Abstract
Establishment of the stable provirus is an essential step in retroviral replication, orchestrated by integrase (IN), a virus-derived enzyme. Until now, available structural information was limited to the INs of human immunodeficiency virus type 1 (HIV-1), avian sarcoma virus (ASV) and their close orthologs from the Lentivirus and Alpharetrovirus genera. Here, we characterized the in vitro activity of the prototype foamy virus (PFV) IN from the Spumavirus genus and determined the three-dimensional structure of its catalytic core domain (CCD). Recombinant PFV IN displayed robust and almost exclusively concerted integration activity in vitro utilizing donor DNA substrates as short as 16 bp, underscoring its significance as a model for detailed structural studies. Comparison of the HIV-1, ASV and PFV CCD structures highlighted both conserved as well as unique structural features such as organization of the active site and the putative host factor binding face. Despite possessing very limited sequence identity to its HIV counterpart, PFV IN was sensitive to HIV IN strand transfer inhibitors, suggesting that this class of inhibitors target the most conserved features of retroviral IN-DNA complexes.
INTRODUCTION
Upon entry into a target cell, a retrovirus undergoes reverse transcription to convert its diploid positive-strand RNA genome into a linear double-stranded cDNA molecule. Integration of the cDNA into the host cell genome is orchestrated by IN (1–3). This virus-derived enzyme engages both termini of the cDNA molecule-recognizing attachment (att) sites located at the ends of long terminal repeats (LTRs) to carry out two reactions: (i) 3′-processing, a single-strand endonucleolytic reaction that exposes 3′-OH groups of the invariant CA dinucleotides, and (ii) strand transfer that inserts the 3′-termini of the viral cDNA into the chromosomal DNA. The CA dinucleotides found in the immediate vicinity of the 3′-ends of cDNA from all retroviruses and LTR-retrotransposons are essential for IN function (4). Most commonly, two or three nucleotides are removed from both 3′-termini of retroviral cDNA during 3′-processing. Because in Spumavirus the polypurine tract (PPT) directly abuts the U3 sequence, reverse transcription does not add additional nucleotides to the U3 (left) end of the cDNA, and, as a consequence, only the right (U5) end is processed (5,6).
Based on structural and mechanistic similarities, retroviral INs belong to a diverse superfamily of metal-dependent nucleotidyl strand transferases, which notably includes ribonuclease (RNase) H enzymes and prokaryotic transposases (7). The catalytic mechanism of the RNase H superfamily of enzymes has been elucidated by a series of detailed crystal structures (8,9). Generally, reactions catalyzed by nucleotidyl transferases proceed via SN2 nucleophilic substitution at the phosphorus atom of the scissile phosphodiester bond. Unlike tyrosine or serine recombinases, retroviral INs and transposases use a water molecule as nucleophile during endonucleolysis, and the 3′-OH group of the viral cDNA during strand transfer. The active sites of these enzymes contain three or four essential acidic residues that participate in coordination of a pair of Mg2+ or Mn2+ cations. The tightly coordinated metal cations act as Lewis acids, one serving to position the incoming nucleophile (water or a 3′-OH group) and the other to activate the leaving 3′-OH group, both stabilizing the hypothetical pentacovalent phosphorus intermediate (10). An elegant ‘ping-pong’ mechanism wherein the roles of the metal cations switch between catalytic cycles (3′-processing and strand transfer in case of retroviral INs) has been proposed by Yang and colleagues (8). In retroviral IN, a pair of Asp and a Glu residue comprise the catalytic D,DX35E motif (11). Reported efforts to crystallize HIV-1 or ASV INs in a Mg2+-bound form have so far resulted in structures with a single Mg2+ cation coordinated by the Asp residues (12–14), although a pair of metal cations is expected to be present during catalysis (8,15).
Retroviral INs share a common three-domain organization: the catalytic core domain (CCD) flanked by smaller N- and C-terminal domains (NTD and CTD). The NTD is a compact three-helical bundle stabilized by a Zn2+ cation, coordinated via His and Cys residues of the invariant HHCC motif (16,17). Comprised solely of β-strands, the SH3-like CTD is rich in Lys and Arg residues and has nonspecific DNA-binding activity (18–21). The architecture of the functional IN–DNA complex and the precise roles of the NTD and the CTD are yet to be elucidated. Although the basic mechanism of phosphodiester bond cleavage and joining by the RNase H superfamily of enzymes is relatively well understood, structural information detailing catalysis by retroviral INs would be invaluable for antiretroviral drug development.
Productive integration requires insertion of both cDNA ends into chromosomal DNA. In the process, referred to as concerted or full-site integration, a pair of strand transfer events take place across the major groove in target DNA. As a result, the integrated viral cDNA is initially flanked by short single-stranded gaps, which have to be repaired to produce short duplications of the target DNA sequence. The duplication size is genus-specific and equals 4 bp for Spumavirus (5). Although the enzymatic activities of retroviral INs can be readily observed in vitro, concerted integration typically requires laborious optimizations of reaction conditions, the predominant strand transfer product often being uncoupled single-end (half-site) integration (22–25). Interestingly, side-by-side comparison of three lentiviral INs revealed that the propensity of INs to carry out concerted integration in vitro is enzyme-specific (26), suggesting that synaptic complex assembly might generally require a protein co-factor(s).
Retroviral integration is not entirely random, and genus-specific biases on the level of local target DNA sequence and genomic scales have been reported (27). HIV-1 and other members of Lentivirus strongly favor integration within active transcription units, whereas Moloney murine leukemia virus (MoMLV), a Gammaretrovirus, preferentially integrates in the vicinity of transcription start sites and CpG islands (28–34). Less distinctive patterns were observed for other genera. The prototype foamy virus (PFV), a member of Spumavirus, slightly disfavors integration within transcription units, displaying only a weak correlation with local gene expression activity (35,36). Integration site selection of Lentivirus was shown to depend on LEDGF/p75 (37). This chromatin-associated host protein interacts directly with lentiviral INs (38–40) and is required for efficient HIV–1 replication (41–43). In addition to affecting the levels of HIV-1 integration, ablation of LEDGF/p75 expression in target cells grossly affected the distribution of the residual integration sites (43–45). Given that even the distant relatives of retroviruses, yeast LTR-retrotransposons, target their integration via direct interaction between their INs and chromatin-associated host factors (46,47), it seems unlikely that this mechanism would not be conserved throughout the Retroviridae family. Even though non-lentiviral targeting factors have yet to be identified, based on the Lentivirus-LEDGF/p75 paradigm, they can be predicted to be genus-specific and to engage the CCDs of their cognate INs (40). Although Lentivirus, which notably includes human immunodeficiency viruses (HIV–1 and -2), is the most clinically relevant genus, comparative analyses of divergent retroviral INs will be essential to uncover conserved as well as genus-specific aspects of retroviral DNA integration. Furthermore, nonlentiviral INs can provide convenient model systems for structural studies of retroviral integration. In this study we describe the in vitro enzymatic properties of a spumaviral IN and present a detailed three-dimensional structure of its CCD.
MATERIALS AND METHODS
DNA
Oligonucleotides used in this work were: PC179, 5′-GAGCCCCGGG AAAGGATATCCCAAACAATATAC; PC180, 5′-GCGCGGATCCTCATTCATTT TTTTCCAAATGATCCATTG; PC192, 5′-GGCGGGTACCAGAAAGCAGGTA GCTTGCAGTGG; PC193, 5′-GGCGGGTACCCGAAGAACTCCAGCATG AGATCC; PC370, 5′-GTAAAAGGATTTGTATATTAGC; PC371, 5′-CCCATTGA CGCAAATGGGCGGTAG; PC376, 5′-GAGCCCCGGGTCATCTGG TCCTATTCT AAGACCAGATAG; PC377, 5′-GCGTGGATCCTCAGGTGGATGGATGGTATA AAGAAGTAC; PC401, 5′-TACTTTGCAGGGCTTCCCAACC; PC402, 5′-CGAAATGACCGACCAAGCGACG; PC503, 5′-GAGCCCCGGGTGT AATACCAAAAAACCAAACCTGGATG, EV53, 5′-AGGATAATCAATATA CAAAATTCCATGACAAT; EV54, 5′-ATTGTCATGGAATTTTGTATATTGA TTATCCT; EV55, 5′-AGGATAATCAATATACAAAATTCCATGACA; EV57, 5′-TAATCAATATACAAAATTCCATGACA; EV58, 5′-ATTGTCATGGAATTT TGTATATTG ATTA; EV59, 5′-CAATATACAAAATTCCATGACA; EV60, 5′-ATTGTCATG GAATTTTGTATATTG; EV61, 5′-TATACAAAATTCCATGACA; EV62, 5′-ATTGTCATGGAATTTTGTATA; EV63, 5′-ATACAAAATTCCA TGACA; EV64, 5′-ATTGTCATGGAATTTTGTAT; EV65, 5′-TACAAAATTC CATGACA; EV66, 5′-ATTGTCATGGAATTTTGTA; EV67, 5′-ACAAAA TTCCATGACA; EV68, 5′-ATTGTCATGGAATTTTGT; EV79, 5′-AAAATTCCAT GACA; EV80, 5′-ATTGTCATGGAATTTT; EV81, 5′-AAATTCCATGACA; EV82, 5′-ATTGTCATGGAATTT; EV85, 5′-CAAAATTCCATGACA; EV86, 5′-ATTGT CATGGAATTTTG.
