Abstract
Here we describe a one-step method to create precise modifications in the genome of Saccharomyces cerevisiae as a tool for synthetic biology, metabolic engineering, systems biology and genetic studies. Through homologous recombination, a mutagenesis cassette containing an inverted repeat of selection marker(s) is integrated into the genome. Due to its inherent instability in genomic DNA, the inverted repeat catalyzes spontaneous self-excision, resulting in precise genome modification. Since this excision occurs at very high frequencies, selection for the integration event can be followed immediately by counterselection, without the need for growth in permissive conditions. This is the first time a truly one-step method has been described for genome modification in any organism.
INTRODUCTION
With ever-mounting emphasis on creating designer organisms for efficient conversion of biomass to fuels, chemicals, drugs, enzymes and therapeutic proteins, tools for engineering genomes and understanding regulatory networks are becoming increasingly important. Therefore, several methods have been described to assemble large genomic fragments (1–3) or delete them (4,5); but there are few efficient methods available to perform precise genomic modifications. The advantage of methods enabling accurate mutagenesis is that they can be easily adapted for deletions, insertions and replacements. Apart from the classical plasmid-based pop-in/pop-out based methodology (6), only two other distinct strategies have been described, to our knowledge, in yeast for precise genomic modification using linear DNA (7,8). These have been improved and modified in subsequent works (9–12); and while both strategies are excellent, they both suffer from certain disadvantages. The first strategy described by Langle-Rouault and Jacobs has a low success rate, with 13–45% efficiency of initial mutagenesis cassette integration and 29–35% efficiency for removal of said cassette (7). Therefore, use of this method necessitates extensive screening for clones with the desired mutation. Delitto perfetto addresses the low-efficiency issues, but adds a few steps in the form of replica-plating and a second transformation to remove the mutagenesis cassette (8).
To address the limitations of these methods, we developed a new method called mutagenic inverted repeat assisted genome engineering (MIRAGE) that can efficiently generate chromosomal mutations in a single transformation step. This method relies on incorporating an inverted repeat (IR) of selection marker(s) in the mutagenesis cassette, which after introduction in the chromosome, along with the desired mutation, is spontaneously excised. This excision event leading to the loss of the selection marker is highly frequent and does not require growth in permissive medium to occur. As a result, counterselection can be performed immediately after selection to obtain the desired chromosomal mutation. To demonstrate its utility, we have created several point mutations, as well as deletions in the yeast chromosome.
MATERIALS AND METHODS
Strains, plasmids and reagents
Saccharomyces cerevisiae W303a was obtained from Open Biosystems (Huntsville, AL). Phusion DNA polymerase, restriction enzymes, T4 DNA ligase and plasmids pRS406, pRS403 (New England Biolabs, Beverly, MA), and pRSFDuet-1 (Novagen, Gibbstown, NJ) were used for mutagenesis cassette preparation. Escherichia coli DH5α (Invitrogen, Carlsbad, CA) was used for plasmid propagation. All chemicals were obtained from Fisher Scientific (Pittsburgh, PA). Yeast growth media used were 2xYPAD (2% yeast extract, 4% peptone and 4% dextrose supplemented with 0.2 g/l adenine hemisulfate and 50 μg/ml kanamycin), synthetic complete (SC), synthetic complete lacking uracil (SC−Ura), synthetic complete lacking tryptophan (SC−Trp) and synthetic complete supplemented with 1 g/l 5-fluoroorotic acid (SC+FOA).
Mutagenesis cassette preparation
Using the primers listed in Supplementary Table 1, ∼1.1 kb ura3 was amplified from pRS406, or ∼2.2 kb ura3-his3 from pDIRe3. Fragments were digested with a combination of DpnI and either EcoRI or MfeI, and ligated overnight at 16°C. These were subsequently digested with a combination of EcoRI and MfeI, and the ∼2.2 kb product (ura3-IR) or ∼4.5 kb (ura3-his3-IR) was gel purified. Supplementary Figure 1 shows DNA products from each step on an agarose gel. This product was directly transformed into heat-shock competent S. cerevisiae.
