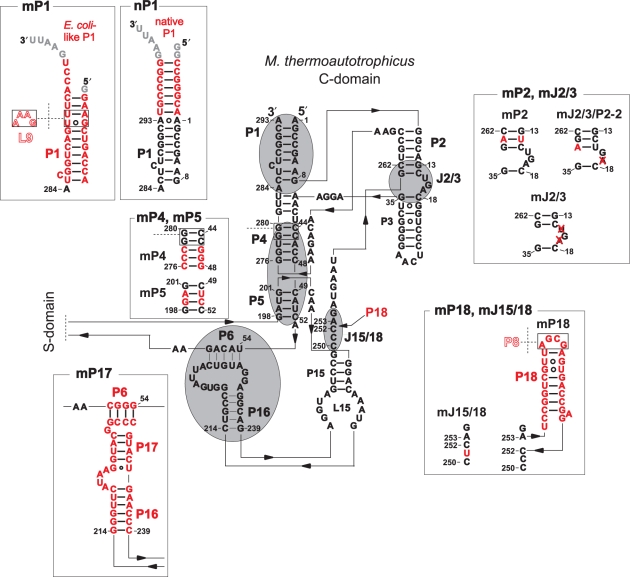
With its substantial increase in RNA-alone activity compared to MM RNA (Table 1), we assumed ME RNA to be more sensitive toward activating changes in the archaeal C-domain. We thus generated an initial set of ME derivatives that were mutated towards the bacterial type A RNase P RNA consensus (28) by mutations ranging from single nucleotide exchanges to exchange of defined modules (Figure 2). RNA-alone and holoenzyme assays were conducted as specified in Table 1, and activities of the ME variants illustrated in Figure 2 are summarized in Table 2.
Figure 2.
Secondary structure presentation of the C-domain from M. thermoautotrophicus according to Massire et al. (20) with mutations introduced during this work. Regions subjected to mutation are circled and highlighted in grey; sequences of the respective mutants (indicated by the prefix ‘m’, e.g. mP1) are given in the adjacent boxes with the altered nucleotides coloured in red. Throughout the paper, combinations of mutations are indicated by a slash inbetween, e.g. ME-mJ2/3/P2 refers to mutation mJ2/3 as well as mutation mP2 within one construct, with the ME in front indicating that the construct possesses an M. thermoautotrophicus C-domain and an E. coli S-domain. Grey nucleotides at the 5′- and 3′-ends indicate non-natural additions present in the T7 in vitro transcripts. Nucleotide numbering is that of M. thermoautotrophicus ΔH P RNA.
Table 2.
Activity of ME variants in RNA-alone and holoenzyme assays
| RNase P RNA | RNA-alone |
Holoenzyme |
||
|---|---|---|---|---|
| kobs [min–1 × 10−4] | Change | kobs [min–1 × 10−4] | Change | |
| ME | 67 ± 17 | 1 | 2.0 ± 1.0 | 1 |
| ME-mP1 | 0.4 ± 0.1 | ↓ 176× | 1.1 ± 0.1 | ↓ 2× |
| ME-mP2 | 9.7 ± 1.2 | ↓ 7× | 1.1 ± 0.2 | ↓ 2× |
| ME-mP4 | 2.8 ± 1.8 | ↓ 24× | 1.2 ± 0.1 | ↓ 2× |
| ME-mP5 | 41 ± 5 | ↓ 2× | 1.2 ± 0.1 | ↓ 2× |
| ME-mP17 | Not detectable | 1.1 ± 0.1 | ↓ 2× | |
| ME-mP18 | 39 ± 6 | ↓ 2× | 4.0 ± 0.3 | ↑ 2× |
| ME-mJ15/18 | 250 ± 45 | ↑ 4× | 6.5 ± 0.3 | ↑ 3× |
| ME-mJ2/3 | 7.7 ± 1.2 | ↓ 9× | 52 ± 6 | ↑ 26× |
| ME-mJ15/18/P1 | 0.5 ± 0.1 | ↓ 143× | 1.3 ± 0.2 | ↓ 2× |
| ME-mJ15/18/P2 | 29 ± 7.2 | ↓ 2× | 3.6 ± 0.3 | ↑ 2× |
| ME-mJ15/18/P4 | 160 ± 15 | ↑ 2× | 3.2 ± 0.3 | ↑ 2× |
| ME-mJ15/18/P5 | 100 ± 25 | 1× | 3.2 ± 0.4 | ↑ 2× |
| ME-mJ15/18/P17 | 1.6 ± 1.2 | ↓ 42× | 1.3 ± 0.1 | ↓ 2× |
| ME-mJ15/18/P18 | 3250 ± 400 | ↑ 49× | 12 ± 0.2 | ↑ 6× |
| ME-mJ15/18/2/3 | 33 ± 3.5 | ↓ 2× | 320 ± 30 | ↑ 160× |
| ME-mJ2/3/P2 | 1350 ± 240 | ↑ 20× | 900 ± 200 | ↑ 450× |
| ME-mJ2/3/P2-2 | 480 ± 120 | ↑ 7× | 480 ± 50 | ↑ 240× |
| ME-mJ15/18/2/3/P2 | 5400 ± 910 | ↑ 81× | 930 ± 20 | ↑ 465× |
| ME-mJ15/18/2/3/P2/nP1 | 9880 ± 2520 | ↑ 147× | 3000 ± 500 | ↑1500× |
For assay conditions and experimental details, see legend to Table 1 and Materials and methods section.
RNA-alone activity
Among the initial set of eight variants, only a single variant, with a point mutation in the region that joins P15 and P18 in bacterial P RNA, had increased RNA-alone activity relative to ME RNA (ME-mJ15/18, 4-fold increase; Table 2). The other variants displayed RNA-alone cleavage rates decreased from 2-fold to below the level of detection in the case of ME-mP17. The latter alteration introduced the P6/16/17 elements of E. coli P RNA, based on the idea that this may help to position the L15 loop for interaction with the tRNA 3′-CCA end. The next most prominent impairments were seen for ME-mP1 (176-fold), which introduced the E. coli P1 helix intended to restore the L9–P1 interdomain contact, and for ME-mP4 (24-fold) with three Watson-Crick base pair identities in P4 changed towards the bacterial consensus (Figure 2, Table 2). Likewise, insertion of E. coli P18 (ME-mP18) to restore the L18–P8 interdomain contact failed to improve activity, in line with a previous study (17).
