Abstract
Deoxyuridine triphosphate nucleotidohydrolase (dUTPase) catalyzes the hydrolysis of dUTP to dUMP and PPi. Although dUTP is a normal intermediate in DNA synthesis, its accumulation and misincorporation into DNA is lethal. Importantly, uracil misincorporation is a mechanism of cytotoxicity induced by fluoropyrimidine chemotherapeutic agents including 5-fluorouracil (5-FU) and elevated expression of dUTPase is negatively correlated with clinical response to 5-FU-therapy. In this study we performed the first functional characterization of the dUTPase promoter and demonstrate a role for E2F-1 and Sp1 in driving dUTPase expression. We establish a direct role for both mutant and wild-type forms of p53 in modulating dUTPase promoter activity. Treatment of HCT116 p53+/+ cells with the DNA-damaging agent oxaliplatin induced a p53-dependent transcriptional downregulation of dUTPase not observed in the isogenic null cell line. Oxaliplatin treatment induced enrichment of p53 at the dUTPase promoter with a concomitant reduction in Sp1. The suppression of dUTPase by oxaliplatin promoted increased levels of dUTP that was enhanced by subsequent addition of fluoropyrimidines. The novel observation that oxaliplatin downregulates dUTPase expression may provide a mechanistic basis contributing to the synergy observed between 5-FU and oxaliplatin in the clinic. Furthermore, these studies provide the first evidence of a direct transcriptional link between the essential enzyme dUTPase and the tumor suppressor p53.
INTRODUCTION
Deoxyuridine triphosphate nucleotidohydrolase (dUTPase) is the sole enzyme responsible for the hydrolysis of dUTP to dUMP and pyrophosphate simultaneously providing substrate for thymidylate synthase (TS) and eliminating dUTP from the DNA biosynthetic pathway. Although dUTP is a normal intermediate in DNA synthesis, its extensive accumulation and misincorporation into DNA is lethal in both prokaryotic and eukaryotic organisms as evidenced from knockout models (1,2). Importantly, uracil misincorporation also represents a major mechanism of cytotoxicity induced by the TS-inhibitor class of chemotherapeutic agents including the fluoropyrimidines 5-fluorouracil (5-FU), fluorodeoxyuridine (FUdR) and capecitabine which are broadly used in the treatment of cancers of the gastrointestinal tract, breast and head and neck (3). Inhibition of TS induces a metabolic blockade, depleting thymidylate pools and in some instances promoting the accumulation of intracellular dUTP pools and subsequent misincorporation of uracil into DNA resulting in DNA damage and cell death (4,5). Expression of dUTPase is reported to be an important mediator of resistance to therapeutic agents that target TS both in vitro and in vivo. We previously demonstrated that diminished dUTPase expression greatly enhanced dUTP pool expansion following TS-inhibition, sensitizing yeast cells to the effects of uracil misincorporation while cells overexpressing dUTPase were significantly protected (6). In colon cancer cells, depletion of dUTPase by siRNA resulted in dUTP accumulation and growth arrest (7). Overexpression of dUTPase was also demonstrated to confer resistance to FUdR (8), while we previously reported that depletion of dUTPase by siRNA sensitized both breast and colon cell lines to FUdR through misincorporation of dUTP and enhanced DNA damage (9). Moreover, we also reported the results of a retrospective clinical study negatively correlating elevated nuclear expression of dUTPase with response to 5-FU-therapy in colorectal cancer patients (10). These studies bolster the concept that dUTPase represents an attractive drug target and we recently reported the identification of novel small molecules with dUTPase inhibitory activity (11).
While detailed analyses of dUTPase structure and catalytic activity have been described, the mechanisms that govern human dUTPase gene regulation and expression remain largely unknown (12–14). Previous studies have reported that expression of the nuclear isoform of dUTPase (DUT-N) is primarily cell cycle and proliferation-dependent, whereas the mitochondrial isoform (DUT-M) is constitutively expressed (15). Immunohistochemical staining of normal tissues demonstrated that high DUT-N expression is exclusively observed in replicating cell types whereas cytoplasmic expression is observed in mitochondria-rich tissues (10). However, both colon cancer cell lines (16) and tumor specimens demonstrate dramatic variation, both in magnitude of dUTPase expression and intracellular localization. Furthermore, the correlation between replication status and DUT-N expression was not observed in colon adenocarcinomas (17). Importantly, dysregulation of dUTPase expression is observed in tumor types which are frequently treated with agents that target TS, therefore elucidating the molecular basis driving dUTPase expression is important from both a basic science and clinical standpoint (10,16,17). Functional analyses of various S-phase-specific genes including TS and thymidine kinase (TK) identified several transcriptional elements that are commonly present in their promoter regions including E2F and Sp1 consensus sites. These genes are also characterized by a lack of TATA or CAAT box initiation sites, but are rich in GC boxes. Promoter sequence analysis of the DUT gene reveals putative regulatory motifs including potential binding sites for NF-κB, E2F and Sp1 transcription factors (15). Recently, a genome-wide ChIP-on-chip identified dUTPase in a subset of 127 genes bound by E2F family members (18). Despite the presence of these putative S-phase-specific binding sites in the DUT-N promoter region, functional analysis of this gene has not been previously reported.
Several studies have also reported downregulation of dUTPase during apoptosis (19,20) and that dUTPase expression may be modulated by the tumor suppressor gene p53 (21,22). In response to stress stimuli such as DNA damage, p53 can initiate cell cycle arrest through transcriptional induction of cell cycle inhibitors such as p21cip1/waf1, mediate DNA repair or induce apoptotic cell death. These mechanisms are designed to prevent proliferation of cells containing damaged DNA and reduce the likelihood of tumor formation. Interestingly, mutations in p53 are one of the most common genetic aberrations detected in malignant disease with >50% of colon tumors exhibiting mutation (23).
In prostate cancer cells, dUTPase was one of many genes identified by microarray analysis as significantly repressed following introduction of wild-type p53 (22). In MCF-7 (p53 wild-type) breast cancer cells, microarray analysis also identified dUTPase mRNA within an extensive panel of genes repressed following 5-FU treatment (21). However, the precise mechanism of the downregulation of dUTPase has not been determined and it is unknown as to whether this phenomenon is the result of indirect downstream events induced by p53 itself or its transactivation and repressive gene targets. Furthermore, dUTPase was one of a number of genes identified as upregulated in p53-null mouse embryonic fibroblasts following introduction of the human tumor-derived p53 R175H by subtraction hybridization (24).
As dUTPase is an essential enzyme in maintaining genomic stability, and demonstrates both aberrant intratumoral expression and an association with resistance to 5-FU, we sought to perform the first functional characterization of the promoter and elucidate the mechanisms involved in regulating dUTPase expression. In addition, p53 mutations are widely observed in many cancers and as the fluoropyrimidines remain the mainstay chemotherapeutics in gastrointestinal cancer treatment, characterizing a role for p53 in regulating dUTPase gene expression in tumor cells may be of major clinical significance and may lead to more targeted therapeutic options. In this study, we demonstrate direct roles for Sp1 and E2F-1 in driving basal DUT-N expression and report a direct role for wild type and mutant p53 in repressing and inducing dUTPase promoter activity respectively. Furthermore, we demonstrate the ability of oxaliplatin, an important chemotherapeutic agent primarily used in combination with 5-FU in the treatment of colorectal cancer, to acutely suppress the dUTPase promoter, mRNA and protein expression and enzymatic activity in a p53-dependent manner. We also present evidence that p53 is associated with the endogenous dUTPase promoter in vivo, and propose that interference with transcription machinery at the dUTPase promoter by wild-type p53 represents a plausible mechanism for this transcriptional repression. Finally, suppression of dUTPase by oxaliplatin resulted in the expansion of intracellular dUTP pools and subsequently enhanced the downstream effects of TS-inhibition. This interaction may contribute to the clinical synergy observed between 5-FU and oxaliplatin in the treatment of colorectal cancer.
MATERIALS AND METHODS
Compounds and reagents
5-FU, fluorodeoxyuridine (FUdR), paclitaxel, doxycycline (DOX), G418 and mithramycin A were purchased from Sigma (St Louis, MO). Oxaliplatin was obtained with permission from Sanofi Synthelabo (Bridgewater, NJ).
Cell culture
The human colon cancer cell lines HCT116 p53+/+, HCT116 p53–/– and HCT116 p21–/– cell lines, generated via homologous recombination, were generous gifts from the laboratory of Dr Bert Vogelstein at Johns Hopkins University (Baltimore, MD). HCT116 cell lines were grown in McCoy's 5A. Drosophila SL2 cells (Invitrogen, Carlsbad, CA) were grown in Schneider's Insect media (Sigma) supplemented with 10% heat-inactivated serum. All other media was supplemented with 10% fetal bovine serum (Lonza, East Rutherford, NJ) with penicillin/streptomycin, and sodium pyruvate (Invitrogen). Cells were maintained in a humidified Forma incubator (Thermoscientific, Waltham, MA) at 37°C with 5% CO2. All cell lines were routinely screened for the presence of mycoplasma using the MycoALERT assay (Lonza, Rockland, ME).
Generation of overexpression constructs
The pPac0 and pPac-Sp1, pN3 empty vector (EV) and pN3-Sp1 were a kind gift from Dr Guntram Suske from the Institut Fur Molekularbiologie und Tumorforschung (Marburg, Germany). Chemically synthesized primers incorporating restriction endonuclease sites were used to generate PCR fragments corresponding to the DUT-N and p53 wild-type coding sequences from the HCT116 p53+/+ cell line. Cloning was performed as previously described (25). cDNA corresponding to the p53 R175H mutant was generated using site-directed mutagenesis of p53 wild type: 5′-CGT TGT GAG GCA CTG CCC CCA CC-3′. Briefly, The cDNA fragments were gel purified using the QIAquick gel purification kit (QIAGEN, Valencia, CA) and ligated into the pre-digested pTRE-Tight vector (BD Clontech, Mountainview, CA) for DUT-N, and pCI-Neo for the p53 cDNA using standard methods. Recombinant DNA was transformed into the E. coli host strain DH5α, DNA isolated and analyzed by restriction digest and sequenced for orientation and integrity of the gene insert. Functionality of the nuclear dUTPase (pTre-Tight:DUT-N), p53WT (pCI-Neo:p53WT) and the p53 R175H mutant (pCI-Neo:p53MUT) constructs were confirmed by transient transfection and Western blotting (9).
Generation of the HCT116 pTet-Off cell line
HCT116 p53+/+ cells were seeded at a density of 1 × 106 on a 10 cm plate for 24 h, transfected using ExpressfectTM (Denville Scientific, Metuchen, NJ) with 2 µg of the pTet-Off plasmid (BD Clontech). Six hours post-transfection, complexes were removed, cells were washed once with PBS and fresh medium was added. After 24 h, cells were trypsinised into six 10 cm plates for 24 h before media containing G418 (800 µg/ml) was added. After selection for approximately 14 days, colonies were isolated and expanded. Potential pTet-Off cells were tested for tetracycline-repressible expression by transient transfection with the pTre-Tight:Luc (BD Clontech) in media containing 10% tet-approved FBS (BD Clontech) in the presence and absence of DOX (500 ng/ml).
Overexpression of dUTPase
HCT116 pTet-Off cells were seeded on 6 cm plates and 3 h after plating the cells were washed with PBS and growth media containing 10% tet-approved FBS (BD Clontech) added. Cells were transfected after 24 h with 2 µg pTre-Tight:DUT-N for 6 h, washed in PBS and fresh media added. Twenty-four hours post-transfection, media containing the appropriate cytotoxic agent was added. Cells were harvested for protein after 48 h incubation and inducible expression of dUTPase confirmed using Western blotting and enzyme activity assay.
dUTPase promoter constructs
The 1.2 kb region of the dUTPase promoter upstream of the transcriptional start site was previously reported to encompass all putative transcription factor-binding sites with high correlation to reported consensus sequences (15). This region was amplified by PCR using chemically synthesized primers (5′- GGT TCC CAC TGC GTT TCT G; 3′-CTC TCC TCT TCC CCC GGT G) and the generated 1.231 kb fragment cloned into the pGL3-Basic immediately upstream of the firefly luciferase gene and was termed the ‘full length’. Promoter truncations were performed using the following primers followed by re-ligation: pGL3:608 (5′-CGA AGC CGC GGT ACT CTC C-3′), pGL3:162 (5′-GAA GGG CTT CAA ACC CAA AAT-3′), pGL3:65 (5′-GAA ATT TCG GTT TTG GCG C-3′).
