Abstract
Stable RNAs are modular and hierarchical 3D architectures taking advantage of recurrent structural motifs to form extensive non-covalent tertiary interactions. Sequence and atomic structure analysis has revealed a novel submotif involving a minimal set of five nucleotides, termed the UA_handle motif (5′XU/ANnX3′). It consists of a U:A Watson–Crick: Hoogsteen trans base pair stacked over a classic Watson–Crick base pair, and a bulge of one or more nucleotides that can act as a handle for making different types of long-range interactions. This motif is one of the most versatile building blocks identified in stable RNAs. It enters into the composition of numerous recurrent motifs of greater structural complexity such as the T-loop, the 11-nt receptor, the UAA/GAN and the G-ribo motifs. Several structural principles pertaining to RNA motifs are derived from our analysis. A limited set of basic submotifs can account for the formation of most structural motifs uncovered in ribosomal and stable RNAs. Structural motifs can act as structural scaffoldings and be functionally and topologically equivalent despite sequence and structural differences. The sequence network resulting from the structural relationships shared by these RNA motifs can be used as a proto-language for assisting prediction and rational design of RNA tertiary structures.
INTRODUCTION
Natural RNA molecules can have a high structural complexity, but even the most complicated RNA machines rely on basic modular architectural principles (1,2). The hierarchical nature of RNA is encoded within its sequence. Due to their higher thermodynamic stability, RNA helices defining RNA secondary structure usually form rapidly prior to tertiary interactions (3). In presence of metal ions, these secondary structure elements collapse first into compact intermediate conformers that undergo further rearrangements through a tertiary conformational search that leads to the final tertiary native structure [e.g. (4,5)]. This last step is essentially dependent on the local folding of recurrent and specific sets of nucleotides that specify for the ‘signature’ of modular and recurrent tertiary structure motifs. So far, numerous RNA structural motifs have been identified by comparative sequence and structural analysis of natural RNA molecules studied by NMR and/or X-ray crystallography (6,7). The simplest examples include A-form helices, apical RNA tetraloops, small internal loops and bulges (1,6,7). More complex sequence signatures control the alignment of RNA multi-helix junctions (8), pseudoknots (9) and the packing of helices in larger structures (10–13). While these motifs exemplify the high modularity and hierarchical structure adopted by stable RNAs, they also suggest that a clear relationship can presently be established between an RNA sequence and its tertiary structure in order to solve the folding problem of naked RNA in a reasonably near future (3).
A-form RNA helices consist of classic Watson–Crick base pairs that involve the Watson–Crick edge of each base in a cis orientation. However, nucleotides have three potential hydrogen bonding faces: the Watson–Crick (WC), Hoogsteen (HG) and Shallow Groove (SG) edges, as well as two possible orientations of the nucleotides with respect of one another: cis and trans (14). Overall these arrangements essentially allow for 12 major types of base-pairing interactions (14). In complex RNA structures such as the ribosome, some of these non-canonical base pairs occur much more frequently than others (15). For example, the U:A (WC:HG) trans, the G:A (HG:SG) trans and the A:G (SG:SG) trans bps are among the most abundant non canonical bps found in natural RNA structures (16,17) (Jaeger, unpublished results). Canonical and non-canonical bps are often combined with one another to create small recurrent tertiary submotifs with characteristic conformations such as the A-minor (10,18,19), GA sheared (20) or bulged-G submotifs (21).
Herein, we present the result of an extensive sequence and structure analysis of high-resolution NMR and crystallographic 3D structures of RNA that led to the identification of a highly recurrent and versatile submotif that we call the UA_handle motif. This submotif enters in the composition of several other motifs of greater structural complexity by combining with other small submotifs such as the G/AR submotif (20) and the well-known A-minor (18,19) and U-turn (1,22,23) submotifs. The ‘sequence network’ that results from the sequence and structural relationships existing between most tertiary motifs identified to date highlights the syntax of emerging folding and assembly principles and rules that direct the stacking, orientation and positioning of RNA helices with respect to one another. As such, this network can be seen as an emerging RNA 3D structure proto-language that can facilitate the prediction of the tertiary structure of natural RNA molecules and the rational design of novel RNA architectures (7,24,25). We present the refined prediction of the architecture for the aptamer core of two riboswitches as examples. Additionally, the structural motif network has possible implications for the evolution of natural RNAs.
MATERIAL AND METHODS
NMR and crystallographic atomic structures of RNA molecules from the Protein Data Bank (PDB; http://www.pdb.org) were visually examined using SwissPDBviewer (expasy.org/spdbv) to identify non-canonical bps according to the Leontis and Westhof nomenclature (14). These non-canonical bps were mapped with symbolic characters on secondary structure diagrams [e.g. (26)]. This led to the fast visual identification of recurrent base pairs patterns that correspond to various classes of tertiary structure motifs of RNAs. To determine the underlying structural characteristic of the different categories of UA_handle motifs presented in this study, their atomic structures were extracted from the original PDB files, visualized with PyMOL (pymol.sourceforge.net) and superimposed using LSQMAN (xray.bmc.uu.se/usf/) to calculate their Root-Mean-Square Deviation (RMSD). Only conserved atomic positions in X(1), U(2), A(3) and X(5) were used for RMSD calculation. Within the set of atomic structures analyzed in this study (Table S2), no additional UA_h motif was identified by automatic search using the program FR3D (27). UA_handle motifs were found to enter into the composition of a large subset of recurrent structure motifs of greater structural complexity, totaling almost 20% of the recurrent motifs identified in the ribosome. The conserved, semi-conserved and variable nucleotide set specifying for the sequence signature of each of these motifs was determined by sequence comparative analysis of subset of rRNA sequences for which 2D structure diagrams were available (17,28,29).
RESULTS
Definition of the UA_handle submotif
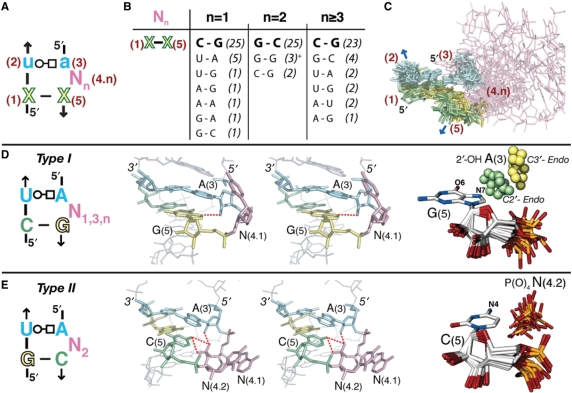
The UA_handle submotif is more widespread than the majority of previously identified RNA motifs (1,6,7). As illustrated in Figure 1, the UA_handle submotif is specified by a minimum of five nucleotides (labeled 1–5). The sequence signature of the UA_handle encodes a structurally conserved ‘core’ consisting of a (WC:HG) trans bp, typically a U(2):A(3), stacked on a classic (WC:WC) cis bp, labeled X(1):X(5) (Figure 1A). A bulge region bridges these two base pairs at position N(4.n), in 3′ of position A(3). It can involve a variable number (n) of nucleotides from one to several hundreds and can adopt very different conformations (Figure 1C). However, structural trends can be identified (Figure 1D and E). For instance, the phosphate backbone in 3′ of A(3) often changes direction, leading to a local parallel orientation of the bulging strand in relation to the opposite strand. In stable RNAs, this motif is essentially involved in the formation of long-range tertiary contacts with the rest of the molecule by acting as a ‘handle’ for tertiary contacts. Because the most distinctive structural feature leading to these versatile interacting properties is the U:A (WC:HG) trans bp, the whole motif is named UA_handle (UA_h). As exemplified below, the objective criteria specifying for UA_h signatures can be refined further by comparative sequence and structural analysis (Figure 1).
Figure 1.
Type I and type II UA_handle signatures and conformers. (A) The UA_handle is typically formed of a U(2):A(3) (WC:HG) trans bp (in blue) stacked on a classic WC bp, X(1):X(5) [purine in green, pyrimidine in yellow), with one or more bulging nucleotides located 3′ of A(3), (N(4.n) in pink]. For bp annotations, see legend of Figure 2. (B) Sequence variation of X(1):X(5) in function of N(4.n). The table is based on the sequence analysis of 99 atomic structures of UA_h with canonical U(2):A(3) (WC:HG) trans bp. The occurrence of each bp is indicated between brackets. (+) These three G(1):G(5) bps are all from one of the UA_hs of the double T-loop motif. Despite a 2nt bulge, this non-canonical UA_h is of type I. (C) Superposition of canonical UA_handle motifs identified in X-ray structures and listed Table S1. The color code corresponds to the one in (A). The conserved atomic positions in X(1):X(5) and U(2):A(3) were used for the superposition. (D) Sequence signature of type I UA_h: an H-bond can be formed between N7-G(5) and 2′OH-A(3) when the sugar pucker of A(3) is in C2′ endo. (E) Sequence signature of type II UA_h: an H-bond can form between N4-C(5) and the O1 or O2 of P(O)4-N(4.2). As seen in the stereographic image, an additional H-bond can potentially form between 2′OH-A(3) and O2P-N(4.2). For more examples of type I UA_h, see also Figure S1 from Supplementary Data.