To make pCPH6P-HSRV2-INΔ, pSSH6P-HSRF2-INFL, and pCPH6P-HSRV2-INCCD, DNA fragments PCR-amplified using Pfu DNA polymerase (Stratagene), pcHSRV2 (48) template and primer pairs PC179/PC180, PC503/PC180 or PC376/PC377, respectively, were digested with XmaI and BamHI and ligated between XmaI and BamHI sites of pCPH6P-BIV-IN (26) in place of the BIV IN coding sequence.
Production of PFV IN for enzymatic assays
To prepare PFV INΔ (IN residues 22–392, corresponding to PFV POL 773–1143) or full-length PFV IN (IN 1-392, POL 752–1143), Escherichia coli PC2 cells [BL21(DE3), endA::TetR, T1R, pLysS] (26) transformed with pCPH6P-HSRV2-INΔ or pSSH6P-HSRV2-INFL, respectively, were grown in Lennox LB broth in shake flasks at 29°C to an A600 of 0.9–1.0 prior to addition of 0.25 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and 50 μM ZnCl2. Following a 4-h induction at 25°C, bacteria were harvested and stored at –70°C. For purification, thawed bacterial paste was sonicated in HSB (0.5 M NaCl, 50 mM Tris–HCl, pH 7.4) in the presence of 0.5 mM phenylmethlysulfonyl fluoride (PMSF). The lysate, precleared by centrifugation at 20 000 g for 45 min and supplemented with 20 mM imidazole (pH 7.5), was incubated with Ni–NTA agarose (Qiagen) at 4°C for 30 min. The resin was extensively washed in HSB containing 20 mM imidazole and bound proteins were eluted with 200 mM imidazole in HSB and supplemented with 10 mM 1,4-dithiothreitol (DTT). Following removal of the hexahistidine tag by overnight digestion with human rhinovirus 14 (HRV14) 3C protease (49) at 7°C, the protein was diluted with 4 vol of ice-cold 50 mM Tris–HCl, pH 7.4 and loaded onto a 5-ml HiTrap heparin column (GE Healthcare). The IN proteins eluted with a linear gradient of 0.2–1.0 M NaCl in 50 mM Tris–HCl, pH 7.4 were further purified by gel filtration chromatography on a HiLoad 16/60 Superdex-200 column (GE Healthcare) in 200 mM NaCl, 50 mM Tris–HCl, pH 7.4, concentrated to 5–12 mg/ml, supplemented with 10 mM DTT and 10% (v/v) glycerol, flash-frozen in liquid nitrogen and stored at −70°C. Protein concentration was determined using the Bradford assay (Bio-Rad Laboratories) with a BSA standard. Where indicated, molar concentrations refer to the monomer protein forms.
In vitro PFV IN assays and sequence analysis of integration products
Short (32–15 bp) donor DNA substrates were obtained by annealing pairs of HPLC-purified synthetic oligonucleotides (VH Bio, UK). Blunt-ended 32 bp U5 donor was prepared using EV53 and EV54; preprocessed donors: 32 bp, EV55 and EV54; 28 bp, EV57 and EV58; 24 bp, EV59 and EV60; 21 bp, EV61 and EV62; 20 bp, EV63 and EV64; 19 bp, EV65 and EV66; 18 bp, EV67 and EV68; 17 bp, EV85 and EV86; 16 bp, EV79 and EV80; 15 bp, EV81 and EV82. 120-bp and 420-bp, donor DNA substrates were obtained as PCR products using Pfu DNA polymerase, pcHSRV2 template with primer pairs PC370/EV54 and PC371/EV54, respectively. The resulting PCR fragments were purified by electrophoresis through 2% (w/v) agarose and isolated using QIAquick gel extraction kit (Qiagen).
A standard strand transfer reaction contained 1.5 μM PFV INΔ or full-length IN, 0.75 μM donor oligonucleotide DNA substrate, 4 nM (300 ng) supercoiled pGEM9-Zf(–) target DNA (referred to as pGEM), 125 mM NaCl, 5 mM MgSO4, 4 μM ZnCl2, 10 mM DTT, 25 mM Bis–Tris propane, pH 7.45 in a final volume of 40 μl. Conditions were modified as indicated in the ‘Results’ section. Reactions were initiated by addition of IN diluted in 150 mM NaCl, 2 mM DTT, 10 mM Tris–HCl, pH 7.4 and, following incubation at 37°C for 90 min, were stopped by addition of 0.5% (w/v) SDS and 25 mM EDTA. Reaction products, deproteinized by digestion with 20 μg proteinase K for 30 min at 37°C and precipitation with ethanol, were separated in 1.5% agarose gels and visualized by staining with ethidium bromide.
For sequence analysis, concerted integration products isolated from agarose gels were treated with phage 29 DNA polymerase (New England Biolabs) in the presence of 500 μM dNTPs, 5′-phosphorylated using phage T4 polynucleotide kinase (New England Biolabs) and ligated to a blunt-ended linear DNA fragment encoding Tn5 aminoglycoside-3′-O-phosphotransferase (kanamycin resistance) gene. The KanR cassette, flanked by KpnI sites, was obtained as a PCR product using Pfu DNA polymerase with primers PC192/PC193 and pCP15 template (26,50). Escherichia coli XLI-Blue cells, transformed with the ligation products, were selected with 35 µg/ml kanamycin. Plasmids isolated from individual colonies that released the expected ∼1.1 and 3-kb fragments upon digestion with KpnI were sequenced using primers PC401 and PC402, which anneal within the KanR cassette.
Virus preparation and infections
Human embryonic kidney cells 293T and canine osteosarcoma D17 cells were maintained in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 2 mM L-glutamine at 37°C in 5% CO2 humidified atmosphere. The PFV-derived retroviral vector ΔΦ-EGFP expressing enhanced green fluorescent protein (EGFP) was prepared by co-transfection of 293T cells with pΔΦ-SFFV-EGFP (Patton,G.S. and McClure,M., unpublished data), pCiPS, pCiSΔψ, and pCiES (51). Cell culture supernatants were passed through 0.45-µm nitrocellulose filters (Millipore), and the virus concentrated by ultracentifugation was stored in liquid nitrogen. D17 cells seeded in 96-well plates (50 000 cells per well) were infected with ΔΦ-EGFP diluted in DMEM, 10% FCS in the presence of strand transfer inhibitors and/or 0.65% DMSO. To increase transduction efficiency, the virus-cell mixtures were subjected to centrifugation for 3 h at 1000 g at room temperature. Cytotoxicity was observed when GS9137 and MK0518 when used at concentrations exceeding 10 and 100 μM, respectively. Two days postinfection cells were transferred into 24-well plates and were harvested for analysis 5 days postinfection. The percentage of EGFP+ cells and their mean fluorescence intensity (MFI) were determined by fluorescence activated cell sorting (FACS) on a CyAn ADP instrument (DAKO Cytomation). Infection in the absence of strand transfer inhibitors resulted in 49 ± 5% of EGFP+ D17 cells; the EGFP+ population was reduced to 27 ± 4% and 16 ± 4% in the presence of the highest concentration of GS1930 and MK0518, respectively, with a concomitant reduction of the MFI. The percentage of EGFP+ cells times MFI of the EGFP+ cell population was used to generate relative infectivity data. Mean values and standard deviations were determined based on results of triplicate infections.