In a second round, the variant with increased activity, ME-mJ15/18, was combined with each of the other structural alterations. RNA-alone activities of the double mutants either equalled that of ME RNA (within a factor of two) or remained substantially lower (ME-mJ15/18/P1, ME-mJ15/18/P17), with one notable exception: incorporation of E. coli P18 (ME-mJ15/18/P18) significantly increased activity compared to ME and also ME-mJ15/18 (Table 2). If the individual changes (mJ15/18 and mP18) were independent and additive, one would have expected a roughly 2-fold increase in activity. However, the 49-fold increase for ME-mJ15/18/P18 versus ME indicates positive cooperativity between the two structural alterations.
Activity with the bacterial P protein as cofactor
When we assayed P RNA mutants for holoenzyme activity using the E. coli P protein as cofactor (Table 2, right columns), again only a single variant showed a marked increase in activity (26-fold) over ME RNA (ME-mJ2/3, with J2/3 reduced from 3 to 1 nt; Figure 2). The fact that this small alteration caused a 9-fold activity decrease in the RNA-alone reaction, but 26-fold increase in the holoenzyme reaction suggested an effect on P protein binding, which is consistent with current knowledge about the P RNA binding interface of the bacterial P protein (Figure S1). All other individual changes had little effect (within 3-fold) on the holoenzyme reaction.
Combination of the J15/18 mutation with one of the other structural alterations in the second round revealed two variants with substantially improved activity in the holoenzyme reaction: ME-mJ15/18/P18 (6-fold) and ME-mJ15/18/J2/3 (160-fold; Table 2).
Changes in P2–P3 and the triple mutant ME-J15/18/2/3/P2
The activity increase in the holoenzyme reaction owing to deletion of two nucleotides in J2/3 prompted us to combine changes towards the bacterial consensus in J2/3 and P2, aiming at a further improvement of E. coli P protein binding to the ME chimera. Helix-P2, which is part of the bacterial P protein binding site (see Introduction section and Figure S1), consists of 6 bp in MM RNA compared with 7 bp in bacterial P RNAs. We thus constructed P RNA variants that combine the J2/3 mutation with a P2 helix extended by 1 bp. In the first mutant an U–A bp was inserted between the fifth and the sixth base pair (counted from the 5′-end; Figure 2) of the archaeal P2 helix. This increased RNA-alone activity 20-fold (ME-mJ2/3/P2), whereas these alterations individually impaired activity 9-fold (mJ2/3) and 7-fold (mP2; Table 2). In the holoenzyme reaction, the activity gain for variant ME-mJ2/3/P2 was 450-fold over ME RNA, compared with a 26-fold rate increase for variant ME-mJ2/3 and a 2-fold drop for variant ME-mP2. Thus, we observed positive cooperativity effects for the combined mJ2/3/P2 changes in the RNA-alone and holoenzyme assays, indicating that the P2–J2/3–P3 region not only contributes to bacterial P protein binding, but also to folding of the catalytic core.
We further constructed variant ME-mJ2/3/P2-2 with the goal to restore the bacterial consensus with minimal structural alteration, solely deleting 1 nt in J2/3 and introducing a point mutation that creates an additional U–A bp at the end of P2. This variant improved RNA-alone activity of ME RNA 7-fold and raised substrate turnover 240-fold in the holoenzyme reaction (Table 2). The 2–3-fold weaker performance of variant ME-mJ2/3/P2-2 relative to ME-mJ2/3/P2 suggests that a terminal U–A bp at the P2/P3 junction has a slightly destabilizing effect compared with a closing C–G bp.
We then combined the mJ15/18 point mutation with the mJ2/3/P2 alteration to generate variant ME-J15/18/2/3/P2. This further increased activity roughly 4-fold relative to ME-mJ2/3/P2 in the RNA-alone reaction, but showed little additional effect in the holoenzyme reaction (Table 2), for unknown reasons.
Length of P1 and the L9–P1 interdomain contact
In some bacterial P RNAs, a long-range tertiary interaction between loop L9 docking onto helix P1 serves as an interdomain strut of major importance for the global RNA fold (20,29). The mature 5′- and 3′-ends of MM RNA, mapped in a previous study (3), would be too short to allow formation of such an L9–P1 contact. However, some archaeal P RNAs seem to have an extended P1 helix compatible with formation of the L9–P1 contact, and even the MM RNA gene sequence encodes the potential for an extended P1 helix that, if part of the P RNA transcript, may enable formation of the P1–L9 interaction. (17). We thus constructed an MM RNA variant with P1 extended by 7 bp based on the sequence of the P RNA (rnpB) gene of M. thermautotrophicus strain ΔH. This increased the RNA-alone activity of MM RNA 200-fold (Table 3). Introduction of this P1 extension into variant ME-mJ15/18/2/3/P2 to generate ME-mJ15/18/2/3/P2/nP1 further increased activity to 147-fold (RNA-alone) and 1500-fold (holoenzyme) over ME RNA (Table 2).
Table 3.
Activity of MM variants in RNA-alone and holoenzyme assays
| RNase P RNA | RNA-alone |
Holoenzyme |
|||
|---|---|---|---|---|---|
| kobs [min–1 × 10−4] | Change | kobs [min−1 × 10−4] | Change | ||
| MM | 1.7 ± 0.1 | 1 | 2 ± 1 | 1 | |
| MM-nP1 | 340 ± 60 | ↑ 200× | 1.3 ± 0.1 | ↓ 2× | |
| MM-mJ15/18/P18 | 2 ± 1 | 1 | 1.2 ± 0.1 | ↓ 2× | |
| MM-mJ2/3/P2 | 230 ± 50 | ↑ 135× | 7 ± 1 | ↑ 4× | |
| MM-mJ2/3/P2-2 | 100 ± 20 | ↑ 59× | 6.7 ± 0.8 | ↑ 4× | |
| MM-mJ15/18/2/3/P2 | 270 ± 60 | ↑ 159× | 80 ± 10 | ↑ 40× | |
| MM-mJ15/18/2/ 3/P2/nP1 | 740 ± 60 | ↑ 435× | 75 ± 4 | ↑ 38× | |
| MM-Marburg | 3.3 ± 0.6 | Not detectable | |||
| MM-Marburg-nP1 | 310 ± 20 | 2.1 ± 0.9 | |||
| MM-Marburg- mapped ends | 240 ± 20 | 1.0 ± 0.2 | |||
For assay conditions and experimental details, see legend to Table 1 and Materials and methods section. For structural details on P1 elements in RNAs MM-Marburg, MM-Marburg-nP1 and MM-Marburg-mapped ends, see Figure S2B.