Comparative sequence and structural analysis of the UA_handle
The atomic structures and sequences of UA_handle motifs identified in different locations within the high resolution atomic structures (Table S1) were classified according to the number of bulging nucleotides for deriving their corresponding sequence consensus (Figure 1). Among the motifs studied, 35% have one nucleotide in bulge, 35% have three or more bulging nucleotides and 30% present two nucleotides in bulge. The (WC:HG) trans bp is most commonly a U:A (92%), but other base pairs such as C:A (4%), A:A (1%), G:A (1%), C:C (1%) and G:G (1%) can be found at positions 2 and 4 as well. As noticed by Krasilnikov and Mondragon for the T-loop motif (30), this variability indicates that the UA_handle is flexible enough to accommodate less conventional (WC:HG) trans bps, which do not have to be perfectly isosteric to the U:A (WC:HG) trans bp.
There is a strong correlation observed between the number of bulging nucleotides in the UA_handle and the sequence of the X(1):X(5) (WC:WC) cis bp (Figure 1B). In UA_handles with one, three or more nucleotides in bulge, the WC is most often a C:G bp (70%), with X(5) being usually a purine (90%). It is only in a few particular structural contexts that other bps are exceptionally observed (Figure 1B). In contrast, UA_handles with two bulging nucleotides mostly display a G:C bp (83%). This correlation is rooted in the formation of sequence specific H-bond contacts that specifically involve the nucleotide at position X(5). Two different structural types of UA_handles can therefore be distinguished (Figure 1D and E).
In UA_handles of type I (∼70% of identified UA_h), a hydrogen bond can form between atom N7 (and sometimes atom O6) of R(5) and the 2′OH of A(3) (Figure 1D). This interaction is particularly favorable in type I motifs with C2′endo sugar pucker at A(3): the average distance between 2′OH-A(3) and N7-R(5) is 3.22 Å (Table S1). However, in one third of type I UA_handles, the sugar pucker of A(3) is in C3′ endo which prevents this H-bond contact. A possible interpretation is that this H-bond contact might only form transiently due to alternative competing tertiary contacts that may involve the bulging nucleotides. On the other hand, this specific contact might have been overlooked as the resolution of RNA X-ray structures are often not high enough to unambiguously distinguish between C2′ and C3′ endo sugar puckers. In summary, the structural conservation of the H-bond contact 2′OH-A(3):N7-R(5), while rationalizing the sequence conservation of the C(1):G(5) bp, directs the ribose phosphate backbone of the bulge region to point outward. The high conformational flexibility of the bulge region allow orientation of the bulging bases in many different directions proper for long range tertiary interactions, especially when the number of nucleotides in bulge is three or more.
For UA_handles of type II (∼30% of identified UA_h), the cytosine at position X(5) can screen the negative charge of the phosphate from the bulging nucleotide at position N(4.2) (Figure 1E). An H-bond can possibly form between atom N4 of C(5) and one of the phosphate oxygens (O1 or O2) of N(4.2): the average distance between N4-C(5) and PO1-N(4.2) is close to 3.47 Å (Table S1). The sugar pucker of A(3) is usually in C2′ endo, which often allows the formation of one additional H-bond between 2′OH-A(3) and the phosphate of N(4.2). The conserved G(1):C(5) bp favors the two bulging nucleotides to be in a typical A-form conformation: The phosphate of N(4.2) is oriented inwards toward the deep groove of the UA-handle and the bulging bases N(4.1–4.2) are oriented outwards in a conformation particularly suitable for long-range Watson-Crick base pairings (Figure 1E). UA_handles of type II are therefore often found to be involved in the formation of pseudoknots (Figure 2, bottom).
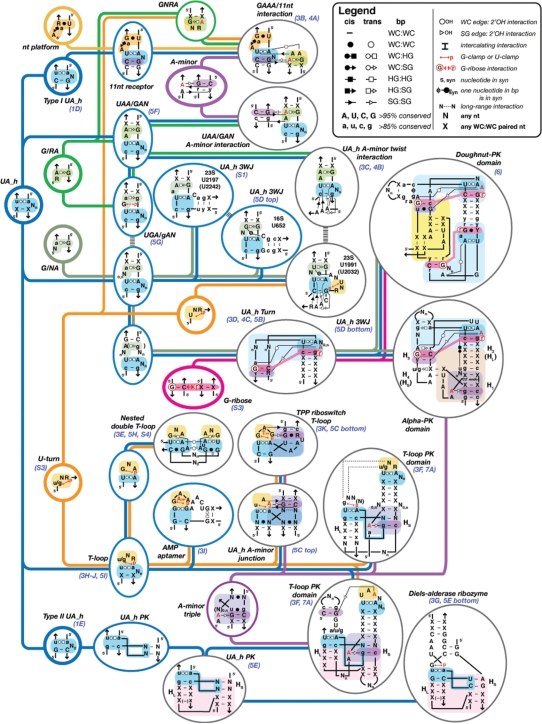
Figure 2.
Hierarchical organizational network of RNA structure motifs built from the UA_h submotif. Motifs are organized from the left to the right according to their increase in structural complexity. Submotifs are minimally sized recurrent set of nucleotides with conserved conformation (circled in bold lines). They are not expected to be stable by themselves as they are almost always associated to other submotifs or nts to create full-fledge stable motifs (circled in blue or gray for secondary and tertiary motifs, respectively.) The most significant structural characteristics of UA_h motifs are indicated on their respective sequence signatures according to the annotation of Leontis and Westhof (74). Visual of the 3D structures of some of these motifs can be found in the figures indicated in blue below the motif name. For bps symbols, see the legend in inset: WC, Watson–Crick edge (circle); HG, Hoogsteen edge (square); SG, shallow groove edge (triangle). For example, the WC:HG trans bp is symbolized by an open circle associated to an open square. The WC:HG cis bp is represented by a plain circle combined to a plain square. Capital letters indicate that the nucleotide is conserved in more than 95% of the cases: small letters indicate that the nucleotide position is conserved in more than 85% of the cases. X, any nucleotide (A, U, C or G) paired to another one through classic WC cis bp; N, any nucleotide (A, U, C or G); R or r, purine; Y or y, pyrimidine; Nn, a sequence of n nucleotides; U or G-clamp, WC edge of a U or G in interaction with a phosphate; syn or S, indicates that a single or paired nucleotide is in syn; Hl and Hs stand for the long and short stems of a pseudo-knot, respectively. 5′ and the arrow symbol indicate 5′ and 3′ ends, respectively. See also Figure S3 (Supplementary Data).
When phylogenetically conserved, type I and type II sequence signatures are the objective criteria for identifying without ambiguity UA_handle conformers within related RNA sequences (Discussion). However, other criteria could be useful to assess the presence of UA_h conformers within an RNA structure in absence of phylogenetic information. In order to identify some of them, the secondary structures of known crystallized 23S ribosomal RNAs were searched for type I and type II sequence signatures, irrespective of their phylogenetic conservation (Figure S2). Only 35% of these signatures are truly folded as typical UA_handle conformers. This suggests that additional sequence and structural constraints at the level of the UA_handle signatures themselves or within their surrounding nucleotides prevent or favor the folding of these signatures into UA_handle motifs. For instance, properly folded UA_handles cannot have a regular helix immediately connecting the 3′ end of U(2) to the 5′ end of A(3). Furthermore, most properly folded UA_handles disfavor a guanine immediately 5′ of X(5) (Figure S2). The presence of a guanine at this position has apparently been counter-selected during evolution as it contributes to the formation of an alternative U:G wobble bp, another prevalent structural feature often found at the extremities of RNA helices (12). While both bulging adenine and guanine could potentially pair with U(2) and compete with the formation of the U(2):A(3) (WC:HG) trans bp, the estimated free energy of formation of a U(2):A WC bp at the end of a helix is not as favorable as the one of a U(2):G wobble bp (31). Additionally, bulging adenines are often stabilized by tertiary interactions (18). This can therefore explain why adenines have been evolutionarily preferred versus guanines for the bulge position immediately 5′ of X(5) in properly folded UA_handle signatures (Figure S2).
Like the U-turn motif, the UA_handle motif is unlikely to be very stable by itself (32). Additional surrounding sequence elements are usually present for conditioning the proper folding of the UA_handle signature into a UA_handle conformer (Figure 2). As such, the UA_handle can be seen as a minimally sized submotif that acts as a building block for the buildup of larger RNA motifs. Understanding how the UA_h combines with additional sets of nucleotides to generate more complex motif signatures is therefore essential for the assessment of any putative UA_h motif identified within an RNA sequence (see below).
The UA_handle as a modular building block for motifs of greater structural complexity
The UA_handle, along with the U-turn, the A-minor and the G/AR submotifs, are the most abundant submotifs identified to date. Among the UA_handles that we have identified so far in NMR and X-ray structures, 52 are submotifs found at distinct locations within ribosomal RNAs, ribozymes, aptamers and riboswitches (Table S2). Among these, ∼90% correspond to UA_handles that enter into the composition of motifs of larger structural complexity (Table S2), some of which have been described in the literature to date. For instance, the UA_handle is present in the ‘11 nt receptor’ motif (19,33), the ‘UAA/GAN’ motif (34), the ‘T-loop’ motif (30,32,35) and ‘G-ribose’ related motifs (9,36). Additionally, it is also part of multi-helix junctions and pseudo-knotted domains from the ribosome (8,9) and various ligands and substrates binding sites of natural and selected aptamers (37–39) and ribozymes (40).