Yeast transformation and reversion
Fifteen milliliters of 2xYPAD was inoculated with 0.45 ml overnight yeast culture and shaken at 30°C and 250 r.p.m. for 4 h. Cells were made competent by centrifugation at 4000 r.p.m., followed by two washes in sterile, deionized water and one wash in 0.1 M lithium acetate (LiAc). Half of the cells were pelleted and resuspended in transformation mixture, comprising of 240 μl 50% (w/v) PEG 3350 (polyethylene glycol), 36 μl 1.0 M LiAc, 50 μl 2 mg/ml salmon-sperm ssDNA and 34 μl of the IR mutagenesis cassette (200–900 ng). After vortexing cells for 60 s, they were incubated at 42°C for 40 min, after which they were pelleted, resuspended and recovered in 1 ml 2xYPAD for 3 h in a 30°C shaker. Cells were then washed once in SC−Ura and plated evenly on three SC−Ura plates. We routinely obtained 2–40 colonies per 2–6 × 106 viable cells using this procedure. After 3–4 days, colonies grown to ∼3 mm in diameter were resuspended in sterile, deionized water and incubated at room temperature for ∼3–4 h. They were then streaked on a SC+FOA plate and incubated at 30°C. Spontaneous reversions occurred at the frequency of 10−5 to 10−6.
Verification of chromosomal modifications
Insertion event was verified by growing transformants in 2 ml SC−Ura and isolating their genomic DNA using Wizard Genomic DNA Purification Kit (Promega, Madison, WI). Lack of a PCR product after amplification indicated a deletion. A control reaction was also performed to ensure that lack of product was not due to PCR failure. Integration of MIRAGE cassette was also verified by PCR amplification between the ura3 terminator and primers annealing upstream and downstream of locus of interest (Figure 3b). IR excision revertants were picked from SC+FOA plates and grown overnight in 2 ml media. Genomic DNA was isolated and PCR was performed using external primers annealing upstream and downstream of the deleted gene. Trp1+ revertants were identified by re-streaking colonies from SC+FOA plates onto SC−Trp plates and incubating for 2–3 days. Point mutations in adh4, adh7 and gpd1 were verified by DNA sequencing (Supplementary Figure 3).
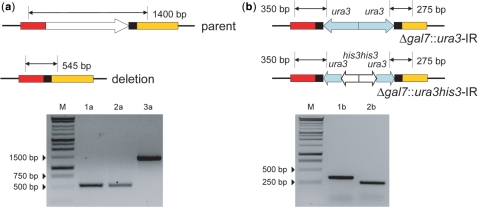
Figure 3.
(a) Expected PCR product sizes before and after deletion of gal7 using MIRAGE with a 25 bp DR. 1a, 2a—Scarless gal7 locus after precise excision of the mutagenic cassette; 3a—negative control, parent. (b) Verification of gal7 replacement with either ura3-IR or ura3-his3-IR.
Construction of pDIRe3
PCR amplified cassettes of ura3 and his3 from pRS406 and pRS403, respectively, were spliced together using overlap–extension PCR. After digestion with EcoRI and BamHI, the cassette was ligated into linearized pRSFDuet-1 and transformed into DH5α. Transformants were then selected for kanamycin resistance.
RESULTS
Design of the MIRAGE method
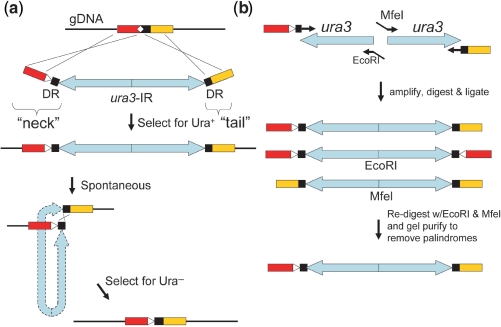
MIRAGE uses a novel type of mutagenesis cassette containing an IR flanked by ‘neck’ and ‘tail’ sequences (Figure 1a). Each repeat of the IR encodes for marker(s) that can be used for selection and counterselection. The outmost 45 bp (base pairs) in the ‘neck’ and ‘tail’ flanks direct the integration of the mutagenesis cassette at the desired location by homologous recombination. The 3′-end of the ‘neck’ contains a 25-bp sequence identical to the 5′-end of the ‘tail’, forming a 25-bp direct repeat (DR). The desired mutation is incorporated in the ‘neck’ primer used for cassette preparation, as shown (white triangle, Figure 1a). Integration of the mutagenesis cassette in the chromosome is therefore accompanied by deletion of the undesired allele. Due to its inherent instability in genomic DNA (13) the IR catalyzes spontaneous self-excision between the DR, resulting in a one-step precise modification of the genome.