Crystallization and structure determination of the PFV IN CCD
A selenomethionine (SeMet) derivative of the IN CCD (corresponding to residues 860–1058 of PFV POL) was produced in E. coli B834 cells (Novagen) transformed with pCPH6P-HSRV2-INCCD. Bacteria were grown in Athena minimal medium (AthenaES, Baltimore, MD) supplemented with 120 μg/ml ampicillin, 34 μg/ml chloramphenicol, and 40 μg/ml SeMet (Molecular Dimensions) to an A600 of 0.5, and protein expression was induced with 0.25 mM IPTG for 5 h at 25°C. Cells were sonicated in 0.5 M NaCl, 1 mM DTT, 0.5 mM PMSF, 50 mM HEPES-NaOH, pH 8.0. Precleared lysate supplemented with 50 mM imidazole was injected into a 5-ml His-Trap FF column (GE Healthcare), and bound protein was eluted with a linear 50–500 mM gradient of imidazole in 0.5 M NaCl, 1 mM DTT, 50 mM HEPES-NaOH, pH 8.0. The hexahistidine tag was removed by digestion with HRV14 3C protease and the protein was further purified by gel filtration through a HiLoad 16/60 Superdex-200 column in 0.2 M NaCl, 1 mM DTT, 20 mM Tris–HCl, pH 7.5. Crystals of SeMet-derivatized PFV IN CCD were obtained in hanging drops at 18°C by mixing 2 μl protein (1.8 mg/ml) and 2 μl reservoir solution containing 1.0 M ammonium formate, 0.2 M MgCl2, 5 mM DTT and 0.1 M MES-NaOH, pH 6.5. The crystals were cryoprotected in reservoir solution supplemented with 20% (v/v) glycerol for several seconds and were then frozen in liquid nitrogen by rapid immersion.
Diffraction data from the SeMet-derivatized PFV IN CCD crystals were collected on beamline BM14 at the European Synchrotron Radiation Facility (Grenoble, France) and processed using MOSFLM (52) and SCALA (53). Analysis of integrated data using POINTLESS (54) indicated that the crystals belong to space group P6122 or P6522. Solvent content analysis suggested a single protein chain per asymmetric unit (solvent content of 41%). Initial attempts to determine the structure by molecular replacement (MR) in PHASER (55) using search models based on HIV-1 or ASV IN CCD structures (7,13,56) and a native 2.5-Å data set, resulted in a plausible solution in space group P6122. Although significant model bias did not allow refinement of the structure, the MR solution reproduced the expected dimeric CCD arrangement via a 2-fold crystallographic symmetry axis with good overall crystal packing. The MR phases were utilized in the calculation of an anomalous Fourier map using the SeMet data set collected at the Se absorption peak (λ = 0.97837 Å) and a 8.5-σ peak was observed at a position that was only ∼4 Å from the Cβ atom of the methionine residue of the MR solution. The position of the Se atom (fractional coordinates 0.77428, 0.63843, 0.40618) was confirmed by SHELXD (57). The first crude experimental phases were calculated and subsequently refined using the heavy atom position by single-wavelength anomalous dispersion (SAD) method as implemented in SHARP (58). This was followed by 120 cycles of solvent flattening from 5.0 to 2.2 Å in SOLOMON (59). The map was drastically improved using a solvent mask based on the MR solution for the last 80 cycles of solvent flattening and phase extension. The structure factors and phases were used as input to ARP/wARP (60) for automated model building. The final model containing 139 protein residues, 48 water molecules and one Mg atom was built using iterative cycles of real space refinement in COOT (61), simulated annealing/restrained refinement in PHENIX (62) and restrained refinement in REFMAC (63) against a high-energy remote dataset collected to 2.20 Å at λ = 0.95373 Å on the same crystal.
Coordinates
The coordinates of the PFV IN CCD have been deposited in the Protein Data Bank (accession code 3DLR).
RESULTS
Recombinant PFV IN is competent for concerted integration in vitro
In the context of viral replication, PFV IN is released from the 1143-residue POL precursor by site-specific proteolysis between Asn-751 and Cys-752 (cleavage site: 748YVVN↓CNTK755) (64). Based on earlier work of Flügel and Pahl (65,66), our expression construct for production of enzymatically-competent IN was comprised of PFV POL residues 773–1143, including 40 residues preceding the conserved HHCC motif. This protein, referred to as PFV INΔ, was used in most experiments described in this work. Both full-length PFV IN and INΔ were remarkably soluble under relatively low ionic strength conditions. In stark contrast to most retroviral INs, these proteins could be concentrated to over 10 mg/ml in solutions containing just 200 mM NaCl in the absence of detergents. Truncating PFV IN to the start of the predicted α-helical Zn-binding domain, by removing 30 amino-terminal residues, severely impaired protein solubility (data not shown). Concordantly, a similar deletion has been previously shown to affect enzymatic activity of PFV IN (66). Based on elution from calibrated gel filtration columns, INΔ as well as full-length PFV IN behaved as monodisperse monomeric species (data not shown).
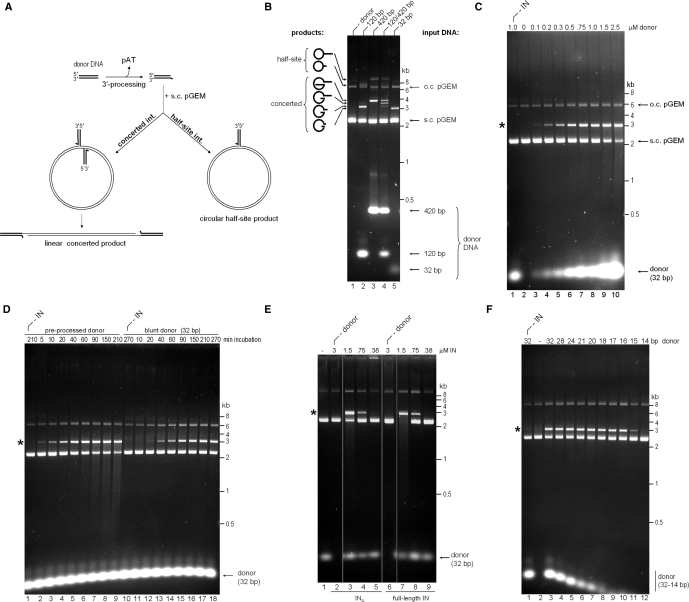
Using supercoiled target DNA in combination with short mimics of viral cDNA ends (referred to as donor DNA substrates) permits discrimination between half-site and concerted integration events (24,25). Concerted integration of pairs of donor DNA molecules results in a gapped linear product, while half-site integration produces tailed open circular DNA species (Figure 1A); both types of products can be conveniently separated in agarose gels. Earlier studies suggested that donor DNA molecules of several hundred base pairs are required to observe concerted HIV-1 integration (24,25). Therefore, we initially tested relatively long (120 or 420 bp) donor DNA substrates carrying the natural blunt foamy virus U5 terminal sequence. Incubation of the 120-bp or 420-bp donor DNAs with 1.5 μM PFV INΔ in the presence of supercoiled plasmid DNA (pGEM) resulted in accumulation of distinct product bands migrating at ∼3200 bp and 3800 bp, respectively (Figure 1B, lanes 2 and 3). These migration positions were consistent with concerted integration of pairs of 120- and 420-bp donor DNA molecules into the 2912-bp target plasmid. Also apparent were less prominent products migrating above the open circular form of target DNA, which correspond to half site reaction products. Importantly, no products were formed when donor DNA was omitted from the reaction (lane 1), demonstrating that the recombinant protein preparation was free of contaminating endonuclease activity. When both types of donor DNA substrates were present in the same reaction, three product bands migrating at ∼3200, 3500 and 3800 bp were observed (Figure 1B, lane 4). This result confirmed that a pair of donor DNA molecules was utilized in formation of the main product. Surprisingly, efficiency of this reaction was not drastically reduced when a blunt 32-bp donor DNA was used (lane 5), suggesting that PFV INΔ is able to efficiently synapse shorter donor DNA molecules.
Figure 1.
Strand transfer activity of recombinant PFV IN. (A) Schematic of strand transfer reactions using circular DNA target. Concerted integration results in a gapped linear product, and the half-site process yields tailed relaxed circles. (B) Concerted integration activity of PFV INΔ in vitro. The enzyme (1.5 μM) was incubated with supercoiled pGEM target DNA in the absence (lane 1) or presence of 100 nM donor DNA (lanes 2–5). Blunt DNA fragments of varying lengths mimicking the U5 PFV cDNA terminus served as donors: 120-bp (lane 2), 420-bp (lane 3), an equimolar mixture of 120-bp and 420-bp donors (50 nM each, lane 4), or a 32-bp oligonucleotide (lane 5). Reaction products separated in 1.5% agarose gels were detected with ethidium bromide. Migration positions of concerted and half-site reaction products, open circular (o.c.) and supercoiled (s.c.) pGEM target DNA forms, donor DNA and a DNA size ladder are indicated. (C) PFV INΔ strand transfer activity in the presence of 0–2.5 μM blunt 32-bp donor DNA (lanes 2–10, as indicated). IN was omitted in lane 1. Here and on the remaining panels, the star indicates the migration position of the concerted integration product. (D) Time course of PFV INΔ strand transfer reactions in the presence of preprocessed (lanes 1–9) and blunt (lanes 10–18) 32-bp donor DNA. IN was omitted in lanes 1 and 10. (E) Comparison of strand transfer activities of full-length PFV IN and INΔ. Supercoiled pGEM target DNA was incubated with 0.38–3 μM INΔ (lanes 2–5) or full-length PFV IN (lanes 6–9) in the absence (lanes 2 and 6) or presence (lanes 3–5, 7–9) of preprocessed 32-bp donor substrate; IN was omitted in lane 1. (F) Effect of donor DNA length. Strand transfer reactions using PFV INΔ and preprocessed donors of 32–14 bp, as indicated atop the gel. The enzyme was omitted in lane 1 and donor DNA was omitted in lane 2.