As shown in Figure 2, the UA_handle combines with other minimally sized recurrent submotifs such as the dinucleotide platform submotif (19,41,42), the G/AR and G/AN submotifs (20) and the U-turn (1,22,23) to form terminal, internal loops or junctions motifs. These motifs can then interact further through A-minor submotifs (18,19), G-ribose motifs (9,36), WC bps and/or nucleotide intercalating interactions (Figure S3) to form even more complex motifs that specify for local helical turn regions, multi-helix stacking arrangements, pseudo-knots and other long-range tertiary interactions. Therefore, the knowledge of the sequence space associated to each structural motif can aid in deciphering whether a sequence is likely to adopt a particular conformation at a 3D level.
To distinguish whether or not a UA_handle signature is likely to fold as a UA_handle conformer the sequence signatures of larger structural motifs incorporating the UA_handle should be taken into account, as these define the syntax in which the UA_handle signature can be used within higher order motifs. Remarkably, a minimal change of syntax at the level of a motif sequence can have a dramatic effect on its outcome at a tertiary structure level (Figure 2). For example, a single nucleotide insertion at the sequence level of an internal loop motif can determine whether this sequence folds as a UAA/GAN motif or 11 nt motif (e.g. ccUAAg/uGAagg versus CCUAaG/U_AaGG, highly conserved nt positions are in capitals). In addition, conserved G:C bps in the surrounding of a UA_handle signature can indicate the presence of G-ribose or typeI/II A-minor submotifs (9,18,19,36) that could allow identification of ‘UA_handle turns’ (UA_h turn) or GAAA/11 nt receptor interactions, respectively (Figures 2). The UA_h turn motif, in particular, can constrain its immediate surrounding helical elements in such a way that additional long-range base pairings can form, leading to the formation of recurrent pseudo-knotted topological domains of 50–65 nt (Figure 2). In the case of the ‘alpha-PK domain’, the length of the helical stems H2, H3 and H4 are conserved and the nucleotides at the junction of H1 and H2 corresponds to the sequence signature of a conserved ‘A-minor triple helix’ motif that forces H1 to stack in continuity of H2. In the case of the ‘doughnut-PK domain’, the topological domain is constrained by the sequence signature of two UA_h turns that favor the formation of a kinked long-range base pairing (Figure 2). Therefore, a careful examination of the set of nucleotides adjacent to UA_handle signatures within the 2D structure of an RNA can lead to the prediction of particular structure motifs with minimal ambiguity.
While the T-loop motif was recently re-defined as a pentaloop (30), it is now apparent that most T-loops identified so far in crystallographic structures should be seen as the full-fledged combination of a UA_handle and the U-turn of UNR or GNR sequences (32,35) (Figure 2). The sequence signature of the UA_handle is not absolutely required to be contiguous. For example, the ‘nested double T-loop’ motif that is formed of two interacting classic T-loops, has its 5′ and 3′ ends localized between positions X(1) and U(2) (Figures 2, 3E and S4). In this specific case, the UA_handle submotif results from long-range interactions. Typically, apical T-loop motifs are characterized by one, two or three bulging nucleotides and follow the type I and type II UA_handle rules (Table S2). T-loops are always found involved in either long-range interactions or as the recognition site of small ligands (Figure 3). In fact, SHAPE chemical probing of the tRNA (43) as well as NMR data of the AMP/ATP aptamer in absence of AMP (37) indicate that T-loop motifs are not particularly stable in absence of RNA or ligand partners. These observations are consistent with molecular dynamic characterizations of the T-loop motif in isolation, with the UA_handle portion being significantly more stable than the U-turn (32). The T-loop motif can sometimes be part of more elaborate structure motifs when one or two helical elements are present in the bulge. For instance, the prevalent ‘A-minor junction’ motif (Geary and Jaeger, unpublished data) which is based on the A-minor triple motif and often involve lonepair triloop motifs (44), can sometimes take advantage of the GNR submotif from a T-loop to form the A-minor interaction, resulting to the ‘UA_h A-minor junction’ motif (Figure 2). One of the TPP-binding sites of the TPP riboswitch is structurally equivalent to the UA_h A-minor junction. In this natural aptamer, the classic type-I A-minor interaction is substituted by an almost isosteric G:G (SG:SG) trans bp (Figures 2 and 3) (38,39). Another example of a T-loop-like motif is the AMP/ATP aptamer mentioned above (Figures 2 and 3I), which presents a helix in its bulge region (37). In this in vitro selected aptamer, the U:A (WC:HG) trans bp is substituted by a structurally similar G:G (WC:HG) trans bp.
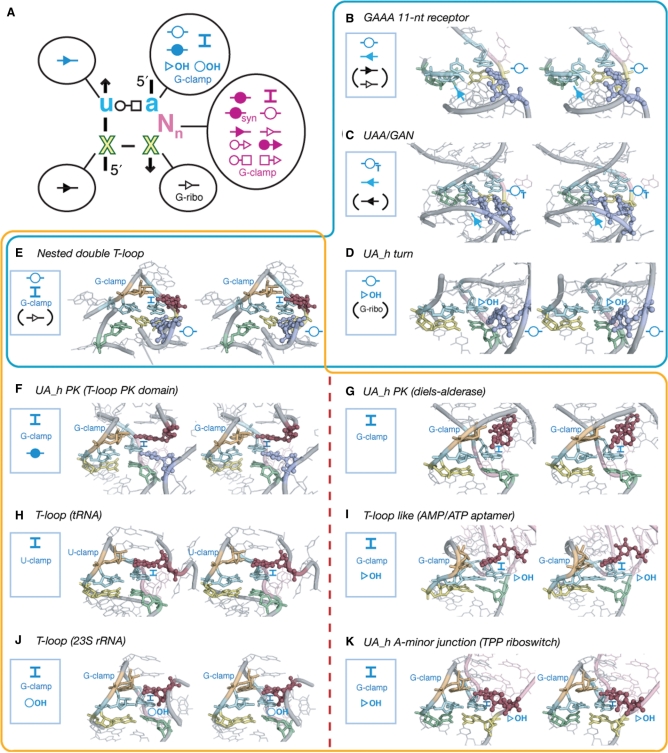
Figure 3.
The UA_handle motif is involved in various modes of tertiary interactions. (A) 2D diagram summarizing the interactions that the different nt positions of the UA_handle can form. For the legend, see Figures 2 and S3. Interactions are colored according to the nucleotide position involved in the UA_h. (B–K) Examples of various interactions involving the U(2):A(3) bp. Bps and tertiary interactions are indicated according to the color code and annotations in (A). Bps indicated between brackets are not shown on the stereo images. (B) GAAA/11 nt motif, A(s.1) is in violet (see Figure 4 and text); (C) UAA:GAN motif (23S; Ec U1578), A(s.1) and A(s.2) are in violet; (D) UA_handle turn (23S; Tt U1621), A(s.1) is in violet; (E) Nested double T-loop (23S; Hm U481); (F) UAh_PK motif (23S; Hm U2069); (G) UA_h_PK motif bound to DielsAlder reaction product (Diels-Alderase ribozyme); (H) T-loop motif (tRNAPhe), notice the U-clamp instead of G-clamp (Figure S3); (I) T-loop like motif bound to AMP (AMP/ATP aptamer); (J) T-loop motif (23S; Hm U1388); (K) T-loop motif bound to TPP (TPP riboswitch). Motifs boxed in blue share a WC:WC trans bp involving A(3). Motifs boxed in orange share a G-clamp or U-clamp. On the right side of the red dotted line are motifs from riboswitches and ribozyme recognizing ligands. Notice the similar modality of nt (or ligand) recognition between the tRNAPhe T-loop and ATP aptamer T-loop-like and between the TPP riboswitch T-loop and the 23S RNA T-loop.
UA_handle motifs of type II, with two nucleotide bulges, are frequently involved in the formation of a pseudoknot motif that we called the ‘UA_h PK’ (Figures 2 and 5E). This motif is not only observed multiple times in the ribosome, but also appears in the artificially evolved Diels-Alderase ribozyme (40) (see Table S2 and Figures 2 and 3G). The length of its various helical components are constrained such that its helix HL, which holds the type II UA_h motif, is three to four bp long with either one or no unpaired nucleotide connecting HL to a helix stacked in continuity of HS, respectively. In three instances in the 23S ribosomal RNA, this pseudoknot is found directing the formation of a larger domain, called the ‘T-loop_PK domain’ (Figure 2, Table S2). This domain is a combination of UA_h PK, T-loop and A-minor triple motifs. The T-loop is localized at the end of a 5 bp stem that is stacked and positioned in continuity of Hs through formation of an A-minor triple motif with Hs. The T-loop is typically in interaction with the nucleotides localized 5′ of position A(3) from the UA_handle PK. In the 23S rRNA, two of the T-loop PK domains form an intercalating nucleotide interaction (Figure S3) while the third one presents an A-minor interaction, structurally different from the previous ones but functionally equivalent in term of stabilizing potency (see also Discussion section).