Site-directed mutagenesis
Targeted site-specific mutagenesis of Sp1- and E2F-binding sites in the dUTPase promoter were performed using the GeneEditor in vitro Site-Directed Mutagenesis System (Promega, Madison, WI) according to manufacturer's instructions. Briefly, the triple Sp1 mutant (pGL3:Sp1Δ) was generated by double base mutation from GG→TT (underlined) within the Sp1 site indicated in bold, using the following chemically synthesized primers, 5′-GCT GGC TTG AAA TTT CGG TTT TG-3′, 5′-CTC GTC CCG GGG AGG TTC GGT GGG TGG GGC GG-3′, 5′-TGG TTC GGG GCT GGC TTG AAA TTT CGG TTT TG-3′ in three consecutive rounds of mutagenesis. The E2F mutant (pGL3:E2FΔ) was generated by double base mutation from CG→AT (underlined) within the E2F site indicated in bold using 5′-GAA ATT TCG GTT TTG GAT CTC TCC CTG CGG C-3′ (Supplementary Figure 1). The integrity of each site-specific mutant was confirmed by direct DNA sequencing.
Antibodies and western blotting
At specified time points, cells were collected and analyzed by western blot as described previously (9). Blots were probed overnight at 4°C with the following antibodies from Santa Cruz Biotechnology (Santa Cruz, CA) at specified dilutions: anti-p53 (1 : 2000), anti-p21 (1 : 1000), anti-Sp1 (1 : 2000), anti-E2F-1 (1 : 1000) and anti-β-actin (1 : 4000). Affinity purified anti-dUTPase was used at 1 : 500 as previously described (26).
Growth inhibition assay
Cells were seeded in 96-well plates at 3×103 and exposed to 5-FU, FUdR or oxaliplatin, alone and in combination for 72 h. Growth inhibition was measured as previously described (9) using CellTiter 96® AQueous One Solution (Promega). Absorbance was measured using a SpectraMax 190 microplate reader (Molecular Devices, Sunnyvale, CA) at 490 nm. Fraction affected (FA) was calculated from the percent growth inhibition using the following equation: (100 − percent growth inhibition)/100. Statistical significance was determined using a two-way ANOVA (Graphpad, San Diego, CA).
dUTP accumulation assay
HCT116 p53+/+ cells were treated with specified concentrations of 5-FU, FUdR, paclitaxel and oxaliplatin or combinations for indicated times, harvested and resuspended in PBS. Cells were analyzed for nucleotide pool content using the assay developed by Sherman and Fyfe (27) modified to detect levels of TTP and dUTP by pre-incubating cellular extracts with recombinant dUTPase (9,28). The amount of radioactive incorporation, measured in the presence of dUTPase represented the TTP pool only, while untreated extracts represented both the dUTP and TTP pools. The amount of dUTP accumulation was determined by subtracting the results of extracts treated with dUTPase from the untreated extracts and presented as percentage accumulation in histogram format. The differences between treatment groups were analyzed for statistical significance using a two-tailed unpaired Student's t-test (Graphpad).
dUTPase activity assay
Twenty-five micrograms of total protein was normalized to a 20 µl volume with PBS/protease inhibitor. Relative dUTPase activity was determined as previously described (9), and expressed as a percentage relative to the appropriate untreated time-matched control.
RNA isolation, cDNA synthesis and quantitative real-time RT–PCR (qPCR)
RNA was isolated using Trizol as per manufacturers method (Invitrogen) and subjected to lithium chloride precipitation. cDNA was reverse transcribed using 200 ng total RNA following M-MLV-RT protocol (Invitrogen). qPCR was conducted using the ABI 7500 (Applied Biosystems, Foster City, CA) and PlexorTM multiplex (Promega). Probe sets used for qPCR analysis were as follows: dUTPase 5′-FAMTM-iso-dC-GGG AGA TCA TAT GAG TTA AAT ACA GGC TTT TTT T-3′, 5′-GGT GAC CTG ATG TAA ACA GTG TCT TC-3′; TS 5′-Cal Fluor® Orange 560-iso-dC-CCA TGT CTC CCG ATC TCT GGT-3′, 5′-TGG AAT CCA AGA GAT CTT CCT CTG ATG-3′; 18S 5′-Cal Fluor® Red 610-iso-dC-GCA TCG TTT ATG GTC GGA ACT ACG-3′, 5′-TTG TTG GTT TTC GGA ACT GAG GC-3′. GAPDH is a pre-validated house-keeping gene purchased with the PlexorTM multiplex system and was detected as Cal Fluor® Red 610. Standard curves with >0.95R2 and PCR-efficiency at 100 ± 2% were confirmed for each primer set within the multiplex. Threshold cycle values (CT) were determined from three independently isolated RNA samples and performed in triplicate. dUTPase and TS mRNA expression levels were determined by normalizing against both 18S and GAPDH expression using the 2–ΔΔCT method (29) and calibrated against appropriate time-matched controls. mRNA expression is presented in a histogram as a percentage of the appropriate untreated time-matched control. Statistical significance was determined using a two-tailed unpaired Student's t-test (Graphpad).
Transient transfection and luciferase promoter assay
Drosophila SL-2 cells and HCT116 isogenic lines were seeded in a 24-well plate. Transient transfections were performed using plasmid DNA normalized to 0.25 µg/µl using FugeneTM (Roche, Basel, Switzerland) for SL2 cells and ExpressfectTM (Denville) for HCT116 cells, according to manufacturer's instructions. The dUTPase pGL3 promoter constructs were transfected alone or co-transfected with pCI-Neo:p53WT, pCI-Neo:p53MUT and the corresponding pCI-Neo EV. Additional experiments were performed by co-transfecting the appropriate dUTPase promoter construct with the mammalian pN3-Sp1 or pN3 EV constructs or the Drosophila pPac-Sp1 and pPac0 constructs. All wells were transfected with 0.2 µg of the appropriate construct with co-transfections totaling 0.4 µg DNA. The pRL-TK plasmid was included in all transfections at a ratio of 1 : 10 (control : reporter) to account for transfection efficiency. The pGL3-Basic EV was also tested with all expression constructs and cytotoxic agents to discount non-specific effects. Six hours post-transfection, cells were washed with PBS and incubated in either fresh media or media containing a cytotoxic agent at the appropriate concentration. After an additional 30 h, cells were washed once with PBS, lysed and quantified as per western blotting methodology to account for variation in cell number. Twenty-five micrograms of total protein was normalized to 20 µl and analyzed using the dual-luciferase reporter assay (Promega) in a TD 20/20 luminometer (Turner Biosystems, Sunnyvale, CA). Three independent promoter experiments were performed, each transfection in triplicate and the mean of two firefly luminescence readings recorded, subsequently the reaction was quenched and Renilla luciferase measured. Co-transfection experiments investigating the effects of Sp1 overexpression were subject only to correction for protein concentration due to non-specific effects of Sp1 on the pRL-TK vector. Histograms are presented as normalized relative luciferase, or fold-change compared to control as appropriate. The differences between comparative transfections were analyzed for statistical significance using a two-tailed unpaired Student's t-test (Graphpad).
RNAi
Two million cells were plated for 24 h and subsequently transfected using Lipofectamine RNAiMax (Invitrogen) alone (Mock) or with 100 nM scrambled control siRNA (Scr) or 100 nM of validated StealthTM DuoPak siRNA targeting Sp1 (Invitrogen). For western blotting, cells were harvested after 72 h for analysis of dUTPase, Sp1 and β-actin protein expression. For dUTPase promoter activity, siRNA-transfected cells were trypsinised after 24 h and re-plated in 24-well plates and allowed to adhere for 24 h before transfection with the pGL3:FL dUTPase promoter for 30 h.
Chromatin immunoprecipitation (ChIP)
Three million cells were plated for 24 h and treated with the appropriate cytotoxic agent for a further 12 h. ChIP was performed using the EZ-ChIP kit (Upstate/Millipore, Temecula, CA) according to the manufacturers’ instructions. Briefly, DNA-protein cross-linking was achieved by addition of 1% formaldehyde to media. Cross-linked chromatin was sonicated on ice to ∼200 bp, verified by gel electrophoresis, purified and incubated with 5 µg of the appropriate ChIP-validated antibody: Sp1 5 µg; (Millipore), E2F-1 5 µg, p53 5 µg (Santa Cruz), by overnight rotation at 4°C, washed and cross-linking reversed. Parallel IPs using the kit-supplied anti-acetyl H3 (Ac-H3) and rabbit IgG as positive and negative controls respectively were performed. For qPCR, recovered IP chromatin and total input DNA were resuspended in 50 µl of TE buffer. qPCR was performed using Dynamo SYBR green (New England BioLabs, Beverly, MA), standard PCR protocol and PCR primers flanking the dUTPase promoter (–39 to –260) generating a 221 bp product encompassing the triple Sp1 sites and E2F site: Upper 5′-CAC CAG AAC TGT GGA CTC GT-3′. Lower 5′-CAG GGA GCG CGC CAA AAC CG-3′ (Figure 1A). Threshold cycle values (CT) were determined in triplicate for each treatment from two independent IP samples, calibrated against input and compared to control IgG using the following equation:
where ΔCT = CT(IP sample) – CT(DNA input).
Figure 1.
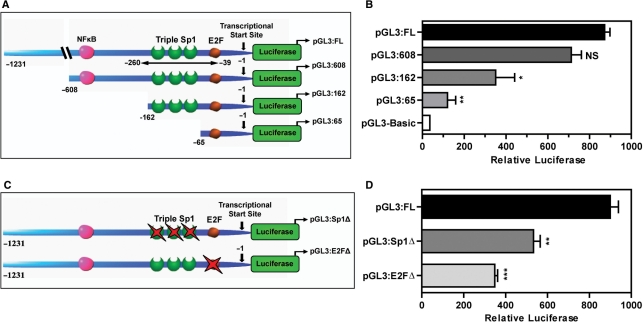
Systematic analysis of dUTPase gene promoter. (A) Schematic diagram illustrating the basic layout of the proximal DUT gene promoter and promoter truncations cloned into the pGL3-Basic promoterless vector encoding the firefly luciferase gene. (B) Relative luciferase activity of dUTPase promoter constructs (0.2 µg) transiently transfected into the HCT116 p53+/+ colon cancer cell line. (C) Schematic diagram illustrating the site-specific triple Sp1 (pGL3:Sp1Δ) and E2F mutant (pGL3:E2FΔ) generated by site-directed mutagenesis as outlined in ‘Experimental Procedures’ section. (D) Relative luciferase activity of dUTPase promoter constructs (0.2 µg) transiently transfected into the HCT116 p53+/+ colon cancer cell line. Transfections were performed as outlined in ‘Experimental Procedures’. Histogram bars represent mean ± SEM of three independent promoter experiments. *P < 0.05, **P < 0.01, ***P < 0.001; NS, not significant.
Changes in promoter enrichment for Ac-H3, Sp1, E2F and p53 following drug treatment are expressed as fold enrichment over control IgG or fold increase compared to untreated time-matched controls where appropriate with differences between ChIPs analyzed for statistical significance using a two-tailed unpaired Student's t-test (Graphpad). Semi-quantitative ChIP analysis was performed using the same primers, standard three-step PCR amplification with products visualized by gel electrophoresis. As a control, IP DNA was subjected to PCR using primers targeting the dUTPase ORF to verify amplification only in ‘input’ DNA.