In the course of optimizing reaction conditions, we observed that increasing the concentration of input donor DNA greatly augmented the yields of concerted PFV DNA integration (Figure 1C, lanes 3–10). Of note, accumulation of the product takes place at the expense of the supercoiled target, which becomes limiting in the reaction. Concerted integration into the linear product accordingly gives rise to background DNA smears (Figure 1C, lanes 9 and 10). Using linear target DNA in the reaction reproduced the expected smear, which was dependent on the presence of both enzyme and donor DNA (Supplementary Figure S1A). Subsequent experiments were performed at the donor:enzyme ratio of 1:2 (0.75 μM donor DNA and 1.5 μM IN).
Although pre-processed HIV-1 donor DNA is a better substrate for half-site strand transfer, it was noted that the efficiency of concerted integration was higher when blunt donor DNA was used (25). In contrast, the PFV enzyme was able to promote the concerted integration of preprocessed 32-bp donor DNA at least as efficiently as the blunt version (Figure 1D). Notable was ∼15–30-min delay in accumulation of products in the reaction with the blunt donor (compare lanes 3 and 13), consistent with the requirement for 3′-processing of the blunt DNA prior to strand transfer. Therefore, at least under the reaction conditions used, the concerted strand transfer activity of PFV INΔ is not functionally coupled to 3′-processing. Importantly, these observations could be readily extended to full-length PFV IN, which displayed similar levels of strand transfer activity when presented with pre-processed oligonucleotide donor DNA (Figure 1E and data not shown). Further optimization of the reaction conditions using PFV INΔ revealed relatively narrow ionic strength (110–125 mM NaCl; Supplementary Figure S1B) and broad pH (pH 6–7.5; Supplementary Figure S1C) optima; strand transfer was abolished when NaCl concentration exceeded 160 mM (Supplementary Figure S1B). The input concentrations of both enzyme and donor DNA could be reduced to 320 and 80 nM, respectively, by supplementing reactions with DMSO and/or the crowding agent polyethelene glycol (PEG) (Supplementary Figure S1D). To determine the minimal length of donor DNA required for concerted strand transfer in vitro, we tested a range of preprocessed oligonucleotide molecules. Strikingly, the substrate could be truncated to just 16 bp without appreciable reduction in PFV INΔ strand transfer activity (Figure 1F and data not shown). Further truncations resulted in a sharp reduction of enzymatic activity (Figure 1F, lanes 11 and 12).
Sequence analysis of the concerted integration products
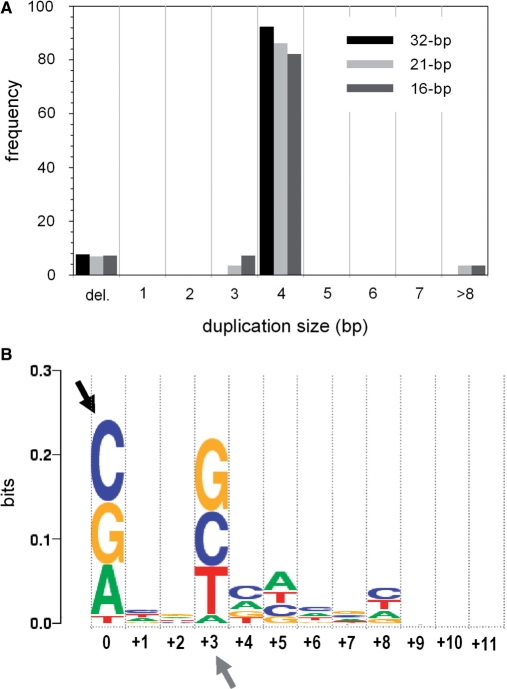
PFV integration results in 4-bp duplication of the target DNA sequence (5). The duplication size sets an important constraint on IN–donor complex assembly: the pair of strand transfer events must target two phosphodiester bonds at a particular distance (∼17 Å in B form DNA) and a specific relative orientation for a pair of in-line nucleophilic substitution reactions. To test the fidelity of PFV INΔ-mediated strand transfer under optimized in vitro conditions, we isolated and cloned concerted integration products from reactions with preprocessed 32-, 21- and 16-bp donor DNA substrates. The products were treated with a strand-displacing DNA polymerase to convert them to nongapped double-stranded linear molecules and ligated to a blunt-ended kanamycin resistance cassette. Upon transformation of competent bacterial cells, ∼70% of recovered kanamycin-resistant plasmids had the expected structure, based on restriction analysis (data not shown). Sequencing of individual clones revealed that over 80% of integration products contained pairs of donor DNA molecules integrated at various positions within pGEM with the expected 4-bp duplication of the target DNA sequence (Figure 2A). A minority of integration products showed 3-bp duplications, large duplications (>8 bp) or deletions. Alignment of all U5-target DNA junctions recovered in our experiments (n = 168) revealed weak biases toward target DNA sequence (Figure 2B). In particular, PFV IN appears refractory to insert donor DNA next to a deoxythymidine (position 0, Figure 2B). Since the majority of recovered integration events were concerted, deoxythymidine is also suppressed in the complementary strand at position +3 (Figure 2B). Overall, the pattern appears similar to the sequence bias observed in vivo (Supplementary Figure S2A). The overall higher GC content of the target plasmid used in our experiments (∼50%) compared to that of the human genome (∼41%) likely accounts for the quantitative differences between the two. Indeed, alignment of a subset of in vivo integration sites (n = 440) with the local GC content between 47 and 53% more closely reproduced the in vitro bias for positions up to +5 (compare Figures 2B and S2B).
Figure 2.
Sequence analysis of concerted integration products. (A) Histograms of duplication size distributions in cloned concerted integration products from reactions mediated by PFV INΔ utilizing pre-processed 32-bp (black bars), 24-bp (light gray), or 16-bp (dark gray) donor DNA substrates. (B) A sequence logo representing nucleotide base frequencies at PFV integration sites. Target DNA sequences (n = 168) as joined to the reactive strand of donor DNA (from all 84 analyzed clones) were used in the alignment. Black arrowhead indicates the nucleotide joined to donor DNA (position 0 of the alignment); gray arrowhead indicates the insertion position of the second donor molecule into the complementary strand (position +3). The height of each logo is proportional to the frequency of the corresponding nucleotide within the alignment, while the height of each stack of logos reflects the level of conservation at each position. The figure was created using WebLogo (83).
PFV IN is sensitive to HIV IN strand transfer inhibitors
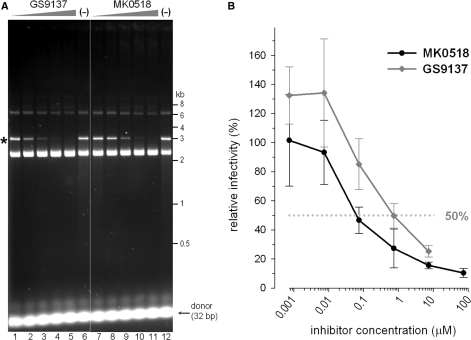
Sequence alignments highlight only ∼15% overall amino acid identity between PFV and HIV-1 INs. Despite being so divergent, the PFV enzyme was sensitive to HIV IN strand transfer inhibitors. Thus, the in vitro strand transfer activity of PFV INΔ was ablated in the presence of >200 nM of either GS9137 (67) or MK0518 (68) (Figure 3A). Concordantly, these compounds were able to suppress transduction of canine D17 cells by a PFV-based retroviral vector (Figure 3B). The drug concentrations required to inhibit PFV infection by 50% under these conditions amounted to ∼800 and 60 nM for GS9137 and MK0518, respectively.
Figure 3.