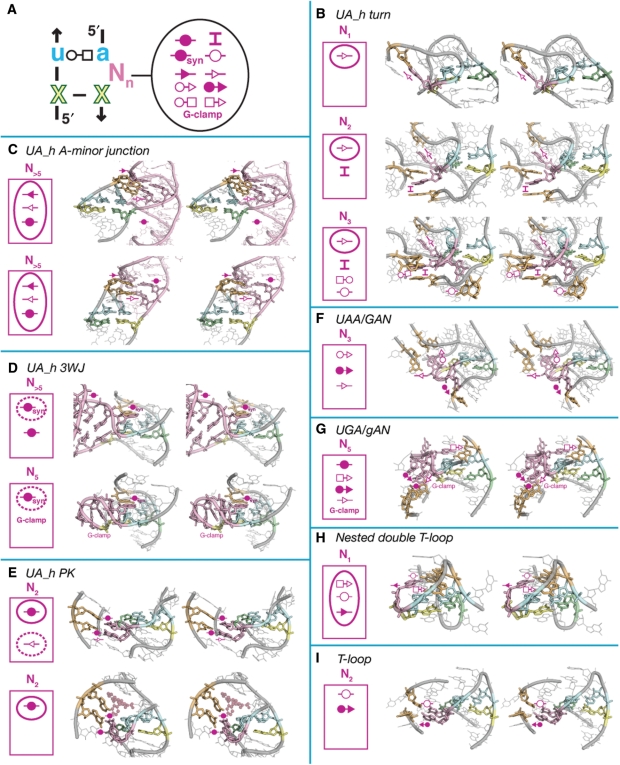
Figure 5.
Examples of typical UA_handle interactions involving the bulging nucleotides N(4.n). (A) Schematic highlighting the various bulge mediated tertiary interactions in UA_handle motifs. (B) UA_h_turn motifs: (top) Hm 23S rRNA U1116 (PDB_ID:1JJ2), (middle) Tt 23S rRNA U1621 (PDB_ID:2J01), (bottom) Hm 23S rRNA U1696 (PDB_ID:1JJ2). (C) UA_h A-minor junction motifs: (top) Tt 23S rRNA U2562 (PDB_ID:2J01), (bottom) from TPP riboswitch (PDB_ID:2GDI). (D) UA_h 3WJ motifs: (top) Ec 16S rRNA U652 (PDB_ID:2AW7), (bottom) Ec 23S rRNA U1991 (PDB_ID:2AWB). (E) UA_h_PK motifs: (top) Hm 23S rRNA U1676 (PDB_ID:1JJ2), (bottom) from Diels-alderase (PDB_ID:1YKV). (F) UAA/GAN motif: Hm 23S rRNA U1457 (PDB_ID:1JJ2). (G) UGA/gAN motif; Tt 23S rRNA U1621 (PDB_ID:2J01). (H) Nested double T-loop: Tt 23S rRNA U475 (PDB_ID:2J01). (I) T-loop motif: Hm 23S rRNA U1388 (PDB_ID:1JJ2). For each examples displayed, the type of interactions involved is indicated in the upper left corner. Conserved and semi-conserved interactions are circled with continuous or dashed lines, respectively. For annotations, see Figures 2 and S3.
The UA_handle as a versatile module for long-range tertiary interactions
Alongside A-minor and GU packing motifs (12,13), the UA_handle can be considered one of the most prevalent motifs promoting formation of long-range tertiary interactions in stable RNAs. UA_handles are mostly found involved in the formation of a great variety of tertiary interactions that can involve the X(1):X(5) WC bp, the U(2):A(3) (WC/HG) trans bp or/and the nucleotides in bulge N(4.n) (Figure 3A and Table S2). The structural nature of UA_handle tertiary interactions can be dependent on the local context of adjacent nucleotides that define motifs of greater structural complexity (Figure 2). On the other hand, UA_handles can also be involved in the formation of less predictable, long-range contacts with nucleotides that are part of distant structural contexts. Despite the remarkable versatility of these tertiary interactions, several interesting structural rules can characterize particular type of UA_handle-based motifs.
Interactions involving the X(1):X(5) bp and/or the U(2):A(3) WC;HG trans bp
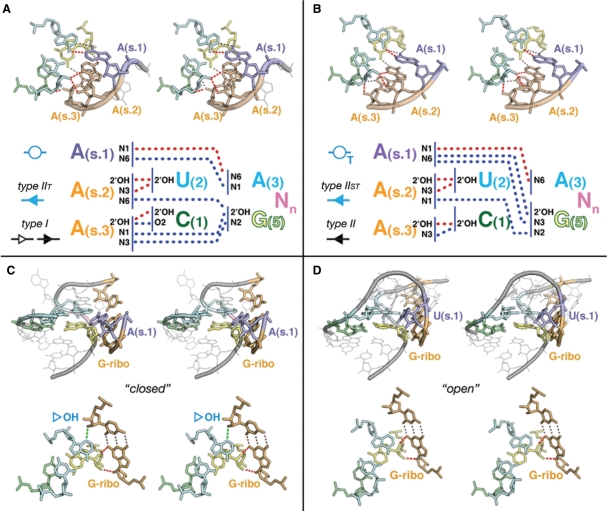
The UA_handle can be directly involved in the formation of A-minor interactions. For instance, the UA_handle of the 11nt receptor motif is involved in the formation of a conserved pattern of nine H-bonds with its cognate GAAA tetraloop (Figures 2 and 4A). U(2) and C(1):G(5) bp are involved in the formation of a type-I/IIT A-minor interaction, also called ‘A-minor tilted’ interaction, with the second (s.2) and third (s.3) adenines of the GAAA loop (19). The first adenine (s.1) forms a WC trans bp with A(3) (Figures 3B and 4B) (19,45).
Figure 4.
Examples of tertiary interactions involving the X(1):X(5) WC bp. (A) the A-minor tilted interaction (type-I/IIT A-minor) from the GAAA/11 nt motif (19): notice the canonical ribose zipper (75) that involves the second and third adenines of the GAAA, in type-I and type-IIT A-minor contacts, respectively (Hm 23S; U_1457). Typical H-bonds pattern for type-I/IIT is indicated. (B) UA_h A-minor twist interaction (type-II/IIST A-minor): this pattern of H-bonds presents the interacting nucleotides in a super-tilted configuration. Notice the cis ribose zipper that involves the second and third adenines, in type-II and type-IIST A-minor contacts, respectively (Bs RNase P; U147). The second adenine in type-IIST allows formation of H-bond, N3(A2): N2(G5), instead type-IIT H-bond, N6(A2):2′OH(G5). Typical H-bond pattern for type-II/IIST is indicated. (C–D) G-ribose (G-ribo) interactions (36): example of ‘closed” and ‘open’ G-ribose interacting motif from UA_h turns. (C) In the ‘closed’ conformation (Hm 23S: U2330), the ‘G ribose’ G:C bp makes three H-bond contacts with the UA_h motif: note the 2′OH-N3(A3) H-bond. (C) In the ‘open’ conformation (Ec 23S: U1019), only two contacts are formed with X(5).
The UAA/GAN motif (34) is a GNRA-like internal loop. Its UA_handle might contribute to the structural stabilization of a G/AA submotif that is involved in A-minor tilted interactions with RNA helices to form the ‘UAA/GAN’ A-minor interaction (Figure 2). This versatile motif can form additional interactions. Its constituent nucleotides, bulging nucleotides included, can be involved in tertiary contacts with surrounding elements in multiple directions spanning more than 280° (Figures 3C, 4B and 5F). Remarkably, the UA-handle components from UAA/GAN and ‘UA_h_3WJ’ motifs can act as receptors for the recognition of three to four consecutive adenines in a way that is very similar to the GAAA/11 nt interaction (Figures 2, 3C and 4B). The resulting type-II/IIST pattern, also called the ‘UA_h A-minor twist’ interaction, is essentially a super-tilted version of the 11 nt/GAAA pattern (Figure 4B). Out of eight conserved H-bonds, four of them are in common with the 11 nt/GAAA pattern (Figure 4A). Through the pivotal interaction between A(s.2) and U(2), the consecutive adenines are oriented almost perpendicular to the UA_handle bp plateaus, allowing A(s.2) and A(s.1) to interact each with the U(2):A(3) and C(1):G(5) bps. For instance, the WC edge of A(s.1) can make a highly tilted A:A WC trans bp with the WC edge of (A3) through one H-bond while also interacting with the SG edge of G(5) (Figures 4B and S3). The UA_h A-minor twist motif is a variant of the A-minor twist motif that occur between stacked adenines and regular helices in the SAM-II riboswitch and glmS ribozyme (46).
Another prevalent pattern of H-bonds is found in the UA_h turn motif (9,36) (Figures 2, 3D, 4C and 4D). Like the ‘kink turn’ (K-turn; Figure 6) (47), this motif allows formation of sharp bends between two adjacent helical elements on their shallow groove side (Figure 4C and D). One helix is capped by a UA handle while the other is capped by a semi-conserved adenine (A.s1) localized 5′ of a conserved G(s.2):C WC bp (Figure 4C and D). Like for the 11nt/GAAA interaction, A(3) is involved in a WC trans bp with position A(s.1) (Figures 3D and 4C). While this base pair seem to be a conserved feature of the UA_h turn, position A(s.1) can seldom be another nucleotide (Figure 4D). The U(2):A(3):A(s.1) triple bp might however adopt an alternative isosteric bp pattern in few motif instances (Figure S5). In all UA_h turns, X(5) is involved in a G-ribose packing interaction with G(s.2) (Figure 4C and D). A conserved bulging adenine, usually immediately in 5′ of X(5), also contributes to the motif stabilization by formation of a type-I A-minor interaction with the bp just below the G(s.2):C bp (Figures 2 and 5B).