Electrophoretic mobility shift assay (EMSA)
Electrophoretic mobility shift analyses were performed as described earlier with minor modification (30). Synthetic double-stranded oligonucleotides (Integrated DNA Technologies, Coralville, IA) corresponding to the –64 to –91 from the DUT-N transcriptional start site and encompassing the triple Sp1 site were labeled with [32P]ATP (Amersham Pharmacia Biotech) according to the Gel-Shift Assay Kit protocol (Promega). For each gel shift reaction, 10 000 cpm of labeled probe were incubated with ∼70 fmol of recombinant Sp1. Where indicated, unlabeled competitor oligonucleotides were incubated for 10 min at room temperature with Sp1 before the addition of labeled probe. Sequences of the oligonucleotides were as follows: DUT Sp1, 5′-GGG CGG TGG GTG GGG CGG GGC TGG CGG G-3′; DUT Sp1Δ, 5′-GTT CGG TGG GTG GTT CGG GGC TGG CTT G-3′.
RESULTS
Functional analysis of the dUTPase promoter
Due to the critical role of dUTPase in maintaining genomic stability, the aberrant expression observed in neoplastic tissues and the reported role of dUTPase in resistance to chemotherapy, we sought to elucidate the transcriptional mechanisms driving dUTPase gene expression. We previously performed a computational sequence analysis of the 5′ region of the human dUTPase gene and identified putative binding sites for NFκ-B, Sp1 and E2F (15) (Supplementary Figure 1A). The presence of Sp1- and E2F-binding sites is consistent with the hypothesis that the dUTPase promoter is regulated in a cell-cycle-dependent manner (31). However, to date there have been no studies investigating the in vivo regulation of human dUTPase at the promoter level. To address this, we first sought to determine the basal DNA regulatory elements of the dUTPase promoter that are required for expression. We utilized a luciferase-based system employing the pGL3-Basic promoterless vector and sub-cloned deletion mutant fragments targeting the putative E2F- and Sp1-binding sites reported to be critical for S-phase gene expression. We generated a series of four dUTPase promoter truncations ranging from the designated ‘full length’ (pGL3:FL) which incorporates the 1.231 kb proximal promoter upstream of the DUT-N transcriptional start site, to the extensive truncation mutant pGL3:65 (Figure 1A) and analyzed luciferase expression in the HCT116 p53+/+ colon cancer cell line model. Truncation of the pGL3:FL to the pGL3:608 resulted in an insignificant decrease of 1.22-fold in promoter activity, suggesting that the 5′ region of the proximal dUTPase promoter may contain relatively minor positive regulatory elements (Figure 1B). A significant reduction in promoter activity of 2-fold was observed between the pGL3:608 and the pGL3:162 which differ by the consensus NFκ-B-binding site. A 2.9-fold reduction was observed when the pGL3:162 was truncated to the pGL3:65 to exclude the triple Sp1 sites. The pGL3:65 that contains only the E2F site demonstrated a significant 7.2-fold reduction in luciferase activity compared to the pGL3:FL. Of note, the activity of pGL3:65 remained 3.4-fold higher than the pGL3-Basic promoterless vector (Figure 1B). These data suggest that consensus sequences for NFκ-B, E2F and to a greater extent Sp1 may positively contribute to dUTPase promoter activity.
Identification of transcription factors involved in dUTPase gene expression
Sp1 and E2F family members are known to be key regulators of S-phase gene promoters (32). Having identified the region of the DUT gene promoter containing the Sp1 and E2F transcription sites to be associated with positive regulation, we sought to determine the relative contributions of both Sp1 and E2F consensus sequences to dUTPase promoter activity. We utilized site-directed mutagenesis of the pGL3:FL to disrupt the triple Sp1 (pGL3:Sp1Δ) and E2F sites (pGL3:E2FΔ) by double nucleotide substitution within each respective site to generate mutant dUTPase luciferase constructs (Figure 1C and Supplementary Figure 1B). We transiently transfected the pGL3:FL, pGL3:Sp1Δ and the pGL3:E2FΔ into the HCT116 p53+/+ cells and analyzed their basal promoter activity. Mutation of the triple Sp1 sites decreased dUTPase promoter activity 1.7-fold while disruption of the E2F site resulted in a 2.6-fold reduction in promoter activity when compared to the pGL3:FL (Figure 1D). This data further establish and confirms the role of Sp1 and E2F sites as positive regulators of dUTPase promoter activity.
The role of Sp1 in dUTPase gene expression
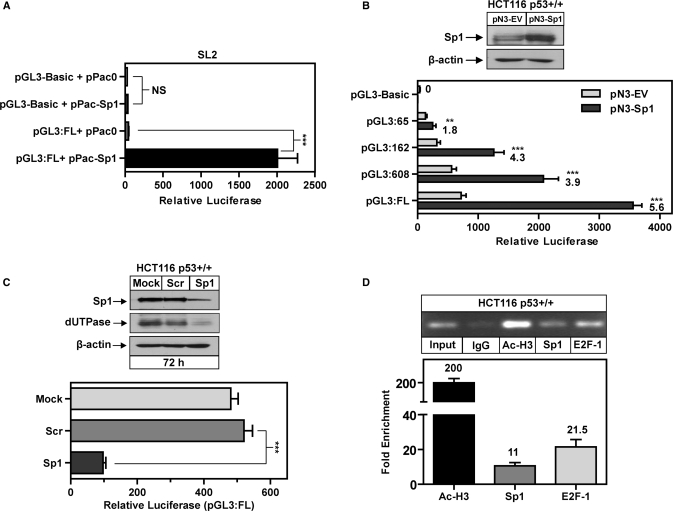
Elevated intratumoral expression of Sp1 is reported to increase expression of genes that promote tumor processes including progression and angiogenesis (33). It is therefore plausible that dysregulation of Sp1 may contribute to increased expression of dUTPase that is observed in tumor tissues. In order to accurately determine the contribution of Sp1 to basal promoter activity, we utilized the well-characterized Sp1-deficient Drosophila SL2 cells as a useful model for analyzing the influence of Sp1-driven promoters in an Sp1-deficient background (32). The pGL3-Basic promoterless vector was subsequently co-transfected with the EV and pPac-Sp1 expression construct, both resulting in minimal luciferase activity. Co-transfection of the pGL3:FL with pPac0 resulted in no significant promoter activity when compared to the co-transfection with pGL3-Basic. However, co-transfection of pGL3:FL and pPac-Sp1 resulted in a 45-fold increase in luciferase activity (Figure 2A). These data demonstrate that the dUTPase promoter requires Sp1 for basal activity and is also highly responsive to Sp1-mediated transcription in an Sp1-deficient model.
Figure 2.
The role of Sp1 in dUTPase gene expression. (A) Relative luciferase activity of dUTPase full-length promoter (pGL3:FL) and pGL3-Basic EV (0.2 µg) co-transfected with pPac0 and pPac-Sp1 (0.2 µg) in Drosophila SL2 cells. (B) Western blot demonstrating overexpression of Sp1 from corresponding promoter lysates following transfection of HCT116 p53+/+ cells with pN3-EV or pN3-Sp1 (0.2 µg), anti-β-actin was used to control for loading; relative luciferase of dUTPase promoter constructs co-transfected with both pN3-EV and pN3-Sp1 (0.2 µg) in HCT116 p53+/+ cells, numbers indicate fold-increase of pN3-Sp1 transfected cells compared to pN3-EV. Transfections were performed as outlined in ‘Experimental Procedures’ Section. Histogram bars represent mean ± SEM of three independent promoter experiments. (C) Top: Western blot following transfection of HCT116 p53+/+ cells with Lipofectamine only (Mock), 100 nM scrambled control siRNA (Scr) and 100 nM of StealthTM DuoPak siRNA targeting Sp1 (Sp1) for 72 h and probed with anti-dUTPase and anti-Sp1. Anti-β-actin was used to control for loading. Bottom: Relative luciferase activity of dUTPase full length promoter (pGL3:FL) transfected (0.2 µg) into HCT116 p53+/+ cells in the presence of scrambled control siRNA or StealthTM DuoPak Sp1-targeted siRNA, histogram bars represent mean ± SEM of three independent promoter experiments. (D) Chromatin immunoprecipitation demonstrating in vivo association of E2F-1, Sp1 and Ac-H3 with the dUTPase promoter during normal cellular proliferation, visualized by semi-quantitative gel electrophoresis and qPCR using primers encompassing the E2F and Sp1 sites (–39 to –260) in the dUTPase promoter as indicated in Figure 1A. **P < 0.01, ***P < 0.001; NS, not significant.
Having confirmed that the dUTPase promoter is responsive to Sp1 in an Sp1-null background, we extended these studies to the HCT116 p53+/+ cells and tested the effect of Sp1 overexpression on the dUTPase promoter. Western blot analysis demonstrated a physiologically relevant 2-fold increase in Sp1 protein expression following transfection with the pN3-Sp1 expression construct (Figure 2B). Importantly, Sp1 overexpression did not induce the pGL3-Basic control discounting any possible non-specific effects. Co-transfection of the pGL3:FL with the pN3-Sp1 construct resulted in a 5.6-fold increase in luciferase activity compared to the pN3-EV. We also tested the Sp1-responsiveness of the promoter truncations and demonstrate that the pGL3:608 truncation is significantly less responsive to Sp1-mediated activation than the pGL3:FL (3.9-fold compared to 5.6-fold respectively), further suggesting that the 5′ region from –608 to –1231 may contain some Sp1-responsive elements. In addition, the pGL3:65 containing only the E2F site was induced 1.8-fold by overexpression of Sp1 suggesting that Sp1 may also mediate dUTPase promoter activation indirectly through E2F consensus sequences (Figure 2B). However, Sp1 overexpression resulted in only a modest increase in luciferase activity of the pGL3:Sp1Δ which did not reach statistical significance (data not shown). These data would suggest that the effects of Sp1 overexpression were primarily limited to the identified triple Sp1 sites.
To confirm the role of Sp1 in dUTPase transcription, we utilized StealthTM RNAi employing two siRNA duplexes targeting the Sp1 mRNA. Transfection with the Sp1-targeted siRNA resulted in knockdown of Sp1 as measured by western blotting at 72 h. Importantly dUTPase protein expression was also markedly reduced at 72 h in cells transfected with Sp1-targeted siRNA when compared to mock- and scrambled control-transfected cells. In addition, depletion of Sp1 also significantly attenuated the activity of the dUTPase full-length promoter by >5-fold in HCT116 p53+/+ cells (Figure 2C).
To further confirm that Sp1 and E2F play a role in driving dUTPase expression in vivo we utilized a highly sensitive and quantitative approach combining ChIP with qPCR and a primer set spanning the triple Sp1 and E2F sites in the dUTPase promoter. We successfully identified Sp1 and E2F-1 transcription factors as enriched 11- and 21.5-fold, respectively, at the dUTPase promoter when compared to non-specific rabbit IgG controls and normalized to input DNA. Acetyl-H3 (Ac-H3), an indicator of active gene transcription was employed as a positive control and demonstrated a highly significant 200-fold enrichment at the dUTPase promoter (Figure 2D). These data provide the first direct evidence that both Sp1 and E2F-1 are actively associated with the dUTPase promoter in vivo and that this region is subject to significant histone acetylation.