Figure 1.
Features of one-step genome modification using MIRAGE. (a) 45 bp flanking ends of ‘neck’ and ‘tail’ target the mutagenesis cassette into the yeast chromosome (gDNA), simultaneously replacing the undesired allele (white diamond) with the desired mutation (white triangle). The ura3-IR confers transformants with uracil prototrophy. Spontaneous and precise excision of the IR occurs within the 25 bp DR, leaving behind only the desired mutation. Since the mechanism of MIRAGE excision is not known, the IR is drawn with hyphenated lines. Omitting the mutation in the ‘neck’ primer will lead to deletion, rather than replacement. (b) Creation of the mutagenesis cassette requires preparation of each ura3 fragment separately. After PCR amplification and digestion with EcoRI and MfeI, the two halves are ligated together. Ligation of compatible EcoRI and MfeI ends abolishes both restriction sites. Redigestion with EcoRI and MfeI, followed by gel purification removes palindromic ligation products.
Since the IR cannot be amplified directly by PCR, each half has to be amplified independently and then ligated to form the mutagenesis cassette (Figure 1b). We used ura3 (encoding for orotidine-5′-phosphate decarboxylase) as our selection/counterselection marker and EcoRI or MfeI restriction sites to form compatible cohesive ends after digestion. After ligation of the two halves EcoRI and MfeI restriction sites are abolished, enabling purification of the mutagenesis cassette from self-ligation products (palindromes). This ∼2.2 kb mutagenesis cassette was then transformed directly into yeast and selected for uracil prototrophy on SC−Ura plates. After 3–4 days of incubation at 30°C, colonies grown to ∼3 mm in diameter were scraped and resuspended in water. After incubation for 3–4 h at room temperature to ensure complete depletion of uracil dropout media within cells, they were immediately selected for uracil auxotrophy on SC+FOA plates. Since the self-excision of the IR occurs at frequencies much higher than point mutations, we found that auxotrophs appeared only upon deletion of the IR within the DR.
Optimizing the DR length
To optimize the integration and excision efficiency, we performed targeted deletion of gal7 (encoding for galactose-1-phosphate uridyl transferase) in our laboratory strain HZ848 (MATα Δura3 trp1-1) since it is easy to verify by PCR and loss of function. The efficiency of targeted mutagenesis in general relies on two major factors: first, integration of the mutagenesis cassette precisely at the desired location in the chromosome, and second, precise removal of the selection marker. In our method, the efficiency of both of these events is dependent on the length of the DR. As illustrated in Supplementary Figure 2, a DR can facilitate incorrect integration of the mutagenesis cassette. To minimize this aberrant event, a shorter DR is preferred. Conversely, the precise removal of the IR relies on recombination between the DR, where longer sequences are desirable. By varying the DR length from 20 to 35 bp in 5-bp increments, we found an optimal balance at 25 bp, for initial integration and final excision efficiency (Figure 2, Table 1). With a longer DR, we found incorrect insertion of the mutagenesis cassette occurred at a higher frequency than the desired targeted integration. The presence of an IR within a longer DR also created an extremely unstable system, decreasing the growth rate in the dropout selection media. This is presumably because the IR is excised at very high frequency, but the exact reason for this phenomenon was not investigated further. At short DR lengths, aberrant excision events take place, likely due to near-precise excision (14) by replication slippage (15). Precise excision of the mutagenesis cassette was initially verified by sequencing PCR amplified genomic locus, and later by observing the size of the PCR product.
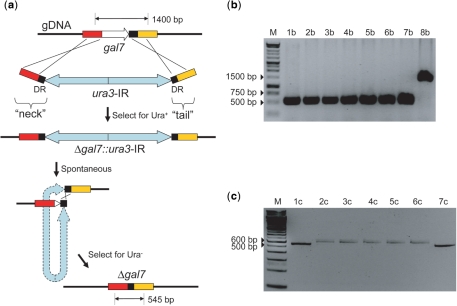
Figure 2.