PFV IN is sensitive to HIV IN strand transfer inhibitors. (A) Inhibition of the in vitro strand transfer activity by GS9137 and MK0518. Preprocessed 32-bp donor DNA substrate was incubated with PFV INΔ and supercoiled target DNA in the presence (lanes 1–5 and 7–11) or absence (lanes 6 and 12) of inhibitors. Final drug concentrations were 0.008 µM (lanes 1 and 7), 0.04 µM (lanes 2 and 8), 0.2 µM (lanes 3 and 9), 1 µM (lanes 4 and 10) and 5 µM (lanes 5 and 11). DMSO was present in all reactions at a final concentration of 1%. (B) Infection of D17 cells by PFV-based retroviral vector in the presence of strand transfer inhibitors MK0518 (black line and circles) or GS9137 (gray line and diamonds). Infectivity in the absence of inhibitors was set to 100%. The dotted gray line represents 50% inhibition.
Crystal structure of the PFV IN CCD
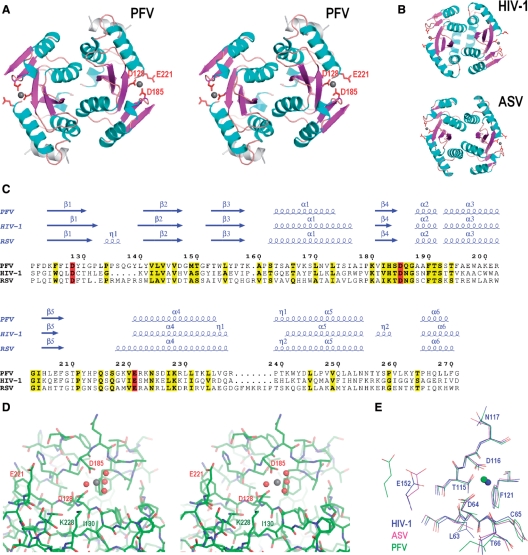
Encouraged by the unique properties of PFV IN and its responsiveness to HIV IN inhibitors, and as a first step toward structural characterization of PFV IN, we determined the crystal structure of its CCD. A fragment of PFV IN (residues 109–307; henceforth the residue numbering corresponds to full-length IN), spanning the predicted location of the CCD, readily produced diffraction-quality crystals, and the structure was solved by the SAD method using a selenomethionine derivative and refined at 2.2 Å resolution (Table 1). The crystals contained one protein chain per asymmetric unit that formed a dimer with a symmetry-related molecule; a total of ∼2000 Å2 solvent accessible area is buried in the dimer interface. Despite very limited sequence conservation between PFV, ASV and HIV-1 INs, the overall structure and topology of their dimeric CCDs are very similar (Figure 4A–C). The CCD spans residues 123–269 of PFV IN (from the beginning of β1 to the end of α6). Apart form a short α-helix comprised of residues 293–304 (shown in gray, Figure 4A), the region C-terminal to the CCD was poorly ordered and is not present in the final model. Of note, the C-terminal α helix (residues 293–304) is involved in the formation of crystal contacts with a symmetry related molecule and displays high anisotropic B factors, indicating mobility within the crystal. Hence, it is unclear if its position in the crystal reflects that in the native protein. In retroviral IN structures reported to date containing both the CCD and the CTD, the interdomain linker region was in α-helical or coiled configuration, or was not observed due to disorder (69–71), suggesting significant flexibility.
Table 1.
Summary of crystallographic statistics
| Data collection | ||
| Dataset | High remote | Peak |
| Wavelength (Å) | 0.95373 | 0.97837 |
| Resolution range (Å) | 42.26–2.20 (2.32–2.20) | 45.00–2.20 (2.32–2.20) |
| Rsyma | 0.105 (0.681) | 0.14 (0.932) |
| Rpima | 0.037 (0.233) | 0.031 (0.201) |
| Multiplicity a | 8.9 (9.3) | 21.3 (22.2) |
| I/σa | 17.4 (3.4) | 24.9 (4.9) |
| Completeness (%)a | 99.5 (99.3) | 99.7 (99.6) |
| Refinement | ||
| Resolution (Å) | 42.26–2.20 | |
| Reflections work set | 9540 | |
| Reflections test set | 1042 | |
| R factor (%) | 20.97 | |
| Rfree (%) | 25.94 | |
| No. protein atoms | 1241 | |
| No. water molecules | 48 | |
| R.m.s. bonds (Å) | 0.010 | |
| R.m.s. angles (°) | 1.12 | |
| Ramachandran plot (%) | ||
| Most favored | 93.1 | |
| Favored | 6.1 | |
| Allowed | 0.8 | |
| Disallowed | 0 |
aData in parentheses represent highest resolution shell.
Figure 4.
Crystal structure of the PFV IN CCD. (A) Stereo view of the PFV CCD structure. Secondary structure elements are color-coded: light blue for α-helices, magenta for β-strands, and orange for loops; the C-terminal α-helix comprised of residues 293–304 is painted gray. The catalytic site residues Asp-128, Asp-185 and Glu-221 are shown as red sticks. Gray spheres are Mg atoms. (B) Structures of the HIV-1 and ASV IN CCDs (PDB accession codes 1bl3 and 1vsd, respectively) (C) Structure-based amino acid sequence alignment of PFV, HIV-1, and RSV IN CCDs. Secondary structure elements (α, alpha helix; β, beta strand; η, 310 helix) are shown above the alignment. Residue numbering corresponds to the full-length PFV IN. Residues conserved across the alignment are shown in bold and highlighted in yellow and those of the catalytic D,DX35E motif in red. (D) Stereo view of the PFV IN active site. Protein structure is shown as sticks; the Mg atom and the associated water molecules are gray and red spheres, respectively. Residues discussed in the text are labeled. (E) Superposition of the PFV, HIV-1 and ASV IN active sites. Carbon and Mg atoms (spheres) are colored according to the viral species: PFV, green; HIV-1, blue; ASV, magenta. Residue numbering corresponds to HIV-1 IN. (A), (B), (D) and (E) were created with PyMOL (DeLano Scientific, http://www.pymol.org), and (C) with ESPript (84).
As predicted based on multiple sequence alignments (72), the active site of PFV IN contains three acidic residues Asp-128, Asp-185 and Glu-221, comprising the D,DX35E motif (Figure 4D). A single hexacoordinated magnesium ion with four associated water molecules (W36, W39, W43 and W44) was observed between the carboxylates of Asp-128 and Asp-185 (Figure 4D); the Mg-O distances (1.89–2.45 Å) and O-Mg-O angles (80–100°) observed in the structure confirm near-octahedral coordination, expected for the metal ion. As in ASV and HIV-1 CCDs (12,14), a network of hydrogen bonds stabilizes the Asp-128 and -185 side-chain rotamers and the positions of the associated water molecules. Mutation of PFV IN active site residues Asp-185 (5,72–74) and Glu-221 (72) have been shown to abolish viral infectivity, retrotransposition and/or the enzymatic activity of the recombinant protein. Our crystal structure also explains the phenotype of a more subtle mutation, I130T, which was shown to severely impair PFV IN function (5,73). Ile-130 is a largely buried residue that, via hydrophobic contacts, supports the side chain of Lys-228, which projects into the active site. Substitution of Ile-130 for a smaller Thr residue can be predicted to destabilize the conformation of Lys-228 and thus directly affect the environment within the active site.
Superposition of the PFV, HIV and ASV structures revealed the most conserved elements of the retroviral IN active site, including the location of the Mg2+ cation, the side chain orientations of the catalytic aspartates 128 and 185 (structurally equivalent to HIV-1 IN Asp-64 and Asp-116, respectively) and the side chains of highly conserved Phe-190 (Phe-121 in HIV-1 IN), as well as those of Ser-184 (Thr-115 in HIV-1 IN) and Tyr-129 (Cys-65 in HIV-1 IN) (Figure 4E). The side chain of Glu-221, located in the beginning of α4, was only partially ordered, and clear electron density could not be observed beyond its Cβ atom.
DISCUSSION
Preliminary comparative studies in our laboratory including a panel of divergent INs from several retroviral genera (26) identified PFV IN as a highly soluble protein. PFV IN has been previously shown to be capable of 3′-processing and half-site strand transfer in vitro (65,66). The current study was designed to extend these prior observations and evaluate PFV IN as a model for more detailed structural studies. Here, we demonstrated that this enzyme is highly proficient at concerted integration in vitro in the absence of protein or artificial co-factors. Furthermore, we showed that PFV IN is capable of utilizing preprocessed oligonucleotide donor DNA substrates as short as 16 bp to carry out almost exclusive concerted strand transfer in vitro, under physiological conditions. This is in a sharp contrast to HIV-1 IN that requires unprocessed donor DNA molecules of several hundred base pair and the presence of DMSO and/or PEG for optimal concerted integration in vitro (24,25). In fact, in the absence of DMSO, PEG, and protein co-factors, HIV-1 IN does not display significant strand activity (26,75). The lack of functional coupling of 3′-processing and concerted integration in the PFV system may have a functional significance. Unlike in Lentivirus, where both viral cDNA ends are processed prior to integration, only one cDNA end (U5) is processed in Spumavirus (5,6). Our findings are in excellent agreement with and significantly extend the results of Delelis et al. (76) who observed a specific interaction of PFV IN with short DNA molecules and reported 3′-processing activity with oligonucleotide substrates as short as 15 bp.