Ectopic p53 represses dUTPase promoter activity
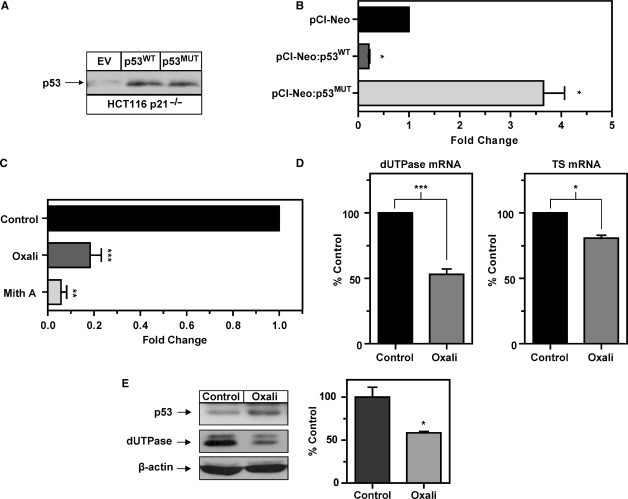
Several reports have identified dUTPase as repressed following ectopic expression or induction of p53 (21,22) and a p53-mutant transactivation gene target (24). Mutations in the p53 tumor suppressor gene are reported in a wide variety of cancers and in >50% of colon cancers (35). Significant evidence exists suggesting that tumor p53 status has direct impact on gene expression, tumor progression and response to chemotherapy (36). Given the critical nature of dUTPase expression in modulating response to the TS-inhibitor class of chemotherapeutics, we sought to test whether dUTPase expression is modulated by ectopic expression of both wild-type p53 (p53WT) and the hotspot R175H p53 mutant (p53MUT). This is one of the most common human tumor-derived mutant p53 proteins, harboring an Arg→His amino-acid substitution resulting in a dominant gain-of-function and has been reported to induce dUTPase in p53 null mouse fibroblast cells (24). We generated overexpression constructs using the pCI-Neo vector and utilized the HCT116 p53+/+ and p53–/– isogenic colon cancer cell lines to determine the precise contribution of p53 on dUTPase promoter activity. For confirmation purposes, p53 expression was verified in the p53+/+ cells and absence noted in the HCT116 p53–/– by western blotting (Figure 3A).
Figure 3.
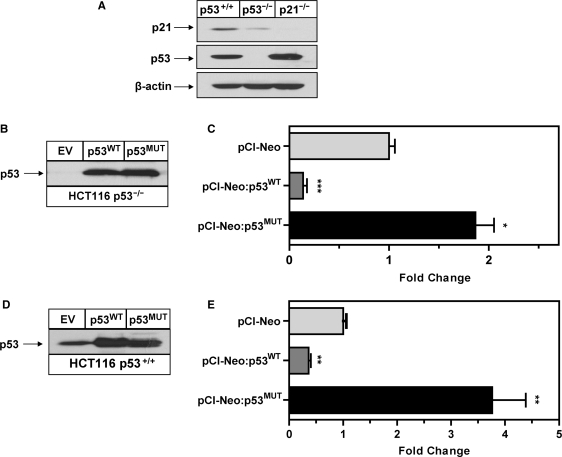
Ectopic p53 represses dUTPase promoter activity. (A) Western blot probed with anti-p53, anti-p21 confirming p53 expression in HCT116 p53+/+ cells and absence of expression in HCT116 p53–/– cells. HCT116 p21–/– cells were included in this Western analysis for direct comparative purposes, anti-β-actin was used to control for loading. (B) Western blot demonstrating equivalent ectopic expression of wild-type p53 and mutant p53 protein following transient transfection with of pCI-Neo:p53WT and pCI-Neo:p53MUT (0.2 µg) in the p53–/– cell line. (C) Fold-change in luciferase activity of dUTPase full-length (pGL3:FL) promoter construct (0.2 µg) co-transfected with pCI-Neo:p53WT and pCI-Neo:p53MUT (0.2 µg) in the p53–/– cell line. (D) Western blot demonstrating equivalent ectopic expression of wild-type p53 and mutant p53 protein following transient transfection with pCI-Neo:p53WT and pCI-Neo:p53MUT (0.2 µg) in the HCT116 p53+/+ cell line. (E) Fold-change in luciferase activity of dUTPase full length (pGL3:FL) promoter construct (0.2 µg) co-transfected with pCI-Neo:p53WT and pCI-Neo:p53MUT (0.2 µg) in the p53+/+ cell line. All transfections were performed as outlined in ‘Experimental Procedures’ Section. Histogram bars represents mean ± SEM of three independent promoter experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
To eliminate any effects from endogenous p53, we utilized the HCT116 p53–/– cells. A titration using pCI-Neo:p53WT and pCI-Neo:p53MUT constructs was performed and indicated that a vector concentration of 0.2 µg per transfection resulted in a physiological relevant increase in p53 protein consistent with the level of induction in p53+/+ cells at 24 h post-DNA damage (data not shown). Co-transfection of the pGL3:FL with pCI-Neo:p53WT and pCI-Neo:p53MUT induced a physiologically relevant expression of the respective p53 proteins detected by western blotting with the pCI-Neo EV control demonstrating no detectable p53 protein (Figure 3B). Co-transfection of the pGL3:FL with pCI-Neo: p53WT resulted in a 7-fold reduction in promoter activity when compared to the pGL3:FL co-transfected with pCI-Neo EV. In contrast, co-transfection of pGL3:FL with pCI-Neo:p53MUT resulted in a 1.9-fold increase in promoter activity when compared to the corresponding EV control (Figure 3C). We then analyzed the effects of p53 overexpression in the p53+/+ cells which contain endogenous levels of p53 protein. Western blot analysis confirmed detection of basal p53 and similar levels of p53 overexpression in these cells compared to the p53–/– cells following transfection with the pCI-Neo:p53WT and and pCI-Neo:p53MUT (Figure 3D). Co-transfection of the pGL3:FL with pCI-Neo:p53WT resulted in a 2.6-fold reduction in promoter activity when compared to the pGL3:FL co-transfected with pCI-Neo EV (Figure 3E). In contrast, co-transfection of pGL3:FL with pCI-Neo:p53MUT resulted in a 3.8-fold increase in promoter activity when compared to the corresponding EV control (Figure 3E). These data indicate that ectopic expression of wild-type p53 can repress dUTPase promoter activity and that ectopic p53 mutant expression can induce dUTPase promoter activity in the HCT116 p53 isogenic cell-line models.
The effects of DNA damage-induced p53 on dUTPase gene expression
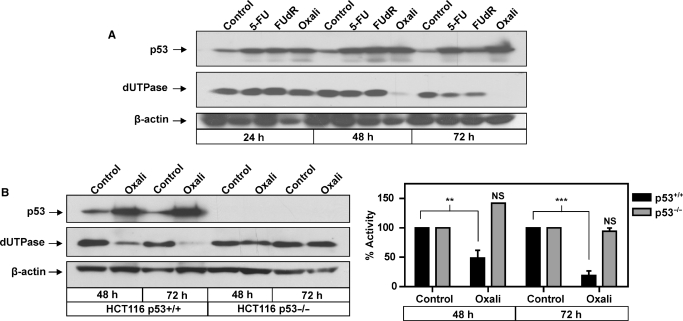
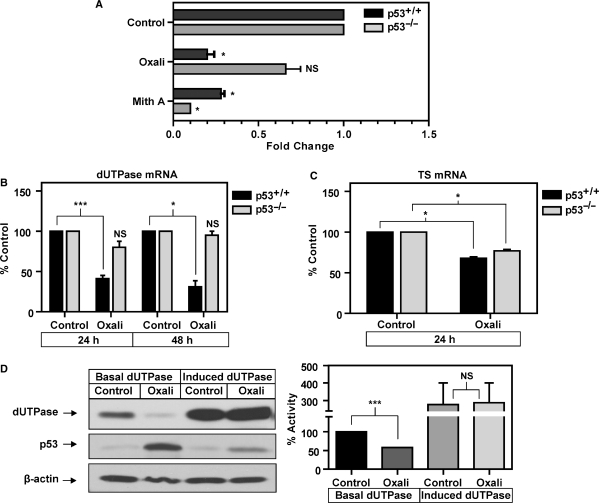
dUTPase expression is reported to be an important determinant of sensitivity to chemotherapeutic agents that target TS. Having determined that ectopic overexpression of both p53WT and p53MUT have direct effects on the dUTPase promoter we sought to determine if DNA-damage-induced p53 can directly modulate dUTPase protein expression. We utilized the TS-targeted chemotherapeutic agents 5-FU and FUdR, both of which are reported to activate p53. In addition, we utilized oxaliplatin, a non-TS-directed DNA-damaging agent and the primary 5-FU combination partner for the treatment of colorectal cancer (37). Western blotting confirmed the induction of p53 at 24–72 h in response to 5-FU, FUdR and oxaliplatin treatment in the p53+/+ cells (Figure 4A). In response to 5-FU and FUdR, dUTPase protein expression remained unchanged until 72 h post-treatment where levels were only moderately reduced. Unexpectedly, 0.5 µM oxaliplatin treatment significantly downregulated dUTPase at 48 h post-treatment and by 72 h dUTPase was further reduced to undetectable levels by western blot analysis (Figure 4A). To investigate the contribution of p53 expression to the novel observation that oxaliplatin downregulates dUTPase, the HCT116 p53+/+ and p53–/– isogenic cell lines were utilized. Western blot analysis demonstrated that p53 was markedly induced in the p53+/+ cells but was not detected or induced in the p53–/– cells following treatment with 1 µM oxaliplatin. In response to oxaliplatin, dUTPase expression and enzymatic activity was significantly downregulated 2- and 5-fold in the p53+/+ cells at 48 and 72 h, respectively. No modulation of expression or activity was detected in the p53–/– cells at 48 or 72 h (Figure 4B). This data suggests that p53 or a downstream p53-target pathway is responsible for the downregulation of dUTPase protein expression following oxaliplatin treatment.
Figure 4.
The effects of DNA damage-induced p53 on dUTPase gene expression. (A) Representative western blot showing timecourse following treatment with the chemotherapeutic agents 5-FU (5 µM), FUdR (1 µM) and oxaliplatin (0.5 µM), probed with anti-p53 and anti-dUTPase, anti-β-actin was used to control for loading. (B) Representative western blot in HCT116 p53+/+ and p53–/– isogenic cells following treatment with 1 µM oxaliplatin for 48 and 72 h. Membrane was probed with anti-p53, anti-dUTPase and anti-β-actin was used to control for loading; histogram represents corresponding cell lysates subjected to dUTPase catalytic activity assay with oxaliplatin-treated samples expressed as a percentage of appropriate untreated time-matched controls set at 100%, bars represents mean ± SEM from two replicates. **P < 0.01, ***P < 0.001; NS, not significant.
p53-mediated downregulation of dUTPase by oxaliplatin is induced through a transcriptional mechanism
It has been reported that proteins involved in cell-cycle progression and DNA synthesis are acutely downregulated in an atypical manner following exit from cell-cycle phases or following induction of DNA damage-induced cell cycle arrest by mechanisms such as ubiquitination and proteasomal degradation (38,39). To confirm that the acute oxaliplatin-induced downregulation of dUTPase protein was due to direct transcriptional repression and not post-transcriptional mechanisms or protein stability, we transfected the dUTPase full-length (pGL3:FL) promoter construct into the HCT116 p53+/+ and p53–/– isogenic cells and analyzed the effects of oxaliplatin. In the p53+/+ cells, oxaliplatin potently attenuated activity of the pGL3:FL promoter by 5-fold with only a modest reduction of 1.5-fold in p53–/– cells (Figure 5A). Mithramycin A, a specific inhibitor of Sp1-driven gene expression that functions by binding to, and inhibiting transcription from GC-rich promoters (33,34) was utilized as a positive control and potently attenuated dUTPase promoter activity by 3.5- and 10-fold in both the p53+/+ and p53–/– isogenic cells respectively, consistent with inhibiting Sp1-mediated dUTPase promoter activity.
Figure 5.
p53-mediated downregulation of dUTPase by oxaliplatin is induced through a transcriptional mechanism. (A) Fold-change in luciferase activity of dUTPase full-length promoter (pGL3:FL) following transfection (0.2 µg) in HCT116 p53+/+ and p53–/– isogenic cells and treatment with 1 µM oxaliplatin and 100 nM mithramycin A compared to pGL3:FL untreated control set at 1. (B) qPCR analysis of dUTPase mRNA in HCT116 p53+/+ and p53–/– isogenic cells following treatment with 1 µM oxaliplatin for 24 and 48 h compared to time-matched controls set at 100%, histogram bars represents mean ± SEM of three independent experiments. (C) qPCR analysis of TS mRNA in HCT116 p53+/+ and p53–/– isogenic cells following treatment with 1 µM oxaliplatin for 24 h compared to time-matched control set at 100%, histogram bars represents mean ± SEM of three independent experiments. (D) Western blot following transfection of HCT116 pTet-off cells with pTre-Tight:DUT-N (1 µg) showing increased expression of dUTPase in the absence of DOX (induced) and basal expression in the presence of 0.5 µg/ml DOX (basal) and subsequent treatment with 1 µM oxaliplatin for 48 h. Anti-p53 was used to confirm p53 induction and anti-β-actin to control for loading; histogram represents corresponding cell lysates subjected to dUTPase catalytic activity assay with treated samples expressed as a percentage of untreated control set at 100%, bars represents mean ± SEM from two independent replicates. *P < 0.05, ***P < 0.001; NS, not significant.