The stimulating effect of extension nP1 on MM RNA-alone activity prompted us to reinvestigate the 5′- and 3′-boundaries of MM RNA by RACE. This was done with the closely related M. thermoautotrophicus strain Marburg instead of the original source strain M. thermautotrophicus ΔH, because we had access to freshly grown cells of the former only. Owing to the switch of strains we performed several control experiments before conducting the RACE experiment: (i) sequencing of the gene encoding the MM-Marburg RNA revealed a few nucleotide changes relative to MM RNA of strain ΔH (Figure 1), but in vitro transcribed MM-Marburg RNA with the short P1 helix as in MM RNA (Figure 1) showed RNA-alone activity very similar to that of MM RNA (Table 3); (ii) introducing the same 7-bp extension as present in MM-nP1 into the MM-Marburg RNA, resulting in variant MM-Marburg-nP1 (Figure S2B), indeed revealed a similar activity increase (ca. 100-fold) as seen for MM-nP1 RNA (Table 3). Having confirmed that MM-Marburg RNA is an appropriate mimic of MM RNA, we carried out 5′- and 3′-RACE (Figure S2A), which unveiled that in vivo transcripts of MM-Marburg RNA, and by inference of MM RNA, carry 5′- and 3′-ends that permit formation of an extended P1 helix (Figure S2B) to provide a receptor site for loop L9.
Construction and analysis of MM variants
Next we addressed the question if the activating structural changes nP1, mJ2/3, mP2 and mJ15/18, initially explored in the context of variant ME, would be beneficial in the natural context of MM RNA as well. Indeed, RNA-alone activity of MM RNA increased 435-fold by introducing these four minor changes (Table 3), while the activity gain was only 38-fold in the holoenzyme reaction (Table 3, columns on the right). Noteworthy, based on the comparison of cleavage rates (kobs) for P RNA MM-mJ15/18/2/3/P2/nP1 (Table 3) and its counterpart ME-mJ15/18/2/3/P2/nP1 (Table 2), the presence of the archaeal S-domain in the MM variant reduced activity 13- and 40-fold in the RNA-alone and holoenzyme reactions, respectively.
Measurement of holoenzyme-substrate affinity
The relatively low catalytic performance of variant MM-mJ15/18/2/3/P2/nP1, particularly in the holoenzyme reaction, led us to analyze substrate affinities of P RNA variants reconstituted with the E. coli P protein. We initially tried direct measurements of affinity between P RNA and protein, but different approaches were unsuccessful in our hands. We thus switched to measuring ternary complex formation between holoenzyme and substrate (Table 4). As complex formation depended on P protein binding to P RNA under the applied low salt concentrations, the assay indirectly measured P RNA–protein affinities.
Table 4.
Affinity of reconstituted RNase P holoenzymes for the ptRNAGly substrate
| RNase P RNA | Kd (nM) | Kd change | kobs change (holoenzymes) |
|---|---|---|---|
| MM | n.d. | 1 | |
| MM-mJ2/3/P2 | n.d. | ↑ 4× | |
| MM-mJ15/18/2/3/P2 | n.d. | ↑ 40× | |
| MM-mJ15/18/2/3/P2/nP1 | n.d. | ↑ 38× | |
| ME | n.d. | 1 | |
| ME-mJ2/3 | 448 ± 30 | ↑ 895× | ↑ 2× |
| ME-mJ2/3/P2 | 47 ± 11 | ↑ 94× | ↑ 450× |
| ME-mJ15/18/2/3 | 48 ± 4 | ↑ 96× | ↑ 160× |
| ME-mJ15/18/2/3/P2 | 4.2 ± 1.2 | ↑ 8× | ↑ 465× |
| ME-mJ15/18/2/3/P2/nP1 | 5.0 ± 1.0 | ↑ 10× | ↑ 1500× |
| EE | 0.5 ± 0.1 | 1 | ↑ 41 000× |
Errors for Kd are standard errors of the curve fit; n.d., non-determinable because of affinities too low for Kd determination by this assay. Kd changes are given as n-fold increase in Kd relative to the E. coli holoenzyme (EE). Note that in our assay for all variants ptRNA binding to P RNA in the absence of protein did not exceed background values (ptRNA alone). Thus, the binding data for the ternary complex indirectly permit to assess P protein binding to P RNA; kobs changes listed for comparison are taken from Tables 2 and 3.
For the original ME chimera as well as MM RNA and all variants thereof ptRNA affinity was too low for Kd determination (Table 4). This changed with variant ME-mJ2/3, for which a Kd of 450 nM could be determined. Additional introduction of the J15/18 mutation increased affinity roughly tenfold. Similarly, the P2 mutation introduced into either the single mutant ME-mJ2/3 or the double mutant ME-mJ15/18/2/3 in either case led to a tenfold increase in affinity. The lowest Kd (4.2 nM) was obtained for the ME-mJ15/18/2/3/P2 RNA; additional incorporation of the nP1 extension did not further reduce Kd (5 nM). Thus, ptRNA affinities obtained with the best ME variants were 10-fold lower than that of the E. coli holoenzyme (EE RNA, Kd = 0.5 nM; Table 4).
For the ME variants, increases in affinity (reductions in Kd) nicely correlated with increases in bacterial holoenzyme activity, with the exception of the nP1 extension which enhanced activity ca. 3-fold when added to variant ME-mJ15/18/2/3/P2, but left substrate affinity essentially unaffected (Table 4). Low substrate affinity of the MM variants explains why activity gains in the bacterial holoenzyme assay were moderate (4–40-fold): since the archaeal S-domain is defective in substrate binding based on our Kd measurements and thus limits the rate of the cleavage reaction, beneficial effects of alterations mJ2/3, mJ15/18 and mP2 in the archaeal C-domain could not be coaxed into activities as high as in the presence of the E. coli S-domain.