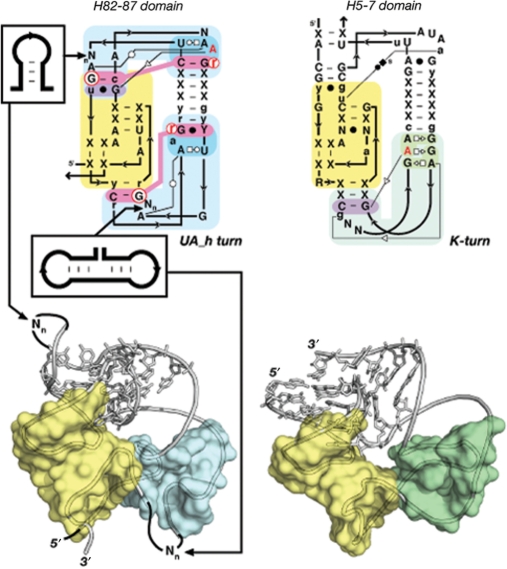
Figure 6.
Principle of functional/topological equivalence. The kink turn [Right: in H5-7 domain (PDB_ID: 1JJ2)] and UA_h turn [Left: in H82–87 domain (PDB_ID:2J01)] can both be components of the doughnut-PK domain identified in the 23S rRNA. Albeit of different sequence signatures and local structures, they are topologically equivalent as they both contribute to the folding of the same topological domain. The doughnut-PK domain can act as scaffolding for structural expansion by accommodating sequence insertions in the UA_h turn (left). For annotations see Figures 2 and S3.
In addition to the 11nt, UAA/GAN, UA_h_3WJ and UA_h turn motifs, the A:A WC trans bp mediated by A(3) is also found in the nested double T-loop motif from the 23S rRNA (Figures 3E and S4). This motif results from the docking of two T-loops through formation of a network of at least 27 H-bond interactions. This intricate array of H-bonds is almost perfectly symmetrical and forms a series of stacked tetraplex bps characterized by a pseudo-symmetry of order 2 (Figure S4A). The two UA_handles interact with one another to form two stacked A:A WC trans bps, involving the two positions A(3) and A(4), respectively. The two GNR components interact with one another to form a bifurcated A:A (HG:HG) trans bp. Additionally, the interface between the U-turn and UA_handle motifs leads to the formation of two A:G (HG:SG) trans bps (Figure S4).
Another prevalent long-range tertiary interaction found in UA_handles is the ‘intercalating nt’ interaction (Figure S3), in which a good example is found in the double nested T-loop (Figure 3E). This type of interaction is always mediated by UA_handle motifs that combine the UA_handle and a ‘G-clamp’ module (more rarely a ‘U-clamp’) (Figure 3E–K). G/U-clamps are particular features of U-turn motifs but are also found in other structural contexts. G-clamps promote an H-bond contact between the WC edge of a G (or U) from one strand and a phosphate group from an opposite strand (Figure S3). Consequently, the two opposite anti-parallel strands are closer from one another than the strands in regular WC bps. Motifs containing G-clamps include T-loops, the T-loop-like AMP aptamer, some UA_hPK motifs, the UA_h A-minor junction from the TPP riboswitch and the UAA/gAN motif (Figures 3E–K and 5G). Typically, all of them mediate the recognition of one long-range nucleotide position (or ligand). This nt is stacked over A(3) that acts as a platform and makes specific H-bond contacts with the G from the G-clamp in 3′ of U(2). The resulting long-range base pair can greatly vary depending the structural context [e.g. (35)]. Therefore, the ‘intercalating nt’ interaction was named after its most common feature, namely its ability to bind distant nucleotides by intercalation (Figure S3).
One of the remarkable features of motifs mediating intercalating nt interactions is that they can be part of the recognition or active site of aptamers, ribozymes or riboswitches. As exemplified by the diels-alderase ribozyme, the AMP aptamer and TPP riboswitch (Figure 3G, I and K), these functional RNA molecules bind their respective substrates or ligands according to structural modalities that are similar to the one observed in UA_handle motifs that promote the folding and stabilization of stable RNAs (Figure 3F, H and J). For instance, it is noteworthy that both tRNA T-loop and AMP aptamer recognize their respective nt target by making a (WC:HG) trans bp allowing the last nt position of the U-turn component to specifically recognize the 2′OH of the nt target. The T-loops from the 23S rRNA (position 1388) and from the TPP riboswitch both recognize their nt or ligand target by forming a (WC:HG) cis bp. These examples demonstrate that, albeit similar, the specific modalities of nt intercalation by UA_handle motif are highly contextual and difficult to predict de novo.
Interactions involving the bulging nucleotides N(4.n)
Of the positions in the UA_handle involved in long-range tertiary interactions, the bulging nt positions are the most versatile. Despite limitations on the length of the bulge due to type I and type II UA_h structural constraints, the conformation of the nucleotides in bulge is extremely variable (Figure 5A). The bulge can interact with its surrounding nucleotides in many different ways that are essentially dependent on the larger structural context of the RNA (see examples Figure 5B and 5F–H). The bulge can also be a point of insertion for additional RNA helical elements leading to the structure of various helical junctions such as the UA_h A-minor junction (Figure 5C), the UA_h 3WJ (Figure 5D) or the AMP aptamer (Figure 3I). Considering the remarkable sequence variability at the level of the bulge, the de novo prediction of its tertiary structure and mode of interaction from its secondary structure is particularly challenging. This is clearly the case for the UAA/GAN motif (Figure 5F) and for most other UA_handle motifs with 1, 3 or more bulging nts.
Several conserved structural trends can nevertheless be identified at the level of the bulge of some UA_handle motifs. For instance, the UA_handle turn has a conserved bulging adenine usually positioned 5′ of X(5) bulge that stabilizes the kink turn by type-I A-minor interaction (Figure 5B). Any other additional bulging nucleotides at the level of the turn essentially contribute to long-range contacts with the surrounding architecture context through specific base-pairings or intercalating interactions (Figure 5B). Their conformations are therefore difficult to anticipate. Note that the phylogenetically conserved UA_h turn motif from the domain three of 23S rRNA of Archaea relies more extensively on long range interactions than the one from bacteria (Figure 5B, middle and bottom).
UA_h A-minor junctions are typically characterized by the insertion of two stacked helical elements at the level of the UA_handle bulge. The base of the helix immediately 5′ of X(5) has at least one conserved G:C base pair that is involved in a classic A-minor interaction with the GNR component of the T-loop (Figure 5C). This tertiary contact therefore dramatically constrains the positioning of the bulging helical elements with respect of the UA_handle. Note that while the helix in 5′ of X(5) is conserved, the one in 3′ of A(3) can be substituted by few unpaired nucleotide like in the TPP riboswitch (Figure 5C). Some UA_h 3WJ motifs have a bulging nucleotide with its base in syn, immediately 3′ of A(3), that is involved in a WC cis bp with the conserved (HG:SG) trans bp component (Figure 5D). This feature is however not observed among all UA_h-3WJ motifs. Another trend not trivial to predict, is that some motifs with UA_h/G-clamp components, like the UAA/gAN and nested double T-loop motifs, make the intercalating nt interaction with one of their bulging nucleotides rather than a distant nucleotide position (Figure 5GH).
In contrast to most previous motifs, type II UA_handles with two bulging nucleotides, usually adopt a classic A-form helical conformation with the bulging strand running locally parallel to the opposite strand. This conformation is particularly suitable for forming classic WC bps (Figure 5E) although this is not always the case (see for example Figure 5I). When paired in a classic WC fashion, this conformation leads to the formation of the UA_h-PK, a pseudoknotted configuration that enters into the composition of larger topological folds (see above and Figures 2 and 5E). Interestingly the pseudoknotted base pairs (Hs) contribute to substrate recognition by creating a stacking platform for one of the diels-alderase substrates (Figure 5E, bottom). In the ribosome, the UA_h-PK usually contributes to the stacking and positioning of an adjacent T-loop hairpin to form the T-loop PK domain (Figures 2 and 7). The two UA_h bulging nucleotides typically make a double-base paired pseudoknot helix [called Hs (Figure 2)] with two complementary nucleotides localized 3′ of this hairpin. A conserved adenine immediately 5′ of the T-loop hairpin can then form a stabilizing A-minor triple interaction with a conserved G:C bp of helix Hs (Figures 2 and 5E, top).
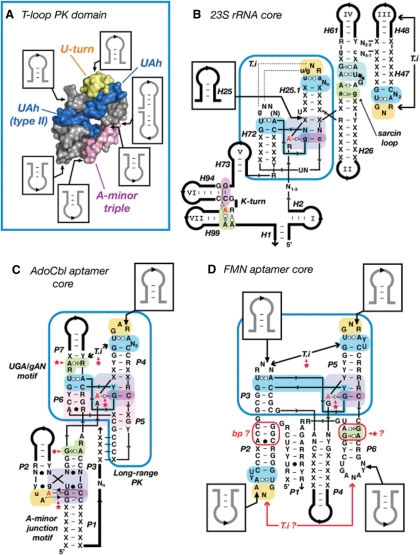
Figure 7.