Mounting experimental evidence indicates that concerted integration is carried out by a tetrameric form of retroviral IN (77,78). While HIV-1 IN exists in a tetramer-dimer equilibrium, with the tetrameric form predominating at concentrations as low as 0.2 μM, its PFV counterpart is mostly monomeric at concentrations below 30 µM (76). In our hands, size exclusion chromatography failed to reveal multimeric forms of PFV IN (data not shown). Surprisingly, it is the latter enzyme that is highly proficient in concerted integration in vitro, while the former, even under most optimized conditions, invariably generates considerable amounts of half-site products. Interestingly, the efficiency of HIV-1 concerted integration negatively correlates with the enzyme concentration, and an optimum of 5–15 nM was highlighted by some investigators (24,79). These observations suggest that a pre-assembled tetramer of IN may not be a precursor for the active synaptic complex, and that the enzyme must exist in a lower multimeric form before interacting with its donor DNA substrate. As one consequence, less prone to multimerize, PFV IN affords studies of the concerted retroviral integration process at higher IN and donor DNA inputs, allowing visualization of the concerted products without resorting to radioactive labeling (Figure 1). Even more importantly, these properties of PFV IN will be extremely helpful in future structural studies of the retroviral synaptic complex. Although PFV IN is predominantly monomeric at concentrations used in our assays, the interaction with DNA likely enhances its propensity to accrue into a functional protomer. Indeed, multimers of PFV IN have been observed in the DNA bound form (76)
Despite possessing a very low level of sequence similarity to HIV INs, the PFV enzyme is sensitive to the HIV IN strand transfer inhibitors MK0518 and GS9137 (Figure 3A). Moreover, the compounds suppressed infectivity of a PFV-based vector at sub-µM concentrations (Figure 3B), albeit their effective concentrations were respectively ∼10- and 1000-fold higher than those required to inhibit HIV-1 to a similar extent (80). Comparison of the PFV IN CCD structure reported here with the available HIV-1 and ASV IN CCD structures (7,13,56) illuminated key features of the IN active site conserved among retroviral genera, including conformations of several side chains, and the location of the Mg atom coordinated by the Asp residues of the D,DX35E motif (Figure 4E). Our results strongly argue that these invariant features of the active site are involved in strand transfer inhibitor binding. Concordantly, mutations at HIV-1 IN positions 66 and 121 have been implicated in drug resistance (81). However, it is important to note that the complete arrangement of the retroviral IN active site will only emerge in the context of an IN–DNA complex. It is well established that the enzymes of the RNaseH superfamily, which includes retroviral INs, utilize a pair of divalent metal cations during catalysis (8,15). Similar to the reported structures of HIV-1 and ASV IN CCDs in their Mg2+-bound forms (12–14), our PFV IN CCD structure contains a single Mg2+ cation coordinated by the Asp residues of the D,DX35E motif. Coordination of the other Mg atom is expected to involve the third catalytic carboxylate (Glu-221 in PFV IN and Glu-152 in HIV-1 IN) as well as ligands donated by a bound DNA substrate (10,15).
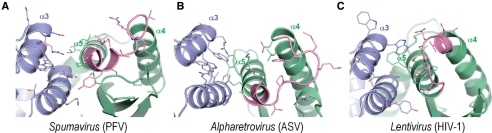
All retroviral genera display distinct preferences toward genomic DNA features that affect their integration frequencies (27). It is very tempting to speculate that as in the case of Lentivirus (43–45), it is the interaction between INs and their cognate chromosomal proteins that determine the observed site distributions. In HIV-1 IN, CCD helices α3, α4 and α5 comprise the principal recognition site for the Lentivirus-specific host factor LEDGF/p75 (40). The organization of the analogous site in the PFV IN CCD structure (Figure 5A) is quite divergent from those of HIV and ASV IN CCDs (Figure 5B and C). The differences include relative packing of the helices, the orientation of the loop connecting α4 and α5 as well as amino acid composition. Overall, the putative host factor binding face of PFV IN is less hydrophobic in content than those of HIV-1 or ASV INs. However, it remains to be determined whether Spumavirus utilizes an IN-binding host factor to help determine the frequency of integration, or, as recently proposed, relies on the ability of its Gag protein to interact with core histones (82).
Figure 5.
Comparison of the putative co-factor binding faces of Spumavirus (PFV) (A) and Alpharetrovirus (ASV) (B) IN CCDs to the LEDGF-binding face of the Lentivirus (HIV-1) IN CCD (C). Dimeric CCD structures are shown as cartoons; protein chains are painted blue and green; helices α3, α4 and α5 are indicated, α4/5 connectors (40) are shown in magenta. Side chains of residues involved or potentially involved in co-factor binding are shown as sticks. The figure was created using PyMOL.
HIV IN has been studied since the late 1980s, and despite its importance as a target for antiretroviral drug development and enduring efforts in leading laboratories, the structure of the full-length protein either alone or in complex with viral DNA remains elusive. With the development of clinically useful HIV IN inhibitors (67,68), the lack of structural data is even more daunting today. Structural information is urgently needed to fill out missing details of active site geometry and IN domain interactions during integration, as well as for the design of the next generation of drugs and for understanding the mechanisms of drug resistance. Exploration of INs from divergent retroviruses will illuminate both conserved and unique features of these proteins and will aid in elucidating the principal architecture of the retroviral preintegration complex. Our results as well as the complementary findings of others (76) strongly argue that PFV IN presents a highly relevant and elegant model for further structural studies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
P.R. is supported by the UK Medical Research Council grant G0400389 to Susan M. Lea. The project was funded by the UK Medical Research Council grant G0600009 to P.C. Funding for open access charge: UK Medical Research Council.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Reginald Clayton and Geert Meerseman (Tibotec Pharmaceuticals) for a generous gift of the IN inhibitors and helpful discussions; David W. Russell (University of Washington) for providing pΔΦ, pCiPS, pCiSΔψ and pCiES; Gillian S. Patton for constructing pΔΦ-SFFV-EGFP; David Bonsall for help with flow cytometry; Anna Stamp (University of Oxford) for assistance with the preparation of the selenomethione-derivatized protein and for advice on crystallization and data collection; Susan M. Lea (University of Oxford) for providing access to the SHARP server and the computing facilities in her lab; Gerard Bricogne and the Global Phasing Consortium for access to the beta version of autoSHARP suite of programs; Alan Engelman (Dana-Farber Cancer Institute) for critical reading of the manuscript; and the staff of ESRF beamlines ID14-2 and BM14 for assistance during data collection.