Folding analysis of P RNA variants by native PAGE
Previous studies have shown that increased activity of P RNAs often goes along with compacted conformers becoming more prominent in native PAA gels (5,25,29,30). Consistent with this observation, MM RNA showed the most diffuse migrational behaviour in native gels, pointing to a multitude of coexisting conformations (Figure 3A). All other P RNAs analyzed, with a gain in activity compared to that of MM, displayed more distinct bands and some additional compaction, at least after the preincubation step to that allows RNA folding. Replacement of either domain of MM with the counterpart of EE RNA (variants ME and EM) resulted in stabilization of distinct folds, although EM RNA suffered from substantial aggregation as inferred from RNA retained in the gel pocket (Figure 3A). Extension of the P1 helix (MM-nP1) obviously supports tertiary folding (Figure 3B), as distinct conformers appeared relative to MM RNA. Interestingly, RNAs MM-nP1 and MM-M-nP1 differ in that one major conformer appears for the latter compared with two conformers in the case of RNA MM-nP1 (Figure 3B). This finding shows that even minor changes between two related RNAs may have profound effects on conformational equilibria. Gel mobility of variants ME-mJ15/18/2/3/P2/nP1 and MM-mJ15/18/2/3/P2/nP1 differed substantially, with the former resembling ME RNA in showing a single major band and the latter displaying at least two major conformers, similar to MM-nP1 RNA. However, the additional changes mJ15/18, mJ2/3 and mP2 further affected conformation relative to RNA MM-nP1, as inferred from weaker relative intensity of the upper conformer and faster migration of the lower conformer in the case of RNA MM-mJ15/18/2/3/P2/nP1.
Figure 3.
Native 11.25% PAGE analyses to compare the conformations of P RNA variants. (A) P RNAs from M. thermoautotrophicus (MM), E. coli (EE) and E. coli/M. thermoautotrophicus chimeras (EM and ME); (B) MM and MM-Marburg P RNA variants with P1 extension (MM-nP1, MM-M-nP1), as well as corresponding C-domain mutants of ME and MM RNA. Assay conditions were the same as in the holoenzyme activity assay (see legend to Table 1 and Material and methods section). Samples were loaded onto the gel either after storage for 55 min on ice (−) or after preincubation for 5 min at 55°C and 50 min at 37°C (+). For further details, see Materials and methods section.
UV melting profiles of selected P RNA variants
UV melting profiles were analyzed for P RNA variants ME, MM and MM-nP1, as well as the mJ15/18/2/3/P2/nP1 variants of ME and MM RNA. Conditions were 4.5 mM Mg2+ and 100 mM NH4+ (Figure 4), resembling those of our kinetic holoenzyme assay. Variant MM (red curve) had the lowest Tm value (73.1°C), which increased to 74.4°C when helix P1 was extended to its natural length (MM-nP1, black curve). Thus, helix P1 has an overall stabilizing effect, in line with large activity losses of bacterial P RNA variants with disrupted P1 elements (31). In addition, there is only one major unfolding transition for MM-nP1, based on the first derivative of the melting profile (dA260/dT), whereas more unfolding of substructures occurred in MM RNA (shoulders between 60°C and 65°C and at approx. 77°C) aside from the major transition at 73.1°C. Variant ME (blue curve) showed a major unfolding transition around 76°C, but also substantial unfolding of substructures between 50°C and 70°C. In contrast, much less unfolding of substructures at lower temperatures was seen for variant ME-mJ15/18/2/3/P2/nP1 (gray curve), for which the P1 extension might be the major cause, in analogy to the profiles of MM versus MM-nP1 RNA.
Figure 4.
UV melting profiles of selected P RNA variants. The shown curves represent the mean of at least three measurements; assay conditions: 250 nM P RNA, 50 mM MES pH 6.0, 2 mM EDTA, 100 mM NH4OAc, 4.5 mM Mg(OAc)2; for UV absorbance (260 nm) melting profiles (left), the y-axis of every species was normalized to 0.0 − 1.0 (normalized Abs); normalized melting curve shapes were not affected by variation of P RNA concentration, indicating that intramolecular unfolding was measured; ‘d normalized Abs/dT’ (right panel): first derivatives of the melting curves on the left; the measuring program consisted of three steps: step 1 (preincubation): heating of the sample to 55°C at a rate of 10°C/min, incubation for 5 min at 55°C, cooling of the sample to 37°C at a rate of 10°C/min, incubation at 37°C for 35 min; step 2 (measuring of melting profile): heating of the sample from 37 to 90°C at a rate of 0.5°C/min with data recording every 0.1°C; stage 3: immediately after reaching 90°C in step 2, the sample was cooled to 25°C at a rate of 2°C/min and kept at 25°C for 5 min; the next round of measurement was again started with step 1.
It is further instructive to compare variant MM-mJ15/18/2/3/P2/nP1 with MM-nP1. The former (green curve) has a similar Tm and derivative melting profile as RNA MM-nP1, but shows more unfolding of local structures below 65°C than MM-nP1. Thus, introducing the set of minor changes (mJ15/18/2/3/P2) towards the bacterial consensus into the concertedly folding MM-nP1 RNA uncoupled unfolding of some local structures from unstacking of the bulk of RNA structure. Notably, the derivative melting profiles of variants MM-mJ15/18/2/3/P2/nP1 and ME-mJ15/18/2/3/P2/nP1 were identical up to 65°C, but the major unfolding transition for the latter was broader and shifted to higher temperatures (Tm = 76.3°C versus 75.0°C). Therefore, replacement of the archaeal S-domain with the E. coli counterpart increased overall folding stability but reduced the extent of coordinate unstacking.