The T-loop PK domain as a prevalent structural scaffold in natural RNA molecules. (A) Summary of the sequence insertions within the 3D structure of the T-loop PK domain. The constituent sub-motifs are in blue (UA_h), yellow (U-turn) and magenta (A-minor triple). The T-loop PK domain shown is from the 23S rRNA (Hm:301–350; Ec:296–342). (B) Detailed secondary structure diagram of the consensual structural core of the bacterial and Archaea 23S rRNAs deduced from crystallographic structures. Note that the T-loop PK domain (boxed in blue) brings together the various structural domains (I, II, III, IV, V, VI and VII) of the ribosome. (C) Prediction of the architecture of the core of the natural AdoCbl aptamer (62). (D) Prediction of the structural core of the FMN aptamer (62). Positioning of elements P1–P2, P4 and P6 are not well defined compared to P3–P5. The T-loop in P2 should form an intercalating nt interaction with a distant nt that is most likely one of the conserved purines in L6. The GA shared motif in P6 as well as the base pairs in P2 might be involved in one of the riboswitch structural states. (B,C,D) The tertiary interaction (T.i) of the T-loop PK domain (boxed in blue) corresponds to a class of topological equivalence. The conserved T-loop can either recognize another terminal loop (B, D), an UGA/gAN motif (C) or a helix (Figure 2), leading to structurally different but functionally equivalent interactions. Red stars indicate new tertiary interaction predictions [in comparison to (62)]. Thin lines are for zero nt length connectors. Thick lines are for variable length sequence insertions. For annotations, see Figures 2 and S3.
DISCUSSION
RNA tertiary structure as a proto-language
The fact that a limited number of RNA submotifs such as the UA_h, G/RA (20), A-minor (18,19) and U-turn (1,22,23) combine in multiple arrangements to create larger motifs exemplifies the remarkable modularity and hierarchy of natural RNA architectures (6,24,48). The ‘structural motif signature’ network (Figure 2) that results from the sequence and structural relationships existing between UA_h-based motifs identified to date defines the syntax of emerging folding and assembly principles that direct the stacking, orientation and positioning of RNA helices with respect of one another. As such, it is tempting to draw an analogy to the syntax of human languages that takes advantage of syllables (submotifs) that can be combined in various orders, the order of which specifies different words (motifs) that can themselves be joined in numerous ways to produce small sentences or parts of sentences (higher order motifs or RNA domains).
Principles of structural and functional/topological equivalence
The high modularity of RNA results from the fact that its basic building blocks are often interchangeable and therefore equivalent. According to the level of molecular complexity considered, it is possible to distinguish between isosteric equivalence, structural equivalence and functional/topological equivalence. When structural elements can be of different nucleotide compositions but perfectly super-imposable, they are called isosteric (14,49). This is the case of the C:G and A:U WC cis bps from A-form RNA helices. However, perfect isostericity is not absolutely required for structural equivalence. For example, all T-loop motifs can be considered structurally equivalent albeit they might have different bulge conformations or different WC:HG trans bps. Therefore, the sequence signature of a particular structural conformer defines a class of structural equivalence. The principle of functional/topological equivalence is the most interesting one (Figure 6) (50). Once RNA molecules grow in size, the local structural constraints relax for stabilizing RNA domains of great size and higher complexity. This allows for increased versatility at the level of smaller constituent motifs. For example, the kink turn and UA_h turn are two distinct structural motifs characterized by different sequence signatures that can form similar sharp turns between adjacent helices and contribute similarly to the fold of the doughnut-PK domain (Figure 6). Therefore, constituent structural motifs characterized by distinct sequence signatures coding for very different local structures can be considered functionally/topologically equivalent as long as they contribute to the very same functional or topological task within a larger structural context. A-minor interacting motifs are another class of functional/topological equivalence. For instance, GNRA tetraloops, lone-pair motifs, sarcin loops and T-loops are all structurally distinct motifs that promote functional assembly through A-minor interactions (Figure 2). Similarly, the T-loop-PK domain can be stabilized by structurally different but functionally equivalent tertiary interactions (A-minor or intercalating nt) that involve T-loop motifs (Figures 2 and 7). More striking examples of functional equivalence are observed in group I introns (51,52) or RNase P RNAs (53), where PK interactions can sometimes substitute for functionally equivalent A-minor interacting motifs to stabilize the same catalytic core.
Principle of structural scaffolding
Another important principle resulting from RNA modularity is the principle of motifs as structural scaffoldings. Several UA_h motifs are particularly suited for allowing increase of structural complexity by insertion of RNA appendices within their sequence with minimal structural disruption (Figure 7A). The points of insertion of nt and RNA helices are however not random, but constrained by the necessity of retaining the key structural tertiary interactions that characterize the motif. For example, the UA_h submotif offers great scaffolding for inserting sequences at the level of its bulge, leading to motif of greater complexity such as the UA_h 3WJ and UA_h A-minor junctions (Figure 2). The UA_h turn allows insertions of additional helical elements at the level of the kink leading to 3WJ or 4WJ (Figure 2 and 6). Perhaps one of the most striking motif scaffoldings is the T-loop-PK domain that is found at the central core of the 23S rRNA and to which all the constituent domains of the 23S rRNA are connected (Figure 7AB). At least six points of sequence insertions have been identified to allow significant structural expansion of this domain that is also part of the core of several riboswitches (Figure 7CD).
Evolutionary implications
The ‘structural motif network’ also suggests that some RNA structural motifs might have particular evolutionary advantage versus others in the architectural make up of complex functional RNAs. The high occurrence of most UA_h motifs at the end of regular helices is most likely resulting from convergent evolution. As a matter of fact, the motif has been found not only at multiple locations within natural RNA but also in artificial RNA aptamers and ribozymes obtain by SELEX (37,40). Small local RNA motifs might be more prevalent than others within stable RNAs because of their optimal stereo-chemical or biophysical properties (16,17). However, by contrast to other motifs such as terminal UNCG or GNRA tetraloops, the UA_h submotif is not thermostable by itself. UA_h based motifs have most likely been selected by nature for their ability to generate long-range tertiary interactions (Results). For instance, they are particularly prone for folding by induced-fit [see e.g. (54,55)] in presence of interacting partner or ligand. For example, the 11nt receptor (56), the T-loop (32), the UA_h 3WJ (57,58), the ATP-aptamer (37) and the UA_h minor junction of the TPP riboswitch (59,60) have all been observed to be rather flexible and meta-stable in absence of interacting partner while they can form stable tertiary interactions in their presence. This phenomenon is particularly striking for the GAAA/11nt interaction that is one of the most stable natural long-range tertiary interactions of RNA (19). Additionally, UA_h based motifs might also have been evolutionarily selected for their ability to minimize alternative kinetic or thermodynamic traps during RNA folding [see discussion in (19)]. The prevalence of larger domains within ribosomal RNAs and riboswitches, such as the T-loop-PK, alpha-PK and doughnut-PK domains that involve between 45 to 65 nts, is more intriguing. Rather than resulting from convergent evolution, they might be the contingent byproducts of historical events that lead to the increase of structural complexity of natural functional molecules through local duplication of sequence and recombination mechanisms (Zhuang, Geary and Jaeger, in preparation).
RNA structure proto-language and tertiary structure prediction
The ‘UA_h motif network’ is only a portion of a larger RNA proto-language network that should facilitate the development of bioinformatic tools for the prediction of the tertiary structure of natural RNA molecules (7,24,25). Principles of structural equivalence and functional equivalence and structural scaffolding are fundamental for understanding RNA structure evolution and therefore, of prime importance for RNA structure prediction by comparative sequence analysis [e.g. (61)]. For instance, the knowledge of the sequence space of structural motifs can readily be used as algorithmic rules for identifying them within RNA 2D structures. Their specific conformations can constrain significantly the overall positioning of helical elements with respect of one another so that the prediction of the overall 3D architectures of some RNA molecules could become possible.
Based on the set of syntax rules that we unraveled, we have been able to predict the overall 3D architecture of the core AdoCbl (Coenzyme B12) riboswitch aptamer as well as part of the FMN riboswitch aptamer (62) (Figure 7C and D). The AdoCbl aptamer, one of the most abundant riboswitches found in nature (63), is essentially built from the association of a T-loop-PK domain with an A-minor motif domain (Figure 7C). These two domains constrain the positioning of the AdoCbl core helices through formation of key tertiary interactions that involve highly conserved nt positions and bps for which no previous explanations were available. Based on the principle of functional/topological equivalence defined above, it is possible to predict that the T-loop in P4 and the UGA/gAN motif in P7 are likely to interact through an intercalating nt interaction, albeit a precise atomic model can not yet be proposed as the nucleotide target within the UGA/gAN motif cannot be identified without ambiguity. Based on the overall organization of the proposed AdoCbl aptamer and ‘in-line’ probing experiments performed in presence and absence of coenzyme B12 (64,65), ligand binding likely contribute by induce fit to the rigidifying of the P3-P4 junction at the interface of the T-loop-PK and A minor domains (Figure 7C). The two core domains are however likely to be stable by themselves as their structures are unchanged upon ligand binding (64,65). The 3′ end of P1 can then be positioned at a proper distance from L5 to induce formation of the regulatory long-range PK interaction. The conserved GC base pairs in P3 and P5 might play a role in this rigidifying process by possibly interacting with a G/RA submotif at the extremity of P3. Consistent with in-line probing (64,65), the P3–P4 hinge region is likely meta-stable in absence of coenzyme B12, preventing formation of the long-range PK. The FMN riboswitch is another interesting example as it exemplifies well the limit of the predictive power of our rules. According to them, the two T-loops identified in the FMN aptamer core (62) should necessarily be part of tertiary interacting motifs. The T-loop in P5 can be predicted to be involved in a T-loop-PK domain where it forms an intercalating nt interaction with the loop of P3. The distant intercalating nt partner of the T-loop in P2 is however much more difficult to predict because of the highly contextual nature of T-loop mediated interactions (see above). A possible candidate might be one of the conserved purines within L6.