To further investigate the role of p53 in the transcriptional downregulation of dUTPase, we employed the PlexorTM multiplex qPCR platform to simultaneously analyze mRNA expression of both dUTPase and an additional S-phase gene and chemotherapeutic target TS. Analysis of dUTPase mRNA in the p53+/+ cells demonstrated that following treatment with 1 µM oxaliplatin, dUTPase mRNA was downregulated 2.5- and >3-fold at 24 and 48 h when compared to respective time-matched controls. In the p53–/– cells, dUTPase mRNA was not significantly modulated by oxaliplatin at 24 or 48 h when compared to their respective time-matched control (Figure 5B). Simultaneous analysis of TS mRNA demonstrated a modest but significant downregulation at 24 h in both p53+/+ and p53–/– cells when compared to the 24 h time-matched control. The oxaliplatin-induced downregulation of TS was not statistically significant different between the p53+/+ and p53–/–cells (Figure 5C). These data confirm that p53 expression plays a significant role in the oxaliplatin-induced transcriptional suppression of dUTPase mRNA but not TS mRNA.
To eliminate the possibility that dUTPase is a target for atypical protein turnover as a result of oxaliplatin-induced cell-cycle arrest, we overexpressed dUTPase using the pTre-Tight:DUT-N construct under the control of a DOX-repressive minimal CMV promoter and treated cells with oxaliplatin and analyzed dUTPase protein expression and catalytic activity. Following overexpression, dUTPase protein and enzyme activity was increased approximately 4-fold in the absence of DOX when compared to an identical transfection in the presence of 0.5 µg/ml DOX where ectopic expression was repressed. When cells with basal dUTPase expression were treated with 1 µM oxaliplatin for 48 h, accumulation of p53 was confirmed and a >2-fold downregulation in dUTPase protein and activity was observed compared to the respective untreated control by western blot analysis. Cells overexpressing dUTPase from the ectopic minimal CMV promoter also demonstrated p53 accumulation following oxaliplatin treatment, with dUTPase expression levels and catalytic activity demonstrating no significant difference in the presence and absence of 1 µM oxaliplatin (Figure 5D). These data suggest that dUTPase protein expressed from the ectopic promoter was not a target for atypical turnover, decreased stability or transcriptional suppression by oxaliplatin treatment.
p53-mediated downregulation of dUTPase does not require p21
Previous studies have demonstrated that p53-mediated induction of the cell-cycle inhibitor p21 following genotoxic stress resulted in repression of a number of genes (40,41). We therefore used the HCT116 p21–/– cells, possessing p53WT but lacking p21 protein to investigate the possible influence of p53-mediated transactivation of p21 on dUTPase promoter activity and gene expression. Western blot analysis previously confirmed the expression of p53 but lack of expression of p21 in these cells (Figure 3C). Western blot analysis confirmed similar levels of p53 overexpression in these cells as the p53+/+ and p53–/– cells following transfection with the pCI-Neo:p53WT and pCI-Neo:p53MUT (Figure 6A). We utilized the pGL3:FL dUTPase promoter construct and analyzed the effects of wild type and mutant p53 expression on luciferase activity. Co-transfection of the pGL3:FL with pCI-Neo:p53WT resulted in 5-fold reduction in luciferase activity. Interestingly, co-transfection of pGL3:FL with the pCI-Neo:p53MUT resulted in a 3.65-fold induction in luciferase activity similar to results obtained in the p53+/+ cells (Figure 6B).
Figure 6.
p53-mediated downregulation of dUTPase does not require p21. (A) Western blot demonstrating equivalent and physiologically relevant ectopic expression of wild-type p53 and mutant p53 protein following transient transfection with pCI-Neo:p53WT and pCI-Neo:p53MUT (0.2 µg) in the p21–/– cell line. (B) Fold-change in luciferase activity of dUTPase full length (pGL3:FL) promoter construct (0.2 µg) co-transfected with pCI-Neo:p53WT and pCI-Neo:p53MUT (0.2 µg) in the p21–/– cell line. (C) Fold-change in luciferase activity of dUTPase full-length promoter (pGL3:FL) (0.2 µg) following treatment with 1 µM oxaliplatin and 100 nM mithramycin A compared to pGL3:FL untreated control set at 1 in HCT116 p21–/– cells. All histogram bars represents mean ± SEM of three independent promoter experiments, transfections were performed as outlined in ‘Experimental Procedures.’ Numbers on histograms indicate fold-change compared to pGL3:FL. (D) qPCR analysis of dUTPase and TS mRNA in HCT116 p21–/– cells following treatment with 1 µM oxaliplatin for 24 h compared to time-matched control set at 100%. (E) Western blot in HCT116 p21–/– cells following treatment with 1 µM oxaliplatin for 48 h. Membrane was probed with anti-p53 and anti-dUTPase, anti-β-actin was used to control for loading; histogram represents corresponding cell lysates subjected to dUTPase catalytic activity assay with oxaliplatin-treated samples expressed as a percentage of appropriate untreated time-matched controls. Histogram bars represents mean ± SEM from two independent replicates, *P < 0.05, **P < 0.01, ***P < 0.001.
The ability of DNA-damage-induced p53 to repress the dUTPase promoter in the absence of p21 induction was subsequently analyzed. Following treatment with oxaliplatin, the pGL3:FL promoter was repressed 5-fold in the p21–/– cells which is identical to the observed repression in the HCT116 p53+/+ cells. Mithramycin A was used as a positive control and significantly suppressed the pGL3:FL promoter activity 20-fold (Figure 6C). To further establish a direct mechanism of transcriptional repression of dUTPase in the absence of p21, we utilized the PlexorTM multiplex qPCR and analyzed dUTPase and TS mRNA expression following treatment with oxaliplatin. Treatment with 1 µM oxaliplatin for 24 h resulted in a >2-fold reduction in dUTPase mRNA. A small but significant reduction in TS mRNA was observed in the p21–/– cells consistent with the p53+/+ and p53–/– isogenic cells (Figure 6D). Finally, western blotting confirmed that dUTPase protein expression was downregulated in the p21–/– cells following treatment with 1 µM oxaliplatin at 48 h in a manner consistent with the p53+/+ cells (Figure 6E). These data suggest that the oxaliplatin-induced transcriptional repression of dUTPase is independent of the p53-transactivation target p21 and is more likely a direct transcriptional effect of p53 expression.
Oxaliplatin-induced repression of the dUTPase promoter requires functional Sp1 sites
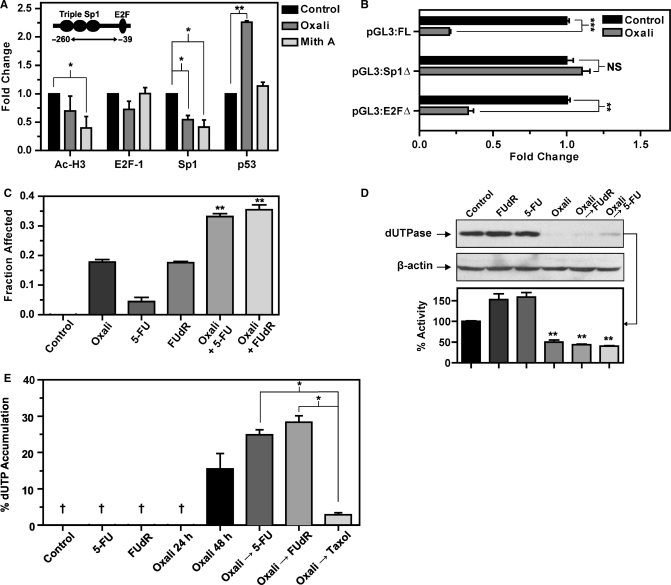
Reports suggested that the transcriptional repression of p53 can be exerted either through direct binding of p53 protein or via its sequestering of transcription factors required for promoter activity. To investigate the downregulation of dUTPase in response to oxaliplatin treatment, we utilized ChIP in conjunction with qPCR to analyze changes at the dUTPase promoter. This approach allows for quantitative analysis of the target region of the dUTPase promoter by normalizing DNA immunoprecipated (IP) with the antibody of interest directly to control IgG IP DNA with a subsequent calibration against ‘input’ DNA. This method accounts for non-specific antibody binding and variations in input DNA concentration and allows for sensitive quantification of alterations at the dUTPase promoter in response to drug treatment. Following treatment with 1 µM oxaliplatin and 100 nM mithramycin A for 12 h, we performed ChIP analysis using antibodies against Sp1, E2F-1, Ac-H3 and p53 in HCT116 p53+/+ cells and compared to untreated controls. We successfully detected basal Ac-H3, E2F-1, Sp1 and p53 enrichment at the dUTPase promoter compared to the IgG control in the region containing the Sp1 and E2F consensus sequences (−39 to −260).
Following treatment with 100 nM mithramycin A, Ac-H3 enrichment was reduced 2-fold consistent with decreased gene transcription whereas Sp1 enrichment was decreased 2.4-fold consistent with mithramycin A displacing Sp1 at the GC-rich dUTPase promoter. E2F-1 enrichment was not modulated in response to mithramycin A. Significantly, enrichment of the tumor suppressor p53 remained unchanged in mithramycin A treated cells (Figure 7A). Treatment with oxaliplatin resulted in a significant 2-fold reduction in Sp1 accompanied by a modest decrease in E2F-1 and Ac-H3 enrichment at the dUTPase promoter. Significantly, enrichment of p53 was increased 2.3-fold at the dUTPase promoter following treatment with oxaliplatin, when compared to the untreated control (Figure 7A). In an attempt to identify the p53-responsive element in the dUTPase promoter, we subsequently transfected the HCT116 p53+/+ cells with the dUTPase Sp1 and E2F site-mutant promoter constructs and compared the repression by oxaliplatin to the full–length construct. Importantly, these site mutant constructs retained significant transcriptional activity in HCT116 cells (Figure 1D). The full-length promoter was repressed 5-fold by oxaliplatin. The E2F site-mutant was repressed by oxaliplatin 3-fold. However, mutation of the triple Sp1 sites completely abrogated the oxaliplatin-induced promoter repression (Figure 7B) suggesting that p53 is acting on the region of the promoter containing the Sp1 sites. To confirm that the mutations within the pGL3:Sp1Δ construct efficiently impair Sp1 binding, we performed EMSA analysis. Using a dUTPase 32P-labelled probe encompassing the triple Sp1-binding sites (–64 to –91), we demonstrate binding of recombinant Sp1 (rhSp1) to both this sequence and a control Sp1 consensus oligo. However, a mutant probe corresponding to the Sp1 site mutations within the pGL3:Sp1Δ construct failed to compete for binding with rhSp1 (Supplementary Figure 3). This data supports the hypothesis that repression of the dUTPase promoter by oxaliplatin requires functional Sp1 sites.
Figure 7.