Kinetic parameters of ME-mJ15/18/2/3/P2/nP1 and its MM counterpart
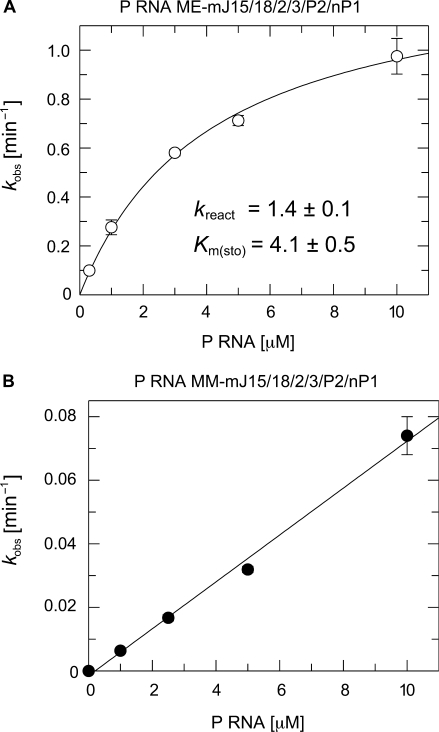
We determined the RNA-alone single turnover kinetic parameters for the best-performing variant ME-mJ15/18/2/3/P2/nP1 to further explore the catalytic capacity of the engineered archaeal C-domain. This analysis revealed a maximum rate (kreact) of approx. 1.4 min−1 and a Km(sto) of about 4 μM for the P RNA ME-mJ15/18/2/3/P2/nP1 (Figure 5A). For comparison, kreact and Km(sto) were determined as 10 min−1 and 0.24 μM, respectively, for E. coli wild-type P RNA under the same conditions (32). Thus, P RNA ME-mJ15/18/2/3/P2/nP1 differs from E. coli P RNA by only a 7-fold lower maximum cleavage rate and a 17-fold higher Km(sto). In contrast, the rate of cleavage by variant MM-mJ15/18/2/3/P2/nP1 showed a linear dependence on P RNA concentration up to 10 μM (Figure 5B), indicative of enzyme concentration being subsaturating and consistent with low substrate affinity in the presence of the archaeal S-domain.
Figure 5.
RNA-alone single turnover kinetics of P RNAs (A) ME-mJ15/18/2/3/P2/nP1 and (B) MM-mJ15/18/2/3/P2/nP1 as a function of the P RNA concentration. Processing assays were performed in the presence of <1 nM 5′-endlabelled ptRNA, 100 mM Mg(OAc)2, 100 mM NH4OAc, 50 mM MES and 2 mM EDTA, at pH 6.0 and 37°C. The kobs values are mean values from at least three independent experiments, with error bars indicating the standard error of the mean. For P RNA ME-m J15/18/2/3/P2/nP1, the single turnover kinetic parameters could be derived. Km(sto) is the single turnover Km (describes the enzyme concentration at which the half maximum rate under conditions of [E] >> [S] is achieved), and kreact is the single turnover Vmax. For further details, see Materials and methods section.
We further tested P RNA ME-mJ15/18/2/3/P2/nP1 for function in B. subtilis and E. coli rnpB mutant strains, also under conditions of simultaneous overexpression of bacterial P proteins to compensate for reduced P RNA-P protein affinity. However, P RNA ME-mJ15/18/2/3/P2/nP1 was unable to rescue the mutant phenotypes (Table S3).
DISCUSSION
We have shown here that a few mutations toward the bacterial P RNA consensus substantially activate the C-domain of archaeal P RNA from Methanothermobacter, in the absence and presence of the bacterial RNase P protein. All variants were screened at two Mg2+ concentrations, 100 mM in the RNA-alone reaction and 4.5 mM in the holoenzyme reaction. Activity measurements at the two Mg2+ concentrations provide snapshots, as the individual variants may have different Mg2+ optima. Such potential differences in Mg2+ requirements imply that the structural changes introduced into the archaeal C-domain may exert their effects on folding, substrate binding, catalysis or binding of the RNase P protein at least partly through changes in binding of crucial Mg2+ ions, which has to be considered when interpreting the data. In a recent study on the RNA-alone reaction catalyzed by E. coli RNase P RNA, Kirsebom and coworkers demonstrated that the Mg2+ requirements with respect to cleavage increased when substrate contacts to the C-domain (involving substrate functional groups near the cleavage site) as well as contacts to the S-domain (involving the T-stem/loop region of the substrate) are weakened (33). The improvement in substrate positioning seen for variant ME-mJ15/18/2/3/P2 in our probing experiments (see below) may thus well include contributions from improved Mg2+ binding to P RNA regions that mediate contacts to the substrate. Along these lines, we showed previously for E. coli and B. subtilis P RNAs with mutations in the substrate 3′-CCA binding region that in vitro processing defects in the holoenzyme reaction were exacerbated when Mg2+ concentrations were lowered from e.g. 4.5 mM to 2 mM (34,35). Likewise, P RNAs with structural deletions (36) or substrates with ribose 2′-substitutions at strategic positions were reported to cause an increase in the Mg2+ requirement of P RNA-alone reactions (37,38). On the basis of these findings it is reasonable to predict that low activity P RNA variants in the study presented here will tend to perform relatively better at higher Mg2+ concentrations compared to the variants with improved catalytic performance.
A central finding of our study was that ribozyme activity of M. thermoautotrophicus P RNA is much higher (100 to 200-fold) than previously assumed owing to the presence of an extended P1 helix (Table 3, Figure 2). This P1 extension also stabilizes the global fold as inferred from native PAGE analysis (Figure 3) and UV melting profiles (Figure 4). A likely explanation is that the extension creates a receptor site for loop L9. Since the L18–P8 interdomain contact is lacking, the archaeal RNA may strongly depend on the L9–P1 contact for interdomain orientation of its type A architecture. The presence of an extended P1 helix is also likely for other archaeal P RNAs (Figure S3). Interestingly, a common motif of archaeal P1 elements is an A:A or C:A mismatch at identical position in type A and type M archaeal P RNAs proposed to interact with the third G residue of a conserved 5′-GAGA L9 tetraloop (17). We have recently demonstrated that a special L9–P1 interaction, occurring in some bacterial P RNAs from thermophiles, represents a major determinant for thermostability of the P RNAs, for RNase P holoenzyme activity at physiological Mg2+-concentrations and for conformational compaction (29). Of note, some bacterial P RNAs harbouring this thermostable P1–P9 module also lack the L18–P8 interaction and have apparently compensated for its absence by strengthening the L9–P1 interdomain contact (29,32). We recently reported that the P1–L9 interaction is degenerate in E. coli-like mesophilic type A RNase P RNAs, and thus unlikely forms at all, because mutations intended to disrupt this contact remained without effect on E. coli P RNA function in vitro as well as in vivo (29). This finding explains why variant ME-mP1, designed to restore the E. coli-like L9–P1 contact when its degeneracy was still unknown, failed to improve RNA-alone activity (Table 2). However, it remains unclear why the impairment of activity was this severe (176-fold).