As demonstrated by these two examples, sequence signatures for structural motifs can be extremely useful for predicting the native 3D architecture of stable RNAs, especially when comparative sequence analysis data are readily available. Nevertheless, even in the remote case where only one sequence is known for a particular RNA molecule, one can possibly use the UA_h motif network to identify putative candidates for UA_h conformers and other related motifs in order to reduce the number of possible secondary structures associated to this sequence. Additionally, the sequence constraints and rules can be working hypotheses for identifying putative UA_h motifs that can be used for modeling, prior to refinement and interpretation, after refinement, of X-ray crystal structures.
Approximately 65% of the UA_h sequence signatures that have been identified in the 2D structures of the 23S rRNA from Escherichia coli and Thermus thermophilus do not correspond to UA_h conformers in the final native state of the ribosomal RNA (Figure S2). This is not surprising when considering that most unfolded UA_h signatures are not phylogenetically conserved. However, in some molecular instances, sequence signatures might also code for transient conformational states that can facilitate the kinetic folding process or dynamics of RNAs [(66); Zhuang, Shea and Jaeger, unpublished results]. In the future, it might therefore be interesting to look more carefully at phylogenetically conserved motif sequence signatures that do not correspond to the expected conformers in the native X-ray structure of an RNA.
RNA structure proto-language and rational design
The structural syntax defined herein can readily be applied to the rational design of self-assembling RNA and scaffoldings with potential applications in synthetic biology and nano-medicine (24,25,67,68). The sequence signatures of particular tertiary motifs corresponding to well-defined conformers can be implemented within artificial RNA sequences to control both 3D shape and self-assembling interfaces. RNA architectonics, the methodology behind this concept, has already demonstrated that self-assembling nano-particles (19,69,70) (Severcan, Geary, Jaeger, manuscript in preparation), nano-fibers (71,72) and 2D arrays (73) can be generated from a very limited number of well-characterized structural motifs (24). In the future, it is anticipated that the increased number of rules and RNA motifs derived from the present analysis will contribute to the design of more complex RNA architectures of arbitrary 3D shapes that could ultimately reach the structural (and functional) complexity of the ribosome. This task will however require more extensive experimental characterizations of the chemical and biophysical properties of RNA structural building blocks.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was funded by the National Institutes of Health (R01 GM079‱604-01) to L.J. E.J.V. was a 2006 undergraduate recipient of a RISE research internship award in Science and Engineering sponsored by the Material Research Laboratory and the California NanoSystems Institute at UCSB. Funding for open access charge: National Institutes of Health GM079604-01.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Noller HF. RNA structure: reading the ribosome. Science. 2005;309:1508–1514. doi: 10.1126/science.1111771. [DOI] [PubMed] [Google Scholar]
- 2.Holbrook SR. RNA structure: the long and the short of it. Curr. Opin. Struct. Biol. 2005;15:302–308. doi: 10.1016/j.sbi.2005.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tinoco I, Jr, Bustamante C. How RNA folds. J. Mol. Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 4.Gesteland RF, Cech TR, Atkins JF. The RNA World. 3rd. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
- 5.Woodson SA. Metal ions and RNA folding: a highly charged topic with a dynamic future. Curr. Opin. Chem. Biol. 2005;9:104–109. doi: 10.1016/j.cbpa.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 6.Hendrix DK, Brenner SE, Holbrook SR. RNA structural motifs: building blocks of a modular biomolecule. Q. Rev. Biophys. 2005;38:221–243. doi: 10.1017/S0033583506004215. [DOI] [PubMed] [Google Scholar]
- 7.Leontis NB, Lescoute A, Westhof E. The building blocks and motifs of RNA architecture. Curr. Opin. Struct. Biol. 2006;16:279–287. doi: 10.1016/j.sbi.2006.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lescoute A, Westhof E. Topology of three-way junctions in folded RNAs. RNA. 2006;12:83–93. doi: 10.1261/rna.2208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steinberg SV, Boutorine YI. G-ribo motif favors the formation of pseudoknots in ribosomal RNA. RNA. 2007;13:1036–1042. doi: 10.1261/rna.495207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doherty EA, Batey RT, Masquida B, Doudna JA. A universal mode of helix packing in RNA. Nat. Struct. Biol. 2001;8:339–343. doi: 10.1038/86221. [DOI] [PubMed] [Google Scholar]
- 11.Gagnon MG, Steinberg SV. GU receptors of double helices mediate tRNA movement in the ribosome. RNA. 2002;8:873–877. doi: 10.1017/s135583820202602x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mokdad A, Krasovska MV, Sponer J, Leontis NB. Structural and evolutionary classification of G/U wobble basepairs in the ribosome. Nucleic Acids Res. 2006;34:1326–1341. doi: 10.1093/nar/gkl025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sponer JE, Reblova K, Mokdad A, Sychrovsky V, Leszczynski J, Sponer J. Leading RNA Tertiary Interactions: Structures, Energies and Water Insertion of A-Minor and P-Interactions. A Quantum Chemical View. J. Phys. Chem. B. 2007;111:9153–9164. doi: 10.1021/jp0704261. [DOI] [PubMed] [Google Scholar]
- 14.Leontis NB, Stombaugh J, Westhof E. The non-Watson–Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JC, Gutell RR. Diversity of base-pair conformations and their occurrence in rRNA structure and RNA structural motifs. J. Mol. Biol. 2004;344:1225–1249. doi: 10.1016/j.jmb.2004.09.072. [DOI] [PubMed] [Google Scholar]
- 16.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 17.Gutell RR, Lee JC, Cannone JJ. The accuracy of ribosomal RNA comparative structure models. Curr. Opin. Struct. Biol. 2002;12:301–310. doi: 10.1016/s0959-440x(02)00339-1. [DOI] [PubMed] [Google Scholar]
- 18.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc. Natl Acad. Sci. USA. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geary C, Baudrey S, Jaeger L. Comprehensive features of natural and in vitro selected GNRA tetraloop-binding receptors. Nucleic Acids Res. 2008;36:1138–1152. doi: 10.1093/nar/gkm1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elgavish T, Cannone JJ, Lee JC, Harvey SC, Gutell RR. AA.AG@helix.ends: A:A and A:G base-pairs at the ends of 16 S and 23 S rRNA helices. J. Mol. Biol. 2001;310:735–753. doi: 10.1006/jmbi.2001.4807. [DOI] [PubMed] [Google Scholar]
- 21.Correll CC, Beneken J, Plantinga MJ, Lubbers M, Chan YL. The common and the distinctive features of the bulged-G motif based on a 1.04 Å resolution RNA structure. Nucleic Acids Res. 2003;31:6806–6818. doi: 10.1093/nar/gkg908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jucker FM, Pardi A. GNRA tetraloops make a U-turn. RNA. 1995;1:219–222. [PMC free article] [PubMed] [Google Scholar]
- 23.Gutell RR, Cannone JJ, Konings D, Gautheret D. Predicting U-turns in ribosomal RNA with comparative sequence analysis. J. Mol. Biol. 2000;300:791–803. doi: 10.1006/jmbi.2000.3900. [DOI] [PubMed] [Google Scholar]
- 24.Jaeger L, Chworos A. The architectonics of programmable RNA and DNA nanostructures. Curr. Opin. Struct. Biol. 2006;16:531–543. doi: 10.1016/j.sbi.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 25.Chworos A, Jaeger L. Nucleic acid foldamers: design, engineering and selection of programmable bio-materials with recognition, catalytic and self-assembly properties. In: Hecht SHI, editor. Foldamers: Structure, Properties, and Applications. New York: Wiley-VCH; 2007. pp. 291–330. [Google Scholar]
- 26.Lescoute A, Westhof E. The interaction networks of structured RNAs. Nucleic Acids Res. 2006;34:6587–6604. doi: 10.1093/nar/gkl963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarver M, Zirbel CL, Stombaugh J, Mokdad A, Leontis NB. FR3D: finding local and composite recurrent structural motifs in RNA 3D structures. J. Math. Biol. 2008;56:215–252. doi: 10.1007/s00285-007-0110-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron and other RNAs. BMC Bioinform. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wuyts J, Perriere G, Van De Peer Y. The European ribosomal RNA database. Nucleic Acids Res. 2004;32:D101–D103. doi: 10.1093/nar/gkh065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krasilnikov AS, Mondragon A. On the occurrence of the T-loop RNA folding motif in large RNA molecules. RNA. 2003;9:640–643. doi: 10.1261/rna.2202703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freier SM, Kierzek R, Jaeger JA, Sugimoto N, Caruthers MH, Neilson T, Turner DH. Improved free-energy parameters for predictions of RNA duplex stability. Proc. Natl Acad. Sci. USA. 1986;83:9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhuang Z, Jaeger L, Shea JE. Probing the structural hierarchy and energy landscape of an RNA T-loop hairpin. Nucleic Acids Res. 2007;35:6995–7002. doi: 10.1093/nar/gkm719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Costa M, Michel F. Frequent use of the same tertiary motif by self-folding RNAs. EMBO. J. 1995;14:1276–1285. doi: 10.1002/j.1460-2075.1995.tb07111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee JC, Gutell RR, Russell R. The UAA/GAN internal loop motif: a new RNA structural element that forms a cross-strand AAA stack and long-range tertiary interactions. J. Mol. Biol. 2006;360:978–988. doi: 10.1016/j.jmb.2006.05.066. [DOI] [PubMed] [Google Scholar]