Deletion of gal7 using MIRAGE to optimize DR length. (a) Expected PCR product sizes for parental and deletion alleles are indicated. Since the mechanism of cassette excision is not known, the IR is drawn with hyphenated lines. (b) Deletion of gal7 using a 25 bp DR. 1b-7b—successful precise deletion of gal7; 8b—negative control, parent. (c) Deletion of gal7 using a 20 bp DR. 1c, 7c—successful precise deletion of gal7; 2c-6c—imprecise deletion of gal7. M, DNA ladder.
Table 1.
Effect of varying DR length on mutagenesis accuracy, based on targeted deletion of gal7
| DR length (bp) | Success rate of correct integration | Success rate of precise excision |
|---|---|---|
| 20 | 66% (4/6) | 28% (2/7) |
| 25 | 50% (3/6) | 100% (3/3) |
| 30 | 25% (1/4) | 100% (3/3) |
| 35 | 0 (0/6) | N.D. |
N.D. = No data
A shorter DR favors desired integration event, but also favors imprecise excision.
More deletions, point mutagenesis, and using a dual selection marker
Having optimized the DR length by demonstrating efficient deletion of gal7, we also used MIRAGE to delete the trp1 locus (encoding for phosphoribosylanthranilate isomerase). Amongst point mutations, we reverted the trp1-1 allele to wild type (i.e. TAG to GAG at the 83rd codon), made silent mutations in adh4 (ACC to ACT at the 451st codon) and adh7 (CAA to CAG at the 149th codon), and created a frameshift mutation in gpd1 (GAT to C at the 391st codon). With a 25-bp DR, we found 50–100% of the colonies had correctly integrated the mutagenesis cassette into the desired location, and all their revertants on SC+FOA plates precisely excised the IR (Figure 3, Table 2). Verification was done by PCR using genomic DNA as template for deletions, whereas tryptophan prototrophy (growth in SC−Trp media) or DNA sequencing confirmed the point mutations.
Table 2.
Success rates of gene deletion and in vivo point mutagenesis experiments
| Correct integration event |
Precise excision event |
|||
|---|---|---|---|---|
| Parental allele | Resulting allele | Success rate | Resulting allele | Success rate |
| GAL7wt | Δgal7::ura3-IR | 50% (3/6) | Δgal7 | 100% |
| trp1-1 | Δtrp1::ura3-IR | 66% (4/6) | Δtrp1 | 100% |
| ADH4wt | adh4 T451::ura3-IR | 60% (3/5) | adh4 T451T | 100% |
| ADH7wt | adh7 Q149::ura3-IR | 50% (1/2) | adh7 Q149Q | 100% |
| GPD1wt | gpd1 D391::ura3-IR | 100% (1/1) | gpd1D391Lfs*27 | 100% |
| trp1-1 | trp1::ura3-IR | 83% (5/6) | TRP1wt | 100% |
Initial integration is not perfect due to the presence of a DR leading to improper integration (Supplementary Figure 1). Excision events were always observed to have precise removal of the IR within the DR. Mutations introduced in adh4 and adh7 were synonymous codon changes (ACC to ACT and CAA to CAG, respectively). In gpd1, codon 391 (GAT) was replaced with C resulting in a substitution and frameshift of the reading frame). The trp1-1 allele is an amber mutation at codon 83. Exact sequences of all sequenced mutant loci are in Supplementary Figure 3.
To extend this method to yeast strains with different ura3 alleles, we created a dual-selection mutagenesis cassette. Instead of using a ura3-IR cassette, we used a ura3-his3-IR of ∼4.5 kb. Since it is unlikely that false positives could occur by simultaneous integration of ura3 and his3 at their own respective loci, colonies could appear only upon integration of the entire mutagenesis cassette into the chromosome. This is indeed what we observed, and using this IR with a double-selection marker we were able to successfully delete gal7 in W303a (MATa ura3-52 his3-11 ade2-1 leu2-3,112 can1-100 trp1Δ2) by first selecting on SC lacking uracil and histidine (SC−UH) plates and then counterselecting on SC+FOA plates (Figure 3). All FOAR clones were found to be His−, as expected from excision of the IR.