A pattern emerging from our reactivation study is that mutations within a single structural element were usually insufficient to improve RNA-alone activity. With the exception of the P1 extension, it took the cooperative effect of at least two alterations to be substantially beneficial for ribozyme activity, such as mP18 plus mJ15/18, mP2 plus mJ2/3 or mP2 plus mJ2/3 plus mJ15/18 (Table 2). Successful double mutants combined introduction (mP18) or alteration (mP2) of a helix and fine adjustments within adjacent joining regions likely involved in proper positioning of these structural elements. The stimulatory effects of combined changes mJ2/3/P2 on RNA-alone activity were, however, unexpected. Yet, since P2 and P3 are part of the catalytic core, length changes in J2/3 may affect coaxial stacking of P2 and P3 and thereby, combined with the P2 extension, induce subtle changes in folding of the entire core comprising the P1/P4/P5 and P2/P3 stacks as well as the respective joining segments.
Surprisingly, in several cases activity increases in the RNA-alone and holoenzyme assays differed substantially. The combined changes in J2/3 and P2 activated RNA-alone activity of variant ME-mJ2/3/P2 20-fold (relative to ME RNA; Table 2) and that of variant MM-mJ2/3/P2 135-fold (relative to MM RNA; Table 3); however, in the holoenzyme reaction activity stimulation was 450-fold for variant ME-mJ2/3/P2, but only 4-fold for P RNA MM-mJ2/3/P2 (Table 2 and 3). A similar situation was seen for the mJ15/18/2/3/P2/nP1 mutation (RNA-alone reaction: 147-fold in the ME, 435-fold in the MM context; holoenzyme reaction: 1500-fold in the ME, 38-fold in the MM context). Such relatively low stimulatory effect on MM variants in the holoenzyme reaction could be attributed to the failure of the archaeal S-domain to confer ternary complex formation at the low metal ion and enzyme concentrations present in the holoenzyme reactions. Indeed, throughout low activities for variants carrying the archaeal instead of the E. coli S-domain (Tables 2 and 3, Figure 5) were generally paralleled by very little formation of holoenzyme-substrate complexes (Table 4). The less severe phenotype of variants harboring the archaeal S-domain in the RNA-alone relative to the holoenzyme reaction can be attributed to the fact that RNA-alone assays were performed at very high P RNA concentrations (10 μM) and high Mg2+ (100 mM), which apparently mitigated the effect of low substrate affinity.
These findings pinpoint the S-domain as the limiting part of the archaeal P RNA. In contrast, the archaeal C-domain with a few mutations not only displayed substantial catalytic proficiency, but also highly productive interaction with the bacterial P protein. It remains to be shown whether minor mutations will be able to partially cure the S-domain's defect, or if larger architectural alterations, for example, the incorporation of additional structural elements such as P13/P14, will be required to restore S-domain function in the absence of archaeal protein cofactors.
The more pronounced activity gain of the mJ15/18/2/3/P2/nP1 mutation in the ME versus MM context (1500-fold versus 38-fold; Table 2 and 3) is of further interest in view of the fact that RNAs MM and ME had equal activities in the holoenzyme reaction (2 × 10−4 min−1; Table 2 and 3). This discrepancy can be explained by interdomain cooperativity between the E. coli S-domain and the engineered archaeal C-domain carrying the mJ15/18/2/3/P2/nP1 mutations. Indeed, our structure probing data indicate changes in the E. coli S-domain between variants ME and ME-mJ15/18/2/3/P2 (see below).
The C to U exchange in J15/18 was the only single change that had a beneficial effect on the RNA-alone reaction by itself (Table 2). The molecular basis of this effect is not immediately clear, since the corresponding nucleotide in the crystal structure of the type A RNase P RNA from Thermotoga maritima is not resolved (21) and type B RNase P RNAs have a purine at this position (28,39). In E. coli P RNA, a 2′-deoxy modification at this U residue and Rp-phosphorothioate as well as inosine modification at the conserved 5′-terminal G of J15/18, two nucleotides upstream of the U residue (Figure 2), interfered with tRNA binding (24,40). This G residue forms a trans Hoogsteen/sugar edge base pair with the 3′-terminal A residue in J5/15 in the crystal structure of type B RNase P RNA from B. stearothermophilus (39). These findings suggest the J15/18 region to be involved in a structurally and functionally important network of interactions.
Comparative probing of P RNAs ME and ME-mJ15/18/2/3/P2 with RNase T1 and Pb2+ ions under native conditions unveiled distinct structural differences between the two RNA variants, surprisingly spanning the entire subregion P6, J6/16, P16, J16/15, P15, J15/18 and J5/15 (Supplementary Figure S4). J16/15 is the site of interaction with the tRNA 3′-CCA end. With respect to J15/18 itself, the 5′-terminal G residue was observed to be more accessible to RNase T1 cleavage in ME versus ME-mJ15/18/2/3/P2 RNA, whereas the site of mutation and the following C residue were more accessible to Pb2+-induced hydrolysis in ME-mJ15/18/2/3/P2 relative to ME RNA (Figure S4). Surprisingly, differences in accessibility were also seen in L11/12 and P11, although these regions are identical in both P RNA variants. This indicates that changes mJ15/18/2/3/P2 in the C-domain have long-range effects on S-domain structure. L11/12 and P11 are close to the T-loop region in the P RNA-tRNA complex model based on the T. maritima P RNA X-ray structure (21). In conclusion, changes mJ15/18/2/3/P2 seem to affect regions that interact with the substrate (J15/16) or which are close to the substrate binding interface (L11/12 and P11). The beneficial effects of these changes on P RNA activity thus seem to stem from improvements in substrate positioning. The structural probing data raise the question if the mutations introduced in P RNA ME-mJ15/18/2/3/P2 will have similar structural and functional consequences when placed into other archaeal C-domains.