- 35.Nagaswamy U, Fox GE. Frequent occurrence of the T-loop RNA folding motif in ribosomal RNAs. RNA. 2002;8:1112–1119. doi: 10.1017/s135583820202006x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steinberg SV, Boutorine YI. G-ribo: a new structural motif in ribosomal RNA. RNA. 2007;13:549–554. doi: 10.1261/rna.387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dieckmann T, Suzuki E, Nakamura GK, Feigon J. Solution structure of an ATP-binding RNA aptamer reveals a novel fold. RNA. 1996;2:628–640. [PMC free article] [PubMed] [Google Scholar]
- 38.Thore S, Leibundgut M, Ban N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- 39.Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Serganov A, Keiper S, Malinina L, Tereshko V, Skripkin E, Hobartner C, Polonskaia A, Phan AT, Wombacher R, Micura R, et al. Structural basis for Diels–Alder ribozyme-catalyzed carbon-carbon bond formation. Nat. Struct. Mol. Biol. 2005;12:218–224. doi: 10.1038/nsmb906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Szewczak AA, Kundrot CE, Cech TR, Doudna JA. RNA tertiary structure mediation by adenosine platforms. Science. 1996;273:1696–1699. doi: 10.1126/science.273.5282.1696. [DOI] [PubMed] [Google Scholar]
- 42.Costa M, Michel F. Rules for RNA recognition of GNRA tetraloops deduced by in vitro selection: Comparison with in vivo evolution. EMBO J. 1997;16:3289–3302. doi: 10.1093/emboj/16.11.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang B, Wilkinson KA, Weeks KM. Complex ligand-induced conformational changes in tRNA(Asp) revealed by single-nucleotide resolution SHAPE chemistry. Biochemistry. 2008;47:3454–3461. doi: 10.1021/bi702372x. [DOI] [PubMed] [Google Scholar]
- 44.Lee JC, Cannone JJ, Gutell RR. The lonepair triloop: a new motif in RNA structure. J. Mol. Biol. 2003;325:65–83. doi: 10.1016/s0022-2836(02)01106-3. [DOI] [PubMed] [Google Scholar]
- 45.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 46.Gilbert SD, Rambo RP, Van Tyne D, Batey RT. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nat. Struct. Mol. Biol. 2008;15:177–182. doi: 10.1038/nsmb.1371. [DOI] [PubMed] [Google Scholar]
- 47.Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO. J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Westhof E, Masquida B, Jaeger L. RNA tectonics: towards RNA design. Fold. Des. 1996;1:R78–R88. doi: 10.1016/S1359-0278(96)00037-5. [DOI] [PubMed] [Google Scholar]
- 49.Lescoute A, Leontis NB, Massire C, Westhof E. Recurrent structural RNA motifs, Isostericity Matrices and sequence alignments. Nucleic Acids Res. 2005;33:2395–2409. doi: 10.1093/nar/gki535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Auletta G, Ellis GF, Jaeger L. Top-down causation by information control: from a philosophical problem to a scientific research programme. J. R. Soc. Interface. 2008;5:1159–1172. doi: 10.1098/rsif.2008.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jaeger L, Michel F, Westhof E. Involvement of a GNRA tetraloop in long-range RNA tertiary interactions. J. Mol. Biol. 1994;236:1271–1276. doi: 10.1016/0022-2836(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 52.Jaeger L, Westhof E, Michel F. Function of a pseudoknot in the suppression of an alternative splicing event in a group I intron. Biochimie. 1996;78:466–473. doi: 10.1016/0300-9084(96)84753-5. [DOI] [PubMed] [Google Scholar]
- 53.Massire C, Jaeger L, Westhof E. Phylogenetic evidence for a new tertiary interaction in bacterial RNase P RNAs. RNA. 1997;3:553–556. [PMC free article] [PubMed] [Google Scholar]
- 54.Williamson JR. Proteins that bind RNA and the labs who love them. Nat. Struct. Biol. 2001;8:390–391. doi: 10.1038/87540. [DOI] [PubMed] [Google Scholar]
- 55.Mitrasinovic PM. On the structural features of hairpin triloops in rRNA: From nucleotide to global conformational change upon ligand binding. J. Struct. Biol. 2006;153:207–222. doi: 10.1016/j.jsb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 56.Davis JH, Tonelli M, Scott LG, Jaeger L, Williamson JR, Butcher SE. RNA helical packing in solution: NMR structure of a 30 kDa GAAA tetraloop-receptor complex. J. Mol. Biol. 2005;351:371–382. doi: 10.1016/j.jmb.2005.05.069. [DOI] [PubMed] [Google Scholar]
- 57.Agalarov SC, Williamson JR. A hierarchy of RNA subdomains in assembly of the central domain of the 30 S ribosomal subunit. RNA. 2000;6:402–408. doi: 10.1017/s1355838200991945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ha T, Zhuang X, Kim HD, Orr JW, Williamson JR, Chu S. Ligand-induced conformational changes observed in single RNA molecules. Proc. Natl Acad. Sci. USA. 1999;96:9077–9082. doi: 10.1073/pnas.96.16.9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mayer G, Raddatz MS, Grunwald JD, Famulok M. RNA ligands that distinguish metabolite-induced conformations in the TPP riboswitch. Angew. Chem. Int. Ed. Engl. 2007;46:557–560. doi: 10.1002/anie.200603166. [DOI] [PubMed] [Google Scholar]
- 60.Lang K, Rieder R, Micura R. Ligand-induced folding of the thiM TPP riboswitch investigated by a structure-based fluorescence spectroscopic approach. Nucleic Acids Res. 2007;35:5370–5378. doi: 10.1093/nar/gkm580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Massire C, Jaeger L, Westhof E. Derivation of the three-dimensional architecture of bacterial ribonuclease P RNAs from comparative sequence analysis. J. Mol. Biol. 1998;279:773–793. doi: 10.1006/jmbi.1998.1797. [DOI] [PubMed] [Google Scholar]
- 62.Barrick JE, Breaker RR. The distributions, mechanisms and structures of metabolite-binding riboswitches. Genome Biol. 2007;8:R239. doi: 10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kazanov MD, Vitreschak AG, Gelfand MS. Abundance and functional diversity of riboswitches in microbial communities. BMC Genomics. 2007;8:347. doi: 10.1186/1471-2164-8-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gallo S, Oberhuber M, Sigel RK, Krautler B. The corrin moiety of Coenzyme B12 is the determinant for switching the btuB riboswitch of E. coli. Chembiochem. 2008;9:1408–1414. doi: 10.1002/cbic.200800099. [DOI] [PubMed] [Google Scholar]
- 65.Nahvi A, Barrick JE, Breaker RR. Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res. 2004;32:143–150. doi: 10.1093/nar/gkh167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Heppell B, Lafontaine DA. Folding of the SAM aptamer is determined by the formation of a K-turn-dependent pseudoknot. Biochemistry. 2008;47:1490–1499. doi: 10.1021/bi701164y. [DOI] [PubMed] [Google Scholar]
- 67.Severcan I, Geary C, Jaeger L, Bindewald E, Kasprzak W, Shapiro BA. Computational and Experimental RNA Nanoparticle Design. In: Alterovitz G, Ramoni M, Mary Benson R, editors. Automation in Genomics and Proteomics: An Engineering Case Based Approach. New York: Wiley; 2009. pp. 193–220. [Google Scholar]
- 68.Guo P. RNA nanotechnology: engineering, assembly and applications in detection, gene delivery and therapy. J. Nanosci. Nanotechnol. 2005;5:1964–1982. doi: 10.1166/jnn.2005.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jaeger L, Westhof E, Leontis NB. TectoRNA: modular assembly units for the construction of RNA nano-objects. Nucleic Acids Res. 2001;29:455–463. doi: 10.1093/nar/29.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khaled A, Guo S, Li F, Guo P. Controllable self-assembly of nanoparticles for specific delivery of multiple therapeutic molecules to cancer cells using RNA nanotechnology. Nano. Lett. 2005;5:1797–1808. doi: 10.1021/nl051264s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nasalean L, Baudrey S, Leontis NB, Jaeger L. Controlling RNA self-assembly to form filaments. Nucleic Acids Res. 2006;34:1381–1392. doi: 10.1093/nar/gkl008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Koyfman AY, Braun G, Magonov S, Chworos A, Reich NO, Jaeger L. Controlled spacing of cationic gold nanoparticles by nanocrown RNA. J. Am. Chem. Soc. 2005;127:11886–11887. doi: 10.1021/ja051144m. [DOI] [PubMed] [Google Scholar]
- 73.Chworos A, Severcan I, Koyfman AY, Weinkam P, Oroudjev E, Hansma HG, Jaeger L. Building programmable jigsaw puzzles with RNA. Science. 2004;306:2068–2072. doi: 10.1126/science.1104686. [DOI] [PubMed] [Google Scholar]
- 74.Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tamura M, Holbrook SR. Sequence and structural conservation in RNA ribose zippers. J. Mol. Biol. 2002;320:455–474. doi: 10.1016/s0022-2836(02)00515-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.