REFERENCES
- 1.Asante-Appiah E, Skalka AM. HIV-1 integrase: structural organization, conformational changes, and catalysis. Adv. Virus Res. 1999;52:351–369. doi: 10.1016/s0065-3527(08)60306-1. [DOI] [PubMed] [Google Scholar]
- 2.Craigie R. Retroviral DNA integration. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington DC: ASM Press; 2002. pp. 613–630. [Google Scholar]
- 3.Lewinski MK, Bushman FD. Retroviral DNA integration—mechanism and consequences. Adv. Genet. 2005;55:147–181. doi: 10.1016/S0065-2660(05)55005-3. [DOI] [PubMed] [Google Scholar]
- 4.Sherman PA, Fyfe JA. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc. Natl Acad. Sci. USA. 1990;87:5119–5123. doi: 10.1073/pnas.87.13.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enssle J, Moebes A, Heinkelein M, Panhuysen M, Mauer B, Schweizer M, Neumann-Haefelin D, Rethwilm A. An active foamy virus integrase is required for virus replication. J. Gen. Virol. 1999;80:1445–1452. doi: 10.1099/0022-1317-80-6-1445. [DOI] [PubMed] [Google Scholar]
- 6.Juretzek T, Holm T, Gartner K, Kanzler S, Lindemann D, Herchenroder O, Picard-Maureau M, Rammling M, Heinkelein M, Rethwilm A. Foamy virus integration. J. Virol. 2004;78:2472–2477. doi: 10.1128/JVI.78.5.2472-2477.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dyda F, Hickman AB, Jenkins TM, Engelman A, Craigie R, Davies DR. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 8.Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 9.Nowotny M, Yang W. Stepwise analyses of metal ions in RNase H catalysis from substrate destabilization to product release. EMBO J. 2006;25:1924–1933. doi: 10.1038/sj.emboj.7601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang W, Lee JY, Nowotny M. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol. Cell. 2006;22:5–13. doi: 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 11.Kulkosky J, Jones KS, Katz RA, Mack JP, Skalka AM. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol. Cell. Biol. 1992;12:2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maignan S, Guilloteau JP, Zhou-Liu Q, Clement-Mella C, Mikol V. Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases. J. Mol. Biol. 1998;282:359–368. doi: 10.1006/jmbi.1998.2002. [DOI] [PubMed] [Google Scholar]
- 13.Goldgur Y, Dyda F, Hickman AB, Jenkins TM, Craigie R, Davies DR. Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium. Proc. Natl Acad. Sci. USA. 1998;95:9150–9154. doi: 10.1073/pnas.95.16.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bujacz G, Jaskolski M, Alexandratos J, Wlodawer A, Merkel G, Katz RA, Skalka AM. The catalytic domain of avian sarcoma virus integrase: conformation of the active-site residues in the presence of divalent cations. Structure. 1996;4:89–96. doi: 10.1016/s0969-2126(96)00012-3. [DOI] [PubMed] [Google Scholar]
- 15.Lovell S, Goryshin IY, Reznikoff WR, Rayment I. Two-metal active site binding of a Tn5 transposase synaptic complex. Nat. Struct. Biol. 2002;9:278–281. doi: 10.1038/nsb778. [DOI] [PubMed] [Google Scholar]
- 16.Cai M, Zheng R, Caffrey M, Craigie R, Clore GM, Gronenborn AM. Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nat. Struct. Biol. 1997;4:567–577. doi: 10.1038/nsb0797-567. [DOI] [PubMed] [Google Scholar]
- 17.Eijkelenboom AP, van den Ent FM, Vos A, Doreleijers JF, Hard K, Tullius TD, Plasterk RH, Kaptein R, Boelens R. The solution structure of the amino-terminal HHCC domain of HIV-2 integrase: a three-helix bundle stabilized by zinc. Curr. Biol. 1997;7:739–746. doi: 10.1016/s0960-9822(06)00332-0. [DOI] [PubMed] [Google Scholar]
- 18.Eijkelenboom AP, Lutzke RA, Boelens R, Plasterk RH, Kaptein R, Hard K. The DNA-binding domain of HIV-1 integrase has an SH3-like fold. Nat. Struct. Biol. 1995;2:807–810. doi: 10.1038/nsb0995-807. [DOI] [PubMed] [Google Scholar]
- 19.Lodi PJ, Ernst JA, Kuszewski J, Hickman AB, Engelman A, Craigie R, Clore GM, Gronenborn AM. Solution structure of the DNA binding domain of HIV-1 integrase. Biochemistry. 1995;34:9826–9833. doi: 10.1021/bi00031a002. [DOI] [PubMed] [Google Scholar]
- 20.Vink C, Oude Groeneger AM, Plasterk RH. Identification of the catalytic and DNA-binding region of the human immunodeficiency virus type I integrase protein. Nucleic Acids Res. 1993;21:1419–1425. doi: 10.1093/nar/21.6.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engelman A, Hickman AB, Craigie R. The core and carboxyl-terminal domains of the integrase protein of human immunodeficiency virus type 1 each contribute to nonspecific DNA binding. J. Virol. 1994;68:5911–5917. doi: 10.1128/jvi.68.9.5911-5917.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang F, Roth MJ. Assembly and catalysis of concerted two-end integration events by Moloney murine leukemia virus integrase. J. Virol. 2001;75:9561–9570. doi: 10.1128/JVI.75.20.9561-9570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aiyar A, Hindmarsh P, Skalka AM, Leis J. Concerted integration of linear retroviral DNA by the avian sarcoma virus integrase in vitro: dependence on both long terminal repeat termini. J. Virol. 1996;70:3571–3580. doi: 10.1128/jvi.70.6.3571-3580.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sinha S, Grandgenett DP. Recombinant human immunodeficiency virus type 1 integrase exhibits a capacity for full-site integration in vitro that is comparable to that of purified preintegration complexes from virus-infected cells. J. Virol. 2005;79:8208–8216. doi: 10.1128/JVI.79.13.8208-8216.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li M, Craigie R. Processing of viral DNA ends channels the HIV-1 integration reaction to concerted integration. J. Biol. Chem. 2005;280:29334–29339. doi: 10.1074/jbc.M505367200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cherepanov P. LEDGF/p75 interacts with divergent lentiviral integrases and modulates their enzymatic activity in vitro. Nucleic Acids Res. 2007;35:113–124. doi: 10.1093/nar/gkl885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bushman F, Lewinski M, Ciuffi A, Barr S, Leipzig J, Hannenhalli S, Hoffmann C. Genome-wide analysis of retroviral DNA integration. Nat. Rev. Microbiol. 2005;3:848–858. doi: 10.1038/nrmicro1263. [DOI] [PubMed] [Google Scholar]
- 28.Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 29.Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 30.Kang Y, Moressi CJ, Scheetz TE, Xie L, Tran DT, Casavant TL, Ak P, Benham CJ, Davidson BL, McCray PB., Jr Integration Site Choice of a Feline Immunodeficiency Virus Vector. J. Virol. 2006;80:8820. doi: 10.1128/JVI.00719-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crise B, Li Y, Yuan C, Morcock DR, Whitby D, Munroe DJ, Arthur LO, Wu X. Simian immunodeficiency virus integration preference is similar to that of human immunodeficiency virus type 1. J. Virol. 2005;79:12199–12204. doi: 10.1128/JVI.79.19.12199-12204.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacNeil A, Sankale JL, Meloni ST, Sarr AD, Mboup S, Kanki P. Genomic sites of human immunodeficiency virus type 2 (HIV-2) integration: similarities to HIV-1 in vitro and possible differences in vivo. J. Virol. 2006;80:7316–7321. doi: 10.1128/JVI.00604-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hacker CV, Vink CA, Wardell TW, Lee S, Treasure P, Kingsman SM, Mitrophanous KA, Miskin JE. The integration profile of EIAV-based vectors. Mol. Ther. 2006;14:536–545. doi: 10.1016/j.ymthe.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 34.Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC, Ecker JR, Bushman FD. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trobridge GD, Miller DG, Jacobs MA, Allen JM, Kiem P, Kaul R, Russell DW. Foamy virus vector integration sites in normal human cells. Proc. Natl Acad. Sci. USA. 2006;103:1498–1503. doi: 10.1073/pnas.0510046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nowrouzi A, Dittrich M, Klanke C, Heinkelein M, Rammling M, Dandekar T, von Kalle C, Rethwilm A. Genome-wide mapping of foamy virus vector integrations into a human cell line. J. Gen. Virol. 2006;87:1339–1347. doi: 10.1099/vir.0.81554-0. [DOI] [PubMed] [Google Scholar]
- 37.Engelman A, Cherepanov P. The lentiviral integrase binding protein LEDGF/p75 and HIV-1 replication. PLoS Pathog. 2008;4:e1000046. doi: 10.1371/journal.ppat.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- 39.Emiliani S, Mousnier A, Busschots K, Maroun M, Van Maele B, Tempe D, Vandekerckhove L, Moisant F, Ben-Slama L, Witvrouw M, et al. Integrase mutants defective for interaction with LEDGF/p75 are impaired in chromosome tethering and HIV-1 replication. J. Biol. Chem. 2005;280:25517–25523. doi: 10.1074/jbc.M501378200. [DOI] [PubMed] [Google Scholar]
- 40.Cherepanov P, Ambrosio AL, Rahman S, Ellenberger T, Engelman A. Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc. Natl Acad. Sci. USA. 2005;102:17308–17313. doi: 10.1073/pnas.0506924102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vandekerckhove L, Christ F, Van Maele B, De Rijck J, Gijsbers R, Van den Haute C, Witvrouw M, Debyser Z. Transient and stable knockdown of the integrase cofactor LEDGF/p75 reveals its role in the replication cycle of human immunodeficiency virus. J. Virol. 2006;80:1886–1896. doi: 10.1128/JVI.80.4.1886-1896.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Llano M, Saenz DT, Meehan A, Wongthida P, Peretz M, Walker WH, Teo W, Poeschla EM. An essential role for LEDGF/p75 in HIV integration. Science. 2006;314:461–464. doi: 10.1126/science.1132319. [DOI] [PubMed] [Google Scholar]