With respect to evolutionary implications of our approach, several findings are remarkable. First, only three minor changes, in J2/3, J15/18 and P2, were required to stimulate RNA-alone activity 80-fold in the ME and 160-fold in the MM context (465- and 40-fold, respectively, in the holoenzyme assay). Second, combining the changes in J2/3 and P2 improved activity in the absence as well as presence of the bacterial P protein. Thus, the latter exerts its function by making use of a structural element crucially important for RNA-alone activity. This may suggest an evolutionary scenario according to which the bacterial P protein was recruited to the catalytic RNA and adapted to it without eliciting significant co-evolution of the RNA subunit. In contrast, the strong dependence of archaeal/eukaryal P RNAs on their protein subunits for enzyme function indicates RNA/protein co-evolution, during which the archaeal P RNA subunit adapted to the protein components concomitant with substantial losses of RNA-alone activity. From a mirror-image perspective, it will be intriguing to explore if, for example, P RNA ME-mJ2/3/P2 may be impaired in its capacity to cooperate with the archaeal C-domain binding proteins Pop5 and Rpp30 (41). Along these lines, the substantial activation of the archaeal C-domain by surprisingly little changes suggests only minor differences in RNA conformation between bacterial and archaeal C-domains, in line with the structure probing results (see above). This may imply that the main function of the archaeal C-domain binding proteins Pop5 and Rpp30 for the C-domain itself is to merely fine-tune the conformation of the catalytic core.
Finally, the maximum single turnover cleavage rate of variant ME-mJ15/18/2/3/P2/nP1 was only 7-fold lower and the Km(sto) only 17-fold higher than the corresponding values measured for E. coli P RNA. It will be interesting to see if restoring the L9–P1 and/or L18–P8 interdomain struts in this ME chimera will further reduce these differences. In conclusion, the profound activation of the archaeal C-domain by minor changes demonstrates the evolutionary closeness of the archaeal and bacterial P RNAs, at least in terms of C-domain structure and function.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the Deutsche Forschungsgemeinschaft (HA 1672/14-1 and GK 1384). Funding for open access charge: DFG/GK 1384.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The gifts of pUC119_T7_M.th._rnpB (James W. Brown, North Carolina State University) and cells of M. thermoautotrophicus strain Marburg (Rolf Thauer, Max Planck Institute for Terrestrial Microbiology, Marburg) are gratefully acknowledged.
REFERENCES
- 1.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 2.Evans D, Marquez SM, Pace NR. RNase P: interface of the RNA and protein worlds. Trends Biochem. Sci. 2006;31:333–341. doi: 10.1016/j.tibs.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Pannucci JA, Haas ES, Hall TA, Harris JK, Brown JW. RNase P RNAs from some Archaea are catalytically active. Proc. Natl Acad. Sci. USA. 1999;96:7803–7808. doi: 10.1073/pnas.96.14.7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kikovska E, Svärd SG, Kirsebom LA. Eukaryotic RNase P RNA mediates cleavage in the absence of protein. Proc. Natl Acad. Sci. USA. 2007;104:2062–2067. doi: 10.1073/pnas.0607326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li D, Willkomm DK, Schön A, Hartmann RK. RNase P of the Cyanophora paradoxa cyanelle: A plastid ribozyme. Biochimie. 2007;89:1528–1538. doi: 10.1016/j.biochi.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 6.Loria A, Pan T. Domain structure of the ribozyme from eubacterial ribonuclease P. RNA. 1996;2:551–563. [PMC free article] [PubMed] [Google Scholar]
- 7.Loria A, Pan T. Recognition of the T stem-loop of a pre-tRNA substrate by the ribozyme from Bacillus subtilis ribonuclease P. Biochemistry. 1997;36:6317–6325. doi: 10.1021/bi970115o. [DOI] [PubMed] [Google Scholar]
- 8.Kirsebom LA. RNase P RNA mediated cleavage: substrate recognition and catalysis. Biochimie. 2007;89:1183–1194. doi: 10.1016/j.biochi.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Schedl P, Primakoff P, Roberts J. Processing of E. coli tRNA precursors. Brookhaven Symp. Biol. 1974;26:53–76. [PubMed] [Google Scholar]
- 10.Gößringer M, Kretschmer-Kazemi Far R, Hartmann RK. Analysis of RNase P protein (rnpA) expression in Bacillus subtilis utilizing strains with suppressable rnpA expression. J. Bacteriol. 2006;188:6816–6823. doi: 10.1128/JB.00756-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biswas R, Ledman DW, Fox RO, Altman S, Gopalan V. Mapping RNA-protein interactions in ribonuclease P from Escherichia coli using disulfide-linked EDTA-Fe. J. Mol. Biol. 2000;296:19–31. doi: 10.1006/jmbi.1999.3443. [DOI] [PubMed] [Google Scholar]
- 12.Tsai HY, Masquida B, Biswas R, Westhof E, Gopalan V. Molecular modeling of the three-dimensional structure of the bacterial RNase P holoenzyme. J. Mol. Biol. 2003;325:661–675. doi: 10.1016/s0022-2836(02)01267-6. [DOI] [PubMed] [Google Scholar]
- 13.Rox C, Feltens R, Pfeiffer T, Hartmann RK. Potential contact sites between the protein and RNA subunit in the Bacillus subtilis RNase P holoenzyme. J. Mol. Biol. 2002;315:551–560. doi: 10.1006/jmbi.2001.5261. [DOI] [PubMed] [Google Scholar]
- 14.Buck AH, Kazantsev AV, Dalby AB, Pace NR. Structural perspective on the activation of RNAse P RNA by protein. Nat. Struct. Mol. Biol. 2005;12:958–964. doi: 10.1038/nsmb1004. [DOI] [PubMed] [Google Scholar]
- 15.Kurz JC, Niranjanakumari S, Fierke CA. Protein component of Bacillus subtilis RNase P specifically enhances the affinity for precursor-tRNAAsp. Biochemistry. 1998;37:2393–2400. doi: 10.1021/bi972530m. [DOI] [PubMed] [Google Scholar]
- 16.Kurz JC, Fierke CA. The affinity of magnesium binding sites in the Bacillus subtilis RNase P x pre-tRNA complex is enhanced by the protein subunit. Biochemistry. 2002;41:9545–9558. doi: 10.1021/bi025553w. [DOI] [PubMed] [Google Scholar]