Oxaliplatin-induced, p53-mediated repression of dUTPase is mediated through Sp1 sites and results in the accumulation of dUTP. (A) Chromatin immunoprecipitation demonstrating in vivo alterations in Sp1 and p53 enrichment at the dUTPase promoter following 12 h treatment with 1 µM oxaliplatin in HCT116 p53+/+ cells. qPCR was performed using primers encompassing the E2F and Sp1 sites in the dUTPase promoter from −39 to −260 as indicated. Histogram bars represent mean ± SE from two independent immunoprecipitations analyzed in triplicate by qPCR. (B) Fold-change in luciferase activity of dUTPase promoter constructs (0.2 µg) treated with 1 µM oxaliplatin compared to untreated control set at 1, histogram bars represents mean ± SE from three independent promoter experiments. (C) Growth inhibition of HCT116 p53+/+ cells treated with 5-FU (0.5 µM), FUdR (0.1 µM) and oxaliplatin (0.5 µM) and combinations. Histogram represents the mean ± SEM fraction of cells affected (FA) following three independent 72 h incubations. (D) Western blot following treatment with 5-FU, FUdR and combinations for 48 h in HCT116 p53+/+ cells, membrane was probed with anti-dUTPase, anti-β-actin was used to control for loading; histogram represents corresponding cell lysates subjected to dUTPase catalytic activity assay with treated samples expressed as a percentage of appropriate untreated time-matched control set at 100%. (E) dUTP accumulation assay measuring intracellular dUTP as a percentage of the total dUTP/TTP pool in HCT116 p53+/+ cells following treatment with the TS-inhibitors 5-FU (0.5 µM), FUdR (0.1 µM) for 24 h and oxaliplatin (0.5 µM) for 24 and 48 h. Combinations were performed by incubating with oxaliplatin (0.5 µM) for 24 h, media was removed, cells were washed and re-incubated in media containing the appropriate TS-inhibitor or the microtubule-stabilizing agent paclitaxel (20 nM) as a non-TS-directed control. Histogram bars represents mean ± SEM from two independent experiments performed in duplicate. † indicates that dUTP was not detected in cell extracts. *P < 0.05, **P < 0.01, ***P < 0.001; NS, not significant.
Oxaliplatin induces accumulation of dUTP alone and in combination with TS-inhibitors in HCT116 p53+/+cells
Oxaliplatin is clinically approved for the treatment of colon cancer and is primarily used in combination with 5-FU or capecitabine where these drugs demonstrate synergistic anti-tumor activity (37). Given the novel observation that a clinically relevant concentration of oxaliplatin (0.5 µM, Figure 4A) can acutely downregulate dUTPase protein in a p53-dependent manner in a colon cancer model, we sought to test the possible clinical implications of this by measuring the accumulation of dUTP, a downstream cytotoxic event induced by chemotherapeutics that inhibit TS. We utilized 5-FU whose effects include TS-inhibition, and incorporation of fluorinated nucleotides into DNA and RNA and the more specific TS-inhibitor FUdR which induces only DNA-directed effects. First, we confirmed that the treatment of HCT116 p53+/+ cells with oxaliplatin and either 5-FU or FUdR for 72 h resulted in enhanced growth inhibition (Figure 7C) and resulted in downregulation of dUTPase protein and enzyme activity (Figure 7D). To verify that the effects on dUTP accumulation with drug combinations were a specific effect of TS-inhibition, we included the microtubule-stabilizing agent paclitaxel as a non-TS-directed control agent. Intracellular dUTP was measured using a polymerase-based nucleotide pool assay which detects both TTP and dUTP from cell extracts, with subsequent incubation with recombinant dUTPase indicating the contribution of dUTP as a percentage of the total dUTP/TTP detected. No dUTP was detected in untreated control cells or following treatment with the sub-cytotoxic concentrations of 0.1 µM FUdR and 0.5 µM 5-FU at 24 h. Following treatment with 0.5 µM oxaliplatin alone for 24 h, there was no detectable dUTP in cell extracts (Figure 7E). However, treatment with 0.5 µM oxaliplatin alone for 48 h resulted in the significant accumulation of dUTP. Specifically, within the total dUTP/TTP pool, 15.4% was dUTP, correlating with the acute reduction in dUTPase expression and activity previously noted. When cells were treated with oxaliplatin for 24 h followed by 0.1 µM FUdR or 0.5 µM 5-FU for a further 24 h, dUTP accumulation was significantly increased to 28% and 25% respectively of the total dUTP/TTP pool. To confirm that this effect was a direct result of downregulation of dUTPase followed by subsequent TS inhibition, we used the control combination of 0.5 µM oxaliplatin for 24 h followed by the non-TS-directed microtubule stabilizing drug paclitaxel (20 nM) for 24 h. This resulted in the detection of <3% of dUTP in the total dUTP/TTP, and would suggest that the increase in dUTP accumulation observed with 5-FU/FUdR and oxaliplatin combinations was a direct result of the TS metabolic blockade in the absence of an active dUTPase enzyme (Figure 7E). These data suggest that suppression of dUTPase by oxaliplatin may enhance the fluoropyrimidine-induced accumulation of dUTP and may contribute to the synergistic interaction between these agents.
DISCUSSION
Expression of dUTPase in malignant cells demonstrates significant variation, both in total expression and subcellular localization which may account for differences in clinical response to the TS-inhibiting class of chemotherapeutics (10). Therefore, elucidating the molecular basis for variable dUTPase expression is of interest from both a basic science and clinical perspective. Using promoter analysis in HCT116 p53+/+ colon cancer cells, the region containing the E2F and triple Sp1 site was found to be sufficient for driving DUT-N promoter activity (Figure 1A and B). Site-directed mutagenesis of the E2F and Sp1 sites clearly demonstrated that both transcription factors positively contribute to basal promoter activity (Figure 1C and D). We also report that the dUTPase promoter is highly responsive to Sp1 overexpression in HCT116 colon cells and silent in the absence of Sp1 and responsive to Sp1 expression in Sp1-deficient Drosophila SL2 cells (Figure 2A). Furthermore, knockdown of Sp1 resulted in the downregulation of dUTPase protein expression and significantly attenuated dUTPase promoter activity (Figure 2C). Finally we confirmed the presence of both Sp1 and E2F-1 transcription factors at the proximal dUTPase promoter in vivo using ChIP and qPCR (Figure 2D). Interestingly, dysregulation of both Sp1 (33,42,43) and E2F-1 (44–47) are widely observed following neoplastic transformation and are reported to contribute to a variety of tumor processes including progression, drug resistance, angiogenesis and metastasis. It is therefore likely that dysregulation of E2F-1 and Sp1 expression may contribute to the wide variation in dUTPase expression levels observed in tumor tissues (10,16,17).
Initial reports identified dUTPase mRNA as modulated by both mutant and wild-type p53 (22,24). However, the precise mechanism by which wild-type p53 exerts its repressive effects on dUTPase has not been described. To our knowledge, this report provides the first evidence demonstrating that dUTPase is directly modulated at the transcriptional level by an in vivo promoter association with p53. Introduction of p53WT resulted in significant repression of the dUTPase promoter (Figure 3C and E). Interestingly, the level of repression in the HCT116 p53–/– cells was significantly greater following introduction of p53WT when compared to the HCT116 p53+/+ cells, suggesting that expression of p53WT already exerts a regulatory influence on the dUTPase promoter in the HCT116 p53+/+ cells. This is further supported by the observation that when the p53MUT is introduced into the p53+/+ cells, the promoter activity is induced to a much greater extent than the p53–/– cells. This phenomenon may be exacerbated due to the artificial nature of the luciferase system, but would indicate the disruption of an unknown mediator involved in dUTPase promoter regulation by the dysregulated interactions of p53MUT. This observation has previously been noted in the TSP50 gene which is negatively regulated by p53, where the authors reported that introduction of mutant p53 significantly increased the TSP50 promoter activity in the three cell lines containing p53WT (48). These data would further suggest that tumoral p53 status and mutation may be important regulators of dUTPase expression and thus may influence chemotherapeutic response.
It is important to consider that ectopic p53 expression is unlikely to accurately simulate p53 activation in response to DNA damage which involves complex post-translation modifications. We therefore utilized chemotherapeutic agents used in the treatment of colorectal cancer which are known to activate p53 including the DNA-damaging agent oxaliplatin and the TS-inhibitors 5-FU and FUdR. Both 5-FU and FUdR resulted in a modest downregulation of dUTPase protein consistent with previous reports (21,49) (Figure 4A). However, for the first time, we identified oxaliplatin as a potent downregulator of dUTPase at the promoter, mRNA and protein levels (Figure 4B and 5A). Treatment of the HCT116 p53–/– cells with the same clinically relevant doses of oxaliplatin failed to modulate the dUTPase promoter or downregulate mRNA and protein expression, indicating a direct role for p53 or a p53-target in the repression (Figure 4B, 5A and B). Moreover, our findings indicate that this acute downregulation preceded complete cell cycle arrest with rapid repression of the dUTPase promoter and downregulation of the mRNA within 24 h. Although both 5-FU and FUdR result in similar levels of p53 induction, the cytotoxicity induced by these agents is of a significantly different nature than oxaliplatin. Specifically, 5-FU and FUdR are reported to initiate rapid S-phase arrest which is associated with thymidylate depletion as a result of TS-inhibition and is independent of p53 (50). The induction of p53 in response to 5-FU is reported to be a later event, possibly due to aberrant detection of uracil and fluoronucleotides in synthesized DNA. Oxaliplatin binds with high affinity to GC-rich DNA forming bulky platinum adducts which block DNA replication, activate p53 and stimulate the nucleotide excision repair pathway (51). Importantly, oxaliplatin treatment in HCT116 cells was previously reported to initiate a p53-dependent cell cycle arrest, indicating an immediate role for p53 in mediating the cellular response to oxaliplatin in the HCT116 colon cancer model (52). A recent report directly compared the p53 response in colon cancer cells treated with 5-FU compared to the direct DNA-damaging agent doxorubicin. The authors noted that 5-FU treatment did not accumulate p53 or result in p53 phosphorylation until 24 h post-treatment. However, doxorubicin treatment resulted in rapid p53 induction through protein stabilization and noted that p53 protein was both rapidly phosphorylated and acetylated within 4 h of treatment (53). Furthermore, p53 signaling is a complex pathway involving multiple post-translational modifications which are specific to its subsequent functions which vary markedly from cell-cycle arrest and DNA repair to apoptosis (54,55). Considering that oxaliplatin is also a direct DNA-damaging agent, it is possible that the mechanism of resultant p53 activation and associated post-translational modifications are markedly different than those observed following treatment with 5-FU and FUdR and this may account for the difference in the ability of these agents to repress dUTPase gene expression. We also note that ectopic dUTPase protein expression from an exogenous CMV-driven promoter was unaffected by oxaliplatin, discounting protein stability or atypical turnover and supporting a direct transcriptional effect (Figure 5D). Simultaneous analysis of TS, another classical S-phase gene revealed only a moderate downregulation following oxaliplatin treatment independent of p53 (Figure 5C), supporting the idea that the presence of a p53 responsive element is a key difference between the regulation of TS and dUTPase.
Previous reports suggested that p53-mediated transactivation of p21 was sufficient and necessary to exert the negative gene regulation of p53 (40). By using a p21 HCT116 null isogenic cell line, we demonstrate that p21 is not required for the transcriptional repression of dUTPase in response to oxaliplatin, further suggesting a direct role for p53 (Figure 6). We subsequently confirmed the presence and enrichment of p53 at the dUTPase proximal promoter following treatment with oxaliplatin. Importantly, this was accompanied by reductions in Sp1 and Ac-H3 (Figure 7A). Although the exact mechanism by which p53 represses the dUTPase promoter is not clear, these observations suggest that p53 interacts with the dUTPase promoter transcriptional machinery in the absence of a p53-specific cis element to suppress gene expression. In support of our observations, additional reports demonstrated p53-mediated transcriptional repression of the protective antioxidant enzyme manganese superoxide dismutase and the anti-apoptotic molecule MCL-1 in the absence of a p53 consensus site and demonstrated this to be mediated through disruption of normal transcription factor binding (56,57). Furthermore, the DNA repair enzyme AP-endonuclease is repressed by p53, in the absence of a cis element through interfering with Sp1 binding (58). This model is supported by our findings which demonstrated increased p53 and reduced Sp1 at the dUTPase promoter following oxaliplatin treatment. Interestingly, treatment with mithramycin A, which directly reduced Sp1 enrichment, did not show enrichment for p53, suggesting that displacing Sp1 or additional Sp-family members at the dUTPase promoter or inhibiting additional Sp1-driven gene expression can abrogate the promoter enrichment of p53. In support of this, our observation that the triple Sp1 site-mutant construct, which importantly still retained 50% transcriptional activity of the full-length construct, was not repressed by oxaliplatin and would further indicate that p53 is exerting its repressive effects through the region of the promoter known to bind Sp1 (Figure 7B). Previous studies have demonstrated that p53 can directly associate with transcription factors in vivo including Sp1 (57,59) and with histone deacetylase 1 (60) and repress transcription from specific promoters. Importantly, it has also been demonstrated that mutant p53 lacks the capacity to interact with HDAC1 and repress transcription (60), representing another possible mechanism for the p53-mutant induction of dUTPase previously reported (24) and our observations in this report showing p53-mutant induction of the dUTPase promoter. Furthermore, the inability of mutant forms of p53 to repress gene transcription may contribute to associated chemoresistance by abrogating apoptosis induced through repression of protective genes.
