Abstract
UMSBP is a CCHC-type zinc finger protein, which functions during replication initiation of kinetoplast DNA minicircles and the segregation of kinetoplast DNA networks. Interactions of UMSBP with origin sequences, as well as the protein oligomerization, are affected by its redox state. Reduction yields UMSBP monomers and activates its binding to DNA, while oxidation drives UMSBP oligomerization and impairs its DNA-binding activity. Kinetics analyses of UMSBP–DNA interactions revealed that redox affects the association of free UMSBP with the DNA, but has little effect on its dissociation from the nucleoprotein complex. A previously proposed model, suggesting that binding of DNA is regulated via the reversible interconversions of active UMSBP monomers and inactive oligomers, was challenged here, revealing that the two redox-driven processes are not interrelated. No correlation could be observed between DNA-binding inhibition and UMSBP oligomerization, upon oxidation of UMSBP. Moreover, while the presence of zinc ions was found to be essential for the interaction of UMSBP with DNA, UMSBP oligomerization occurred through zinc-depleted, unfolded zinc finger domains. Site directed mutagenesis analysis of UMSBP suggested that its unique methionine residue, which can be oxidized into methionine sulfoxide, is not involved in the redox-mediated regulation of UMSBP–DNA interactions.
INTRODUCTION
Kinetoplast DNA (kDNA), the mitochondrial DNA of trypanosomatids, is a giant network of catenated DNA circles. It consists, in the species Crithidia fasciculata, of ∼5000 duplex DNA minicircles of 2.5 kbp and ∼50 maxicircles of 37 kbp that are interlocked topologically to form a DNA network. Two short sequences, the dodecameric universal minicircle sequence (UMS) GGGGTTGGTGTA and the hexameric sequence ACGCCC, which were located at the minicircle's replication origin and implicated with its replication initiation, were conserved in all trypanosomatid species studied (1–4). A UMS-binding protein (UMSBP), a protein that binds specifically the dodecameric UMS sequence and a 14-mer sequence, containing the core hexamer, conserved at the minicircle H-strand replication origin, was purified to apparent homogeneity from Crithidia fasciculata cell extract (5,6). Based on its high affinity to conserved origin sequences (5–7), its intramitochondrial localization to the kinetoflagelar zone (8), where minicircles replication initiation was proposed to occur (9), and its interaction in vivo with kDNA networks (10), UMSBP has been proposed to play the role of a kDNA minicircles initiator protein. A recent study, using RNA interference (RNAi) analysis in Trypanosoma brucei, has revealed the function of UMSBP during the initiation of minicircles replication and in the segregation of the kDNA network (11).
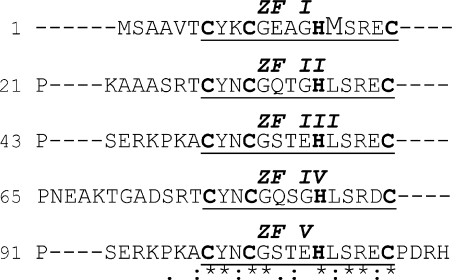
C. fasciculata UMSBP has a potential to form five CCHC-type zinc-finger (ZF) structures. This motif forms a compact zinc finger that has been associated with the binding of single-stranded nucleic acids (12–15). This structure is redox-sensitive, probably since oxidation of the thiol groups in cysteine residues enhances the ejection of the zinc ion, resulting in a conformational change, which impairs the protein binding to the DNA (16). Previous studies have shown that UMSBP binding to the origin sequence, as well as its oligomerization, are affected in vitro by the protein redox state (17). Reduction of UMSBP activates its binding to DNA and promotes UMSBP monomerization, while oxidation inhibits its DNA-binding activity and enhances its oligomerization. These studies have also demonstrated that UMSBP binds UMS only in its monomeric form raising the hypothesis that redox may regulate the action of UMSBP at the replication origin, through the reversible interconversions of active UMSBP monomers and its inactive oligomeric forms (17). Analysis of deletion mutants has revealed that the five zinc finger motifs in UMSBP may differ in their function. While truncation of the zinc fingers residing at the C-terminal region of UMSBP (Figure 4, ZFs III, IV and V) impaired the binding of the DNA ligand but had no effect on its capacity to dimerize, deletion of the protein N-terminal zinc finger (ZFs I) had relatively little effect on the binding of UMSBP to DNA, but significantly inhibited its capacity to dimerize (17). The involvement of ZF II in UMSBP activities has yet to be clarified.