- 17.Harris JK, Haas ES, Williams D, Frank DN, Brown JW. New insight into RNase P RNA structure from comparative analysis of the archaeal RNA. RNA. 2001;7:2. doi: 10.1017/s1355838201001777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown JW, Nolan JM, Haas ES, Rubio MA, Major F, Pace NR. Comparative analysis of ribonuclease P RNA using gene sequences from natural microbial populations reveals tertiary structural elements. Proc. Natl Acad. Sci. USA. 1996;93:3001–3006. doi: 10.1073/pnas.93.7.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Massire C, Jaeger L, Westhof E. Phylogenetic evidence for a new tertiary interaction in bacterial RNase P RNAs. RNA. 1997;3:553–556. [PMC free article] [PubMed] [Google Scholar]
- 20.Massire C, Jaeger L, Westhof E. Derivation of the three-dimensional architecture of bacterial ribonuclease P RNAs from comparative sequence analysis. J. Mol. Biol. 1998;279:773–793. doi: 10.1006/jmbi.1998.1797. [DOI] [PubMed] [Google Scholar]
- 21.Torres-Larios A, Swinger KK, Krasilnikov AS, Pan T, Mondragon A. Crystal structure of the RNA component of bacterial ribonuclease P. Nature. 2005;437:584–587. doi: 10.1038/nature04074. [DOI] [PubMed] [Google Scholar]
- 22.Williams D, Brown JW. In vitro selection of an archaeal RNase P RNA mimics natural variation. Archaea. 2004;1:241–245. doi: 10.1155/2004/903283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Busch S, Kirsebom LA, Notbohm H, Hartmann RK. Differential role of the intermolecular base-pairs G292-C75 and G293-C74 in the reaction catalyzed by Escherichia coli RNase P RNA. J. Mol. Biol. 2000;299:941–951. doi: 10.1006/jmbi.2000.3789. [DOI] [PubMed] [Google Scholar]
- 24.Heide C, Pfeiffer T, Nolan JM, Hartmann RK. Guanosine 2-NH2 groups of Escherichia coli RNase P RNA involved in intramolecular tertiary contacts and direct interactions with tRNA. RNA. 1999;5:102–116. doi: 10.1017/s1355838299981499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wegscheid B, Hartmann RK. The precursor tRNA 3′-CCA interaction with Escherichia coli RNase P RNA is essential for catalysis by RNase P in vivo. RNA. 2006;12:2135–2148. doi: 10.1261/rna.188306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beebe JA, Fierke CA. A kinetic mechanism for cleavage of precursor tRNA(Asp) catalyzed by the RNA component of Bacillus subtilis ribonuclease P. Biochemistry. 1994;33:10294–10304. doi: 10.1021/bi00200a009. [DOI] [PubMed] [Google Scholar]
- 27.Warnecke JM, Held R, Busch S, Hartmann RK. Role of metal ions in the hydrolysis reaction catalyzed by RNase P RNA from Bacillus subtilis. J. Mol. Biol. 1999;290:433–445. doi: 10.1006/jmbi.1999.2890. [DOI] [PubMed] [Google Scholar]
- 28.Massire C. Ph.D thesis. Strasbourg: Université Louis Pasteur; 1999. Développement de logiciels d'aide à la modélisation d'ARN. Application à la composante ribonucléique de la ribonucléase P bactérienne. [Google Scholar]
- 29.Marszalkowski M, Willkomm DK, Hartmann RK. Structural basis of a ribozyme's thermostability: P1-L9 interdomain interaction in RNase P RNA. RNA. 2008;14:1–7. doi: 10.1261/rna.762508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buck AH, Dalby AB, Poole AW, Kazantsev AV, Pace NR. Protein activation of a ribozyme: the role of bacterial RNase P protein. EMBO J. 2005;24:3360–3368. doi: 10.1038/sj.emboj.7600805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlegl J, Hardt W-D, Erdmann VA, Hartmann RK. Contribution of structural elements to Thermus thermophilus ribonuclease P RNA function. EMBO J. 1994;13:4863–4869. doi: 10.1002/j.1460-2075.1994.tb06813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marszalkowski M, Teune JH, Steger G, Hartmann RK, Willkomm DK. Thermostable RNase P RNAs lacking P18 identified in the Aquificales. RNA. 2006;12:1915–1921. doi: 10.1261/rna.242806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brännvall M, Kikovska E, Wu S, Kirsebom LA. Evidence for induced fit in bacterial RNase P RNA-mediated cleavage. J. Mol. Biol. 2007;372:1149–1164. doi: 10.1016/j.jmb.2007.07.030. [DOI] [PubMed] [Google Scholar]
- 34.Wegscheid B, Hartmann RK. The precursor tRNA 3′-CCA interaction with Escherichia coli RNase P RNA is essential for catalysis by RNase P in vivo. RNA. 2006;12:2135–2148. doi: 10.1261/rna.188306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wegscheid B, Hartmann RK. In vivo and in vitro investigation of bacterial type B RNase P interaction with tRNA 3′-CCA. Nucleic Acids Res. 2007;35:2060–2073. doi: 10.1093/nar/gkm005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siegel RW, Banta AB, Haas ES, Brown JW, Pace NR. Mycoplasma fermentans simplifies our view of the catalytic core of ribonuclease P RNA. RNA. 1996;2:452–462. [PMC free article] [PubMed] [Google Scholar]
- 37.Perreault JP, Altman S. Important 2′-hydroxyl groups in model substrates for M1 RNA, the catalytic RNA subunit of RNase P from Escherichia coli. J. Mol. Biol. 1992;226:399–409. doi: 10.1016/0022-2836(92)90955-j. [DOI] [PubMed] [Google Scholar]
- 38.Cuzic S, Hartmann RK. A 2′-methyl or 2′-methylene group at G+1 in precursor tRNA interferes with Mg2+ binding at the enzyme-substrate interface in E-S complexes of E. coli RNase P. Biol. Chem. 2007;388:717–726. doi: 10.1515/BC.2007.095. [DOI] [PubMed] [Google Scholar]
- 39.Kazantsev AV, Krivenko AA, Harrington DJ, Holbrook SR, Adams PD, Pace NR. Crystal structure of a bacterial ribonuclease P RNA. Proc. Natl Acad. Sci. USA. 2005;102:13392–13397. doi: 10.1073/pnas.0506662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hardt W-D, Erdmann VA, Hartmann RK. Rp-deoxy-phosphorothioate modification interference experiments identify 2′-OH groups in RNase P RNA that are crucial to tRNA binding. RNA. 1996;2:1189–1198. [PMC free article] [PubMed] [Google Scholar]
- 41.Tsai HY, Pulukkunat DK, Woznick WK, Gopalan V. Functional reconstitution and characterization of Pyrococcus furiosus RNase P. Proc. Natl Acad. Sci. USA. 2006;103:16147–16152. doi: 10.1073/pnas.0608000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.