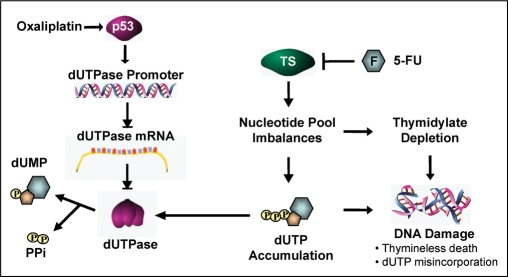
As a single agent, oxaliplatin has limited clinical activity in colorectal cancer and is primarily used in combination with 5-FU or capecitabine resulting in synergistic anti-tumor activity (3,37,61). Attempts to define the molecular basis for this synergistic interaction have demonstrated that oxaliplatin induces modest suppression of both TS mRNA and activity of the 5-FU catabolic enzyme dihydropyrimidine dehydrogenase (DPD) (62,63). In the present study, we confirmed that oxaliplatin induced a modest downregulation of TS mRNA independent of p53 and p21. In addition, we provide the first evidence of a p53-dependent acute downregulation of dUTPase in the HCT116 p53+/+ colon cell line following treatment with a clinically relevant dose of oxaliplatin. When oxaliplatin was combined with a TS-inhibitor, there was a significant increase in the accumulation of dUTP consistent with downregulation of dUTPase and increased effects of TS-inhibition (Figure 7E). Targeted use of oxaliplatin in tumors with elevated levels of dUTPase and wild-type p53 may represent a novel strategy for improving the therapeutic efficacy of TS-directed agents and warrants further investigation (Figure 8).
Figure 8.
Working model of p53-mediated repression of dUTPase transcription and subsequent enhancement of 5-FU-mediated downstream effects. Schematic diagram illustrating the downstream effects of p53-mediated transcriptional repression of dUTPase following treatment with a clinically relevant dose of oxaliplatin. The model proposes that p53 induction following DNA damage may inhibit dUTPase transcription via interaction with or inhibition of transcription machinery. Subsequent treatment with 5-FU results in TS-inhibition and, in the absence of an active dUTPase enzyme, promotes the accumulation of dUTP within cells, promoting increased DNA damage through uracil misincorporation in the presence of a TS-inhibiting agent.
Additional genes with critical roles in cytoprotection and survival including anti-apoptotic, oxidative stress elimination and DNA repair genes have previously been demonstrated to be repressed by p53, promoting the hypothesis that the pro-apoptotic function of p53 is mediated in part through transcriptional repression of cytoprotective genes. It is therefore plausible that the p53-mediated repression of dUTPase may enhance DNA damage and represent one component in the commitment to irreversible cell death and contribute to the tumor suppressing functions governed by p53.
In conclusion, p53 plays a critical role in the transformation and progression of many human malignancies, thus, the effect of p53 on uracil–DNA metabolism may increase our understanding of the complex regulatory pathways governed by p53 and may assist in providing more targeted chemotherapy. Our results have identified and characterized a novel mechanism of p53-dependent transcriptional regulation of an evolutionary conserved and essential enzyme involved in regulating intracellular uracil pools and maintaining genomic stability.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
FUNDING
The National Institute of Health R21 (5R21CA104796-3, 5P30CA14089-33). Funding for open access charge: National Institute of Health R21 (5R21CA104796-3).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr Guntram Suske from the Institut Fur Molekularbiologie und Tumorforschung (Marburg, Germany) for the kind gift of the Sp1 expression vectors and Dr Bert Vogelstein from the John Hopkins University (Baltimore, MD) for the kind gift of the HCT116 isogenic cell lines. We would also like to thank Dr Michael R. Stallcup and Peter J. Baumeister for critically reviewing this manuscript.
REFERENCES
- 1.el-Hajj HH, Zhang H, Weiss B. Lethality of a dut (deoxyuridine triphosphatase) mutation in Escherichia coli. J. Bacteriol. 1988;170:1069–1075. doi: 10.1128/jb.170.3.1069-1075.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gadsden MH, McIntosh EM, Game JC, Wilson PJ, Haynes RH. dUTP pyrophosphatase is an essential enzyme in Saccharomyces cerevisiae. EMBO J. 1993;12:4425–4431. doi: 10.1002/j.1460-2075.1993.tb06127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat. Rev Cancer. 2003;3:330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 4.Curtin NJ, Harris AL, Aherne GW. Mechanism of cell death following thymidylate synthase inhibition: 2'-deoxyuridine-5'-triphosphate accumulation, DNA damage, and growth inhibition following exposure to CB3717 and dipyridamole. Cancer Res. 1991;51:2346–2352. [PubMed] [Google Scholar]
- 5.Webley SD, Welsh SJ, Jackman AL, Aherne GW. The ability to accumulate deoxyuridine triphosphate and cellular response to thymidylate synthase (TS) inhibition. Br. J. Cancer. 2001;85:446–452. doi: 10.1054/bjoc.2001.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tinkelenberg BA, Hansbury MJ, Ladner RD. dUTPase and uracil-DNA glycosylase are central modulators of antifolate toxicity in Saccharomyces cerevisiae. Cancer Res. 2002;62:4909–4915. [PubMed] [Google Scholar]
- 7.Studebaker AW, Lafuse WP, Kloesel R, Williams MV. Modulation of human dUTPase using small interfering RNA. Biochem. Biophys. Res. Commun. 2005;327:306–310. doi: 10.1016/j.bbrc.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 8.Canman CE, Radany EH, Parsels LA, Davis MA, Lawrence TS, Maybaum J. Induction of resistance to fluorodeoxyuridine cytotoxicity and DNA damage in human tumor cells by expression of Escherichia coli deoxyuridinetriphosphatase. Cancer Res. 1994;54:2296–2298. [PubMed] [Google Scholar]
- 9.Koehler SE, Ladner RD. Small interfering RNA-mediated suppression of dUTPase sensitizes cancer cell lines to thymidylate synthase inhibition. Mol. Pharmacol. 2004;66:620–626. doi: 10.1124/mol.66.3.. [DOI] [PubMed] [Google Scholar]
- 10.Ladner RD, Lynch FJ, Groshen S, Xiong YP, Sherrod A, Caradonna SJ, Stoehlmacher J, Lenz HJ. dUTP nucleotidohydrolase isoform expression in normal and neoplastic tissues: association with survival and response to 5-fluorouracil in colorectal cancer. Cancer Res. 2000;60:3493–3503. [PubMed] [Google Scholar]
- 11.Wilson PM, Fazzone W, LaBonte MJ, Deng J, Neamati N, Ladner RD. Novel opportunities for thymidylate metabolism as a therapeutic target. Mol. Cancer Ther. 2008;7:3029–3037. doi: 10.1158/1535-7163.MCT-08-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mol CD, Harris JM, McIntosh EM, Tainer JA. Human dUTP pyrophosphatase: uracil recognition by a beta hairpin and active sites formed by three separate subunits. Structure. 1996;4:1077–1092. doi: 10.1016/s0969-2126(96)00114-1. [DOI] [PubMed] [Google Scholar]
- 13.Toth J, Varga B, Kovacs M, Malnasi-Csizmadia A, Vertessy BG. Kinetic mechanism of human dUTPase, an essential nucleotide pyrophosphatase enzyme. J. Biol. Chem. 2007;282:33572–33582. doi: 10.1074/jbc.M706230200. [DOI] [PubMed] [Google Scholar]
- 14.Varga B, Barabas O, Kovari J, Toth J, Hunyadi-Gulyas E, Klement E, Medzihradszky KF, Tolgyesi F, Fidy J, Vertessy BG. Active site closure facilitates juxtaposition of reactant atoms for initiation of catalysis by human dUTPase. FEBS Lett. 2007;581:4783–4788. doi: 10.1016/j.febslet.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 15.Ladner RD, Caradonna SJ. The human dUTPase gene encodes both nuclear and mitochondrial isoforms. Differential expression of the isoforms and characterization of a cDNA encoding the mitochondrial species. J. Biol. Chem. 1997;272:19072–19080. doi: 10.1074/jbc.272.30.19072. [DOI] [PubMed] [Google Scholar]
- 16.Webley SD, Hardcastle A, Ladner RD, Jackman AL, Aherne GW. Deoxyuridine triphosphatase (dUTPase) expression and sensitivity to the thymidylate synthase (TS) inhibitor ZD9331. Br. J. Cancer. 2000;83:792–799. doi: 10.1054/bjoc.2000.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fleischmann J, Kremmer E, Muller S, Sommer P, Kirchner T, Niedobitek G, Grasser FA. Expression of deoxyuridine triphosphatase (dUTPase) in colorectal tumours. Int. J. Cancer. 1999;84:614–617. doi: 10.1002/(sici)1097-0215(19991222)84:6<614::aid-ijc13>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 18.Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kajimoto S, Horie M, Manabe H, Masuda Y, Shibayama-Imazu T, Nakajo S, Gong XF, Obama T, Itabe H, Nakaya K. A tyrosine kinase inhibitor, beta-hydroxyisovalerylshikonin, induced apoptosis in human lung cancer DMS114 cells through reduction of dUTP nucleotidohydrolase activity. Biochim. Biophys. Acta. 2007;1782:41–50. doi: 10.1016/j.bbadis.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 20.Lee SC, Chan J, Clement MV, Pervaiz S. Functional proteomics of resveratrol-induced colon cancer cell apoptosis: caspase-6-mediated cleavage of lamin A is a major signaling loop. Proteomics. 2006;6:2386–2394. doi: 10.1002/pmic.200500366. [DOI] [PubMed] [Google Scholar]
- 21.Hernandez-Vargas H, Ballestar E, Carmona-Saez P, von Kobbe C, Banon-Rodriguez I, Esteller M, Moreno-Bueno G, Palacios J. Transcriptional profiling of MCF7 breast cancer cells in response to 5-fluorouracil: relationship with cell cycle changes and apoptosis, and identification of novel targets of p53. Int. J. Cancer. 2006;119:1164–1175. doi: 10.1002/ijc.21938. [DOI] [PubMed] [Google Scholar]
- 22.Spurgers KB, Gold DL, Coombes KR, Bohnenstiehl NL, Mullins B, Meyn RE, Logothetis CJ, McDonnell TJ. Identification of cell cycle regulatory genes as principal targets of p53-mediated transcriptional repression. J. Biol. Chem. 2006;281:25134–25142. doi: 10.1074/jbc.M513901200. [DOI] [PubMed] [Google Scholar]
- 23.Voskuil DW, Kampman E, van Kraats AA, Balder HF, van Muijen GN, Goldbohm RA, van't Veer P. p53 over-expression and p53 mutations in colon carcinomas: relation to dietary risk factors. Int. J. Cancer. 1999;81:675–681. doi: 10.1002/(sici)1097-0215(19990531)81:5<675::aid-ijc1>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 24.Pugacheva EN, Ivanov AV, Kravchenko JE, Kopnin BP, Levine AJ, Chumakov PM. Novel gain of function activity of p53 mutants: activation of the dUTPase gene expression leading to resistance to 5-fluorouracil. Oncogene. 2002;21:4595–4600. doi: 10.1038/sj.onc.1205704. [DOI] [PubMed] [Google Scholar]