- 43.Shun MC, Raghavendra NK, Vandegraaff N, Daigle JE, Hughes S, Kellam P, Cherepanov P, Engelman A. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007;21:1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman F. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 2005;11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- 45.Marshall HM, Ronen K, Berry C, Llano M, Sutherland H, Saenz D, Bickmore W, Poeschla E, Bushman FD. Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting. PLoS ONE. 2007;2:e1340. doi: 10.1371/journal.pone.0001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xie W, Gai X, Zhu Y, Zappulla DC, Sternglanz R, Voytas DF. Targeting of the yeast Ty5 retrotransposon to silent chromatin is mediated by interactions between integrase and Sir4p. Mol. Cell. Biol. 2001;21:6606–6614. doi: 10.1128/MCB.21.19.6606-6614.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leem YE, Ripmaster TL, Kelly FD, Ebina H, Heincelman ME, Zhang K, Grewal SI, Hoffman CS, Levin HL. Retrotransposon Tf1 is targeted to Pol II promoters by transcription activators. Mol. Cell. 2008;30:98–107. doi: 10.1016/j.molcel.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moebes A, Enssle J, Bieniasz PD, Heinkelein M, Lindemann D, Bock M, McClure MO, Rethwilm A. Human foamy virus reverse transcription that occurs late in the viral replication cycle. J. Virol. 1997;71:7305–7311. doi: 10.1128/jvi.71.10.7305-7311.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walker PA, Leong LE, Ng PW, Tan SH, Waller S, Murphy D, Porter AG. Efficient and rapid affinity purification of proteins using recombinant fusion proteases. Biotechnology. 1994;12:601–605. doi: 10.1038/nbt0694-601. [DOI] [PubMed] [Google Scholar]
- 50.Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 51.Trobridge G, Josephson N, Vassilopoulos G, Mac J, Russell DW. Improved foamy virus vectors with minimal viral sequences. Mol. Ther. 2002;6:321–328. doi: 10.1006/mthe.2002.0672. [DOI] [PubMed] [Google Scholar]
- 52.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. 1992 [Google Scholar]
- 53.Collaborative Computional Project. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 54.Evans P. Scaling and assessment of data quality. Acta Crystallogr. D. Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 55.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lubkowski J, Dauter Z, Yang F, Alexandratos J, Merkel G, Skalka AM, Wlodawer A. Atomic resolution structures of the core domain of avian sarcoma virus integrase and its D64N mutant. Biochemistry. 1999;38:13512–13522. doi: 10.1021/bi991362q. [DOI] [PubMed] [Google Scholar]
- 57.Sheldrick GM. A short history of SHELX. Acta Crystallogr. A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 58.Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. D. Biol. Crystallogr. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- 59.Abrahams JP, Leslie AG. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D. Biol. Crystallogr. 1996;52:30–42. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- 60.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 61.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 62.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D. Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 63.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 64.Pfrepper KI, Rackwitz HR, Schnolzer M, Heid H, Lochelt M, Flugel RM. Molecular characterization of proteolytic processing of the Pol proteins of human foamy virus reveals novel features of the viral protease. J. Virol. 1998;72:7648–7652. doi: 10.1128/jvi.72.9.7648-7652.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pahl A, Flugel RM. Endonucleolytic cleavages and DNA-joining activities of the integration protein of human foamy virus. J. Virol. 1993;67:5426–5434. doi: 10.1128/jvi.67.9.5426-5434.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pahl A, Flugel RM. Characterization of the human spuma retrovirus integrase by site-directed mutagenesis, by complementation analysis, and by swapping the zinc finger domain of HIV-1. J. Biol. Chem. 1995;270:2957–2966. doi: 10.1074/jbc.270.7.2957. [DOI] [PubMed] [Google Scholar]
- 67.Sato M, Motomura T, Aramaki H, Matsuda T, Yamashita M, Ito Y, Kawakami H, Matsuzaki Y, Watanabe W, Yamataka K, et al. Novel HIV-1 integrase inhibitors derived from quinolone antibiotics. J. Med. Chem. 2006;49:1506–1508. doi: 10.1021/jm0600139. [DOI] [PubMed] [Google Scholar]
- 68.Summa V, Petrocchi A, Bonelli F, Crescenzi B, Donghi M, Ferrara M, Fiore F, Gardelli C, Gonzalez Paz O, Hazuda DJ, et al. Discovery of raltegravir, a potent, selective orally bioavailable HIV-integrase inhibitor for the treatment of HIV-AIDS infection. J. Med. Chem. 2008;51:5843–5855. doi: 10.1021/jm800245z. [DOI] [PubMed] [Google Scholar]
- 69.Chen JC, Krucinski J, Miercke LJ, Finer-Moore JS, Tang AH, Leavitt AD, Stroud RM. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding. Proc. Natl Acad. Sci. USA. 2000;97:8233–8238. doi: 10.1073/pnas.150220297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen Z, Yan Y, Munshi S, Li Y, Zugay-Murphy J, Xu B, Witmer M, Felock P, Wolfe A, Sardana V, et al. X-ray structure of simian immunodeficiency virus integrase containing the core and C-terminal domain (residues 50-293)–an initial glance of the viral DNA binding platform. J. Mol. Biol. 2000;296:521–533. doi: 10.1006/jmbi.1999.3451. [DOI] [PubMed] [Google Scholar]
- 71.Yang ZN, Mueser TC, Bushman FD, Hyde CC. Crystal structure of an active two-domain derivative of Rous sarcoma virus integrase. J. Mol. Biol. 2000;296:535–548. doi: 10.1006/jmbi.1999.3463. [DOI] [PubMed] [Google Scholar]
- 72.Lee HS, Kang SY, Shin CG. Characterization of the functional domains of human foamy virus integrase using chimeric integrases. Mol. Cells. 2005;19:246–255. [PubMed] [Google Scholar]
- 73.Delelis O, Petit C, Leh H, Mbemba G, Mouscadet JF, Sonigo P. A novel function for spumaretrovirus integrase: an early requirement for integrase-mediated cleavage of 2 LTR circles. Retrovirology. 2005;2:31. doi: 10.1186/1742-4690-2-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Heinkelein M, Pietschmann T, Jarmy G, Dressler M, Imrich H, Thurow J, Lindemann D, Bock M, Moebes A, Roy J, et al. Efficient intracellular retrotransposition of an exogenous primate retrovirus genome. EMBO J. 2000;19:3436–3445. doi: 10.1093/emboj/19.13.3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cherepanov P, Surratt D, Toelen J, Pluymers W, Griffith J, De Clercq E, Debyser Z. Activity of recombinant HIV-1 integrase on mini-HIV DNA. Nucleic Acids Res. 1999;27:2202–2210. doi: 10.1093/nar/27.10.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Delelis O, Carayon K, Guiot E, Leh H, Tauc P, Brochon JC, Mouscadet JF, Deprez E. Insight into the integrase-DNA recognition mechanism. A specific DNA-binding mode revealed by an enzymatically labeled integrase. J. Biol. Chem. 2008;283:27838–27849. doi: 10.1074/jbc.M803257200. [DOI] [PubMed] [Google Scholar]
- 77.Li M, Mizuuchi M, Burke TR, Jr, Craigie R. Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J. 2006;25:1295–1304. doi: 10.1038/sj.emboj.7601005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Faure A, Calmels C, Desjobert C, Castroviejo M, Caumont-Sarcos A, Tarrago-Litvak L, Litvak S, Parissi V. HIV-1 integrase crosslinked oligomers are active in vitro. Nucleic Acids Res. 2005;33:977–986. doi: 10.1093/nar/gki241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pandey KK, Bera S, Zahm J, Vora A, Stillmock K, Hazuda D, Grandgenett DP. Inhibition of human immunodeficiency virus type 1 concerted integration by strand transfer inhibitors which recognize a transient structural intermediate. J. Virol. 2007;81:12189–12199. doi: 10.1128/JVI.02863-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hombrouck A, Voet A, Van Remoortel B, Desadeleer C, De Maeyer M, Debyser Z, Witvrouw M. Mutations in human immunodeficiency virus type 1 integrase confer resistance to the naphthyridine L-870,810 and cross-resistance to the clinical trial drug GS-9137. Antimicrob. Agents Chemother. 2008;52:2069–2078. doi: 10.1128/AAC.00911-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shafer RW, Schapiro JM. HIV-1 drug resistance mutations: an updated framework for the second decade of HAART. AIDS Rev. 2008;10:67–84. [PMC free article] [PubMed] [Google Scholar]
- 82.Tobaly-Tapiero J, Bittoun P, Lehmann-Che J, Delelis O, Giron ML, de The H, Saib A. Chromatin tethering of incoming foamy virus by the structural Gag protein. Traffic. 2008;9:1717–1727. doi: 10.1111/j.1600-0854.2008.00792.x. [DOI] [PubMed] [Google Scholar]
- 83.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.