DISCUSSION
Methods enabling genomic point mutagenesis are the most versatile tools in genome engineering since they can be easily adapted to other modifications such as deletions, insertions and replacements (10). A prerequisite for such a method is that the selection/counterselection marker should not leave any scar in the chromosome. This requirement precludes the use of FLP and Cre recombinases that are very popular in performing chromosomal deletions. There have been two distinct strategies described for performing such precise mutations in yeast using PCR products. The first strategy embeds a selection marker within a long DR (7,9). Presence of the DR can cause spontaneous deletion of the selection marker at low frequencies when grown in permissive conditions. Unfortunately, the long DR can cause aberrant integration of the mutagenesis cassette, and the low frequency of excision combined with imprecise excision events, makes the method relatively inefficient. The other strategy based on the delitto perfetto method (8) addresses some issues and is very efficient at introducing point mutations with few false positives. However, this method requires two separate transformation steps, one to delete the undesired allele and a second to introduce the desired mutation, as well as a replica plating step, possibly to maximize efficiency (10,11).
In order to streamline genome engineering, we sought to create a quick and efficient method to create precise chromosomal mutations. To this end we have developed a method called MIRAGE that can create precise alterations in the chromosome in a single transformation step. In this method, the mutagenesis cassette is designed to have an IR of selection marker(s) embedded within a 25-bp DR. Flanking these are regions of homology that target the cassette to the desired site in the chromosome.
By using an IR in the mutagenesis cassette, we have overcome low excision frequency limitations of the strategy introduced by Langle-Rouault and Jacobs (7). This may be due to any combination of four roles that the IR may play—first, it encodes the marker(s) to select/counterselect for the desired genotype. Second, it introduces chromosomal instability that results in its own excision from the chromosome at extremely high frequencies, nullifying the need for growth on permissive medium for expediting its loss. However, to minimize contamination from surrounding non-transformants, colony purification on SC−Ura medium may be performed without any adverse effect. Third, its excision from the chromosome results in a double strand break (11) that may create a hotspot for recombination. And fourth, it may bring the flanking DR in close proximity allowing for very precise homologous recombination between relatively short sequences. Indeed, Langle-Rouault and Jacobs (7) used DR between 50 bp and 60 bp to ensure homologous recombination (albeit with low efficiency) between distant sequences in the chromosome, whereas in our method DR as short as 25 bp are highly proficient at recombination.
Cassette preparation takes 2 days due to the requirement for high-efficiency overnight ligation. However, compared to the two-step methods like delitto perfetto, MIRAGE requires only a single transformation step and no replica plating, making it a quicker method, which is particularly useful when dealing with slow-growing mutants. Additionally, while we have shown selection/counterselection based on Ura/FOA, other markers such as Trp/5-fluoroanthranilic acid (FAA) (16), Lys/aminoadipate (17) or any combination thereof, could also be used.
We have shown that inverted repeat, an unstable genetic element, can be employed as a useful genetic tool to perform one-step modifications to the S. cerevisiae genome with high efficiency. This method does not require the use of helper plasmids, making it widely applicable. Also of note is that yeast seemed to propagate a relatively long IR (∼4.5 kb) as stably as a shorter IR (∼2.2 kb), while actively transcribing the genes contained. This is particularly interesting in the light that E. coli is conferred nonviable when IR of >240 bp is introduced in the chromosome (18). It would also be interesting to test whether this method can be adapted for use in other systems, including mammalian and bacterial cells, since DR bracketed IR should, in theory, induce double-strand breaks in these chromosomes upon excision.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
National Institutes of Health (GM077596 to H.Z.). Funding for open access charge: National Institutes of Health (GM077596).
Conflict of interest: None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Z. Shao, R. P. Sullivan, M. McLachlan, T. Johannes, F. Wen, and J. Du for invaluable discussions and input. We also thank P. A. B. Orlean for providing plasmids and yeast strains.