- 25.Tinkelenberg BA, Fazzone W, Lynch FJ, Ladner RD. Identification of sequence determinants of human nuclear dUTPase isoform localization. Exp. Cell Res. 2003;287:39–46. doi: 10.1016/s0014-4827(03)00048-x. [DOI] [PubMed] [Google Scholar]
- 26.Ladner RD, McNulty DE, Carr SA, Roberts GD, Caradonna SJ. Characterization of distinct nuclear and mitochondrial forms of human deoxyuridine triphosphate nucleotidohydrolase. J. Biol. Chem. 1996a;271:7745–7751. doi: 10.1074/jbc.271.13.7745. [DOI] [PubMed] [Google Scholar]
- 27.Sherman PA, Fyfe JA. Enzymatic assay for deoxyribonucleoside triphosphates using synthetic oligonucleotides as template primers. Anal. Biochem. 1989;180:222–226. doi: 10.1016/0003-2697(89)90420-x. [DOI] [PubMed] [Google Scholar]
- 28.Horowitz RW, Zhang H, Schwartz EL, Ladner RD, Wadler S. Measurement of deoxyuridine triphosphate and thymidine triphosphate in the extracts of thymidylate synthase-inhibited cells using a modified DNA polymerase assay. Biochem. Pharmacol. 1997;54:635–638. doi: 10.1016/s0006-2952(97)00259-1. [DOI] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Mandola MV, Stoehlmacher J, Muller-Weeks S, Cesarone G, Yu MC, Lenz HJ, Ladner RD. A novel single nucleotide polymorphism within the 5' tandem repeat polymorphism of the thymidylate synthase gene abolishes USF-1 binding and alters transcriptional activity. Cancer Res. 2003;63:2898–2904. [PubMed] [Google Scholar]
- 31.Jensen DE, Black AR, Swick AG, Azizkhan JC. Distinct roles for Sp1 and E2F sites in the growth/cell cycle regulation of the DHFR promoter. J. Cell. Biochem. 1997;67:24–31. doi: 10.1002/(sici)1097-4644(19971001)67:1<24::aid-jcb3>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 32.Lin SY, Black AR, Kostic D, Pajovic S, Hoover CN, Azizkhan JC. Cell cycle-regulated association of E2F1 and Sp1 is related to their functional interaction. Mol. Cell. Biol. 1996;16:1668–1675. doi: 10.1128/mcb.16.4.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Safe S, Abdelrahim M. Sp transcription factor family and its role in cancer. Eur. J. Cancer. 2005;41:2438–2448. doi: 10.1016/j.ejca.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 34.Koutsodontis G, Kardassis D. Inhibition of p53-mediated transcriptional responses by mithramycin A. Oncogene. 2004;23:9190–9200. doi: 10.1038/sj.onc.1208141. [DOI] [PubMed] [Google Scholar]
- 35.Lakin ND, Jackson SP. Regulation of p53 in response to DNA damage. Oncogene. 1999;18:7644–7655. doi: 10.1038/sj.onc.1203015. [DOI] [PubMed] [Google Scholar]
- 36.Longley DB, Allen WL, Johnston PG. Drug resistance, predictive markers and pharmacogenomics in colorectal cancer. Biochim. Biophys. Acta. 2006;1766:184–196. doi: 10.1016/j.bbcan.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 37.Rothenberg ML, Oza AM, Bigelow RH, Berlin JD, Marshall JL, Ramanathan RK, Hart LL, Gupta S, Garay CA, Burger BG, et al. Superiority of oxaliplatin and fluorouracil-leucovorin compared with either therapy alone in patients with progressive colorectal cancer after irinotecan and fluorouracil-leucovorin: interim results of a phase III trial. J. Clin. Oncol. 2003;21:2059–2069. doi: 10.1200/JCO.2003.11.126. [DOI] [PubMed] [Google Scholar]
- 38.Fischer JA, Muller-Weeks S, Caradonna S. Proteolytic degradation of the nuclear isoform of uracil-DNA glycosylase occurs during the S phase of the cell cycle. DNA Repair. 2004;3:505–513. doi: 10.1016/j.dnarep.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 39.Fischer JA, Muller-Weeks S, Caradonna SJ. Fluorodeoxyuridine Modulates Cellular Expression of the DNA Base Excision Repair Enzyme Uracil-DNA Glycosylase. Cancer Res. 2006;66:8829–8837. doi: 10.1158/0008-5472.CAN-06-0540. [DOI] [PubMed] [Google Scholar]
- 40.Lohr K, Moritz C, Contente A, Dobbelstein M. p21/CDKN1A mediates negative regulation of transcription by p53. J. Biol. Chem. 2003;278:32507–32516. doi: 10.1074/jbc.M212517200. [DOI] [PubMed] [Google Scholar]
- 41.Shats I, Milyavsky M, Tang X, Stambolsky P, Erez N, Brosh R, Kogan I, Braunstein I, Tzukerman M, Ginsberg D, et al. p53-dependent down-regulation of telomerase is mediated by p21waf1. J. Biol. Chem. 2004;279:50976–50985. doi: 10.1074/jbc.M402502200. [DOI] [PubMed] [Google Scholar]
- 42.Wang L, Wei D, Huang S, Peng Z, Le X, Wu TT, Yao J, Ajani J, Xie K. Transcription factor Sp1 expression is a significant predictor of survival in human gastric cancer. Clin. Cancer Res. 2003;9:6371–6380. [PubMed] [Google Scholar]
- 43.Yao JC, Wang L, Wei D, Gong W, Hassan M, Wu TT, Mansfield P, Ajani J, Xie K. Association between expression of transcription factor Sp1 and increased vascular endothelial growth factor expression, advanced stage, and poor survival in patients with resected gastric cancer. Clin. Cancer Res. 2004;10:4109–4117. doi: 10.1158/1078-0432.CCR-03-0628. [DOI] [PubMed] [Google Scholar]
- 44.Banerjee D, Gorlick R, Liefshitz A, Danenberg K, Danenberg PC, Danenberg PV, Klimstra D, Jhanwar S, Cordon-Cardo C, Fong Y, et al. Levels of E2F-1 expression are higher in lung metastasis of colon cancer as compared with hepatic metastasis and correlate with levels of thymidylate synthase. Cancer Res. 2000;60:2365–2367. [PubMed] [Google Scholar]
- 45.Obama K, Kanai M, Kawai Y, Fukushima M, Takabayashi A. Role of retinoblastoma protein and E2F-1 transcription factor in the acquisition of 5-fluorouracil resistance by colon cancer cells. Int. J. Oncol. 2002;21:309–314. [PubMed] [Google Scholar]
- 46.Banerjee D, Schnieders B, Fu JZ, Adhikari D, Zhao SC, Bertino JR. Role of E2F-1 in chemosensitivity. Cancer Res. 1998;58:4292–4296. [PubMed] [Google Scholar]
- 47.Polager S, Kalma Y, Berkovich E, Ginsberg D. E2Fs up-regulate expression of genes involved in DNA replication, DNA repair and mitosis. Oncogene. 2002;21:437–446. doi: 10.1038/sj.onc.1205102. [DOI] [PubMed] [Google Scholar]
- 48.Xu H, Shan J, Jurukovski V, Yuan L, Li J, Tian K. TSP50 encodes a testis-specific protease and is negatively regulated by p53. Cancer Res. 2007;67:1239–1245. doi: 10.1158/0008-5472.CAN-06-3688. [DOI] [PubMed] [Google Scholar]
- 49.Lankat-Buttgereit B, Lenschen B, Schmidt H, Goke R. The action of Pdcd4 may be cell type specific: evidence that reduction of dUTPase levels might contribute to its tumor suppressor activity in Bon-1 cells. Apoptosis. 2008;13:157–164. doi: 10.1007/s10495-007-0153-x. [DOI] [PubMed] [Google Scholar]
- 50.Longley DB, Ferguson PR, Boyer J, Latif T, Lynch M, Maxwell P, Harkin DP, Johnston PG. Characterization of a thymidylate synthase (TS)-inducible cell line: a model system for studying sensitivity to TS- and non-TS-targeted chemotherapies. Clin. Cancer Res. 2001;7:3533–3539. [PubMed] [Google Scholar]
- 51.Raymond E, Faivre S, Woynarowski JM, Chaney SG. Oxaliplatin: mechanism of action and antineoplastic activity. Semin. Oncol. 1998;25:4–12. [PubMed] [Google Scholar]
- 52.Boyer J, McLean EG, Aroori S, Wilson P, McCulla A, Carey PD, Longley DB, Johnston PG. Characterization of p53 wild-type and null isogenic colorectal cancer cell lines resistant to 5-fluorouracil, oxaliplatin, and irinotecan. Clin. Cancer Res. 2004;10:2158–2167. doi: 10.1158/1078-0432.ccr-03-0362. [DOI] [PubMed] [Google Scholar]
- 53.Ju J, Schmitz JC, Song B, Kudo K, Chu E. Regulation of p53 expression in response to 5-fluorouracil in human cancer RKO cells. Clin. Cancer Res. 2007;13:4245–4251. doi: 10.1158/1078-0432.CCR-06-2890. [DOI] [PubMed] [Google Scholar]
- 54.Kastan MB, Onyekwere O, Sidransky D, Vogelstein B, Craig RW. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- 55.Sakaguchi K, Herrera JE, Saito S, Miki T, Bustin M, Vassilev A, Anderson CW, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pietrzak M, Puzianowska-Kuznicka M. p53-dependent repression of the human MCL-1 gene encoding an anti-apoptotic member of the BCL-2 family: the role of Sp1 and of basic transcription factor binding sites in the MCL-1 promoter. Biol. Chem. 2008;389:383–393. doi: 10.1515/BC.2008.039. [DOI] [PubMed] [Google Scholar]
- 57.Dhar SK, Xu Y, Chen Y, St Clair DK. Specificity protein 1-dependent p53-mediated suppression of human manganese superoxide dismutase gene expression. J Biol. Chem. 2006;281:21698–21709. doi: 10.1074/jbc.M601083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zaky A, Busso C, Izumi T, Chattopadhyay R, Bassiouny A, Mitra S, Bhakat KK. Regulation of the human AP-endonuclease (APE1/Ref-1) expression by the tumor suppressor p53 in response to DNA damage. Nucleic Acids Res. 2008;5:1555–1566. doi: 10.1093/nar/gkm1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Torgeman A, Mor-Vaknin N, Zelin E, Ben-Aroya Z, Lochelt M, Flugel RM, Aboud M. Sp1-p53 heterocomplex mediates activation of HTLV-I long terminal repeat by 12-O-tetradecanoylphorbol-13-acetate that is antagonized by protein kinase C. Virology. 2001;281:10–20. doi: 10.1006/viro.2000.0779. [DOI] [PubMed] [Google Scholar]
- 60.Murphy M, Ahn J, Walker KK, Hoffman WH, Evans RM, Levine AJ, George DL. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev. 1999;13:2490–2501. doi: 10.1101/gad.13.19.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Twelves C. Can capecitabine replace 5-FU/leucovorin in combination with oxaliplatin for the treatment of advanced colorectal cancer? Oncology. 2002;16:23–26. [PubMed] [Google Scholar]
- 62.Yeh KH, Cheng AL, Wan JP, Lin CS, Liu CC. Down-regulation of thymidylate synthase expression and its steady-state mRNA by oxaliplatin in colon cancer cells. Anticancer Drugs. 2004;15:371–376. doi: 10.1097/00001813-200404000-00010. [DOI] [PubMed] [Google Scholar]
- 63.Boisdron-Celle M, Craipeau C, Brienza S, Delva R, Guerin-Meyer V, Cvitkovic E, Gamelin E. Influence of oxaliplatin on 5-fluorouracil plasma clearance and clinical consequences. Cancer Chemother. Pharmacol. 2002;49:235–243. doi: 10.1007/s00280-001-0406-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.