REFERENCES
- 1.Itaya M, Fujita K, Kuroki A, Tsuge K. Bottom-up genome assembly using the Bacillus subtilis genome vector. Nat. Methods. 2008;5:41–43. doi: 10.1038/nmeth1143. [DOI] [PubMed] [Google Scholar]
- 2.Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, Stockwell TB, Brownley A, Thomas DW, Algire MA, et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 3.Bang D, Church GM. Gene synthesis by circular assembly amplification. Nat. Methods. 2008;5:37–39. doi: 10.1038/nmeth1136. [DOI] [PubMed] [Google Scholar]
- 4.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 5.Hirashima K, Iwaki T, Takegawa K, Giga-Hama Y, Tohda H. A simple and effective chromosome modification method for large-scale deletion of genome sequences and identification of essential genes in fission yeast. Nucleic Acids Res. 2006;34:e11. doi: 10.1093/nar/gnj011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scherer S, Davis RW. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc. Natl Acad. Sci. USA. 1979;76:4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Langle-Rouault F, Jacobs E. A method for performing precise alterations in the yeast genome using a recyclable selectable marker. Nucleic Acids Res. 1995;23:3079–3081. doi: 10.1093/nar/23.15.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Storici F, Lewis LK, Resnick MA. In vivo site-directed mutagenesis using oligonucleotides. Nat. Biotechnol. 2001;19:773–776. doi: 10.1038/90837. [DOI] [PubMed] [Google Scholar]
- 9.Akada R, Kitagawa T, Kaneko S, Toyonaga D, Ito S, Kakihara Y, Hoshida H, Morimura S, Kondo A, Kida K. PCR-mediated seamless gene deletion and marker recycling in Saccharomyces cerevisiae. Yeast. 2006;23:399–405. doi: 10.1002/yea.1365. [DOI] [PubMed] [Google Scholar]
- 10.Gray M, Piccirillo S, Honigberg SM. Two-step method for constructing unmarked insertions, deletions and allele substitutions in the yeast genome. FEMS Microbiol. Lett. 2005;248:31–36. doi: 10.1016/j.femsle.2005.05.018. [DOI] [PubMed] [Google Scholar]
- 11.Storici F, Durham CL, Gordenin DA, Resnick MA. Chromosomal site-specific double-strand breaks are efficiently targeted for repair by oligonucleotides in yeast. Proc. Natl Acad. Sci. USA. 2003;100:14994–14999. doi: 10.1073/pnas.2036296100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray M, Kupiec M, Honigberg SM. Site-specific genomic (SSG) and random domain-localized (RDL) mutagenesis in yeast. BMC Biotechnol. 2004;4:7. doi: 10.1186/1472-6750-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordenin DA, Lobachev KS, Degtyareva NP, Malkova AL, Perkins E, Resnick MA. Inverted DNA repeats: a source of eukaryotic genomic instability. Mol. Cell Biol. 1993;13:5315–5322. doi: 10.1128/mcb.13.9.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foster TJ, Lundblad V, Hanley-Way S, Halling SM, Kleckner N. Three Tn10-associated excision events: relationship to transposition and role of direct and inverted repeats. Cell. 1981;23:215–227. doi: 10.1016/0092-8674(81)90286-5. [DOI] [PubMed] [Google Scholar]
- 15.Lobachev KS, Shor BM, Tran HT, Taylor W, Keen JD, Resnick MA, Gordenin DA. Factors affecting inverted repeat stimulation of recombination and deletion in Saccharomyces cerevisiae. Genetics. 1998;148:1507–1524. doi: 10.1093/genetics/148.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toyn JH, Gunyuzlu PL, White WH, Thompson LA, Hollis GF. A counterselection for the tryptophan pathway in yeast: 5-fluoroanthranilic acid resistance. Yeast. 2000;16:553–560. doi: 10.1002/(SICI)1097-0061(200004)16:6<553::AID-YEA554>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 17.Ito-Harashima S, McCusker JH. Positive and negative selection LYS5MX gene replacement cassettes for use in Saccharomyces cerevisiae. Yeast. 2004;21:53–61. doi: 10.1002/yea.1057. [DOI] [PubMed] [Google Scholar]
- 18.Leach DRF, Okely EA, Pinder DJ. Repair by recombination of DNA containing a palindromic sequence. Mol. Microbiol. 1997;26:597–606. doi: 10.1046/j.1365-2958.1997.6071957.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.