Figure 4.
Met16 residue is unique to UMSBP first zinc finger. Amino acids sequence alignment within UMSBP created by CLUSTAL 2.0.8 Multiple Sequence Alignments software. Indicated, are the 5 CCHC motifs (boldface), the 5 CCHC-type zinc finger domains (underline), the unique Met16 residue (enlarged font); and numbers of amino acids. Zinc finger domains (ZF) are numbered starting from the protein N-terminus. The amino acid conservation: (*) identical, (:) conserved amino acids substitutions, (.) semi-conserved acids substitutions.
The functional effect of redox on protein oligomerization has been demonstrated in several experimental systems. Multimerization of the apoptosis signal-regulated kinase-1 (Ask1), induced by hydrogen peroxide and its reduction by thioredoxin, regulates the H2O2-induced c-Jun NH2-terminal kinase (JNK) activation and apoptosis (18). Activation of Escherichia coli Hsp33, a redox-regulated molecular chaperone, requires the presence of reactive oxygen and hydroxyl radicals, which are sensed by the thiol-containing zinc center of the protein. Upon exposure to oxidative stress the protein undergoes a conformational rearrangement and dimerizes, to yield its functionally active structure (19–21). Pre-initiation and replication initiation complex formation in bovine papilloma virus type-1 is dependent on interaction between the transcription factor E2 and the viral initiator E1. This interaction was found to be regulated by redox. Under oxidation conditions, disulphide bond is formed between the two E2 trans-activating domains, which prevents their association with two E1 proteins, hence preventing replication initiation (22).
We have recently reported on the cycling of UMSBP activity throughout the trypanosomatid cell cycle and its tight correlation with the cycling of the protein's redox state (23). Here, we describe the effect of redox on the association of UMSBP with the replication origin sequence and on its dissociation from the nucleoprotein complex. We also demonstrate that unlike its DNA-binding activity, UMSBP oligomerization is not dependent on zinc-containing, folded zinc finger structures. We further challenged a previously proposed model for the regulation of UMSBP (17), demonstrating that binding of UMSBP to the origin sequence is not regulated through the protein oligomerization per se and examined the role of the single methionine residue in UMSBP (Met16) in UMSBP interactions with the origin sequence.
MATERIALS AND METHODS
Purification of UMSBP
Preparation of recombinant UMSBP was carried out following Onn et al. (17). UMSBP coding sequence was cloned into pET22B + expression vector (Novagen). The plasmid was introduced by electroporation into Escherichia coli BL21, and protein expression was induced by the addition of 1 mM isopropyl-d-thiogalactopyranoside (IPTG) for 5 h at 37°C. Cells were harvested and resuspended in buffer L (50 mM potassium phosphate buffer pH 8.0, 300 mM NaCl, 10 mM imidazole–Cl and 20 mM β-mercaptoethanol) and lysed by seven cycles of maximum-power sonication bursts of 30 s each (Misonic Sonicator XL). Triton X-100 was added to a final concentration of 1% (vol/vol), followed by a 30 min centrifugation at 39 000 × g and 4°C. The supernatant was added to Ni-nitrilotriacetate (NTA) beads (Qiagen) and incubated with gentle rotation at 4°C for 1 h. The beads were packed into a column (a 2 ml bed volume of Ni-NTA beads was used for a lysate prepared from a 10 l cell culture) and washed five times with 10 bed volumes of buffer L, containing 600 mM NaCl, followed by 10 bed volumes of buffer L with no NaCl added. Bound UMSBP was eluted from the column using buffer E (50 mM potassium phosphate buffer, pH 8.0, 250 mM imidazole-Cl and 20 mM β-mercaptoethanol). The eluted fraction was loaded onto a phenyl-Sepharose (Pharmacia) column (a 2 ml bed volume of phenyl Sepharose beads was used for an Ni-NTA-eluted fraction prepared from an original lysate of a 10 l cell culture), equilibrated with 50 mM Tris–Cl pH 8.0, 1.3 M ammonium sulfate, 5 mM DTT and washed sequentially with 50 mM Tris–Cl pH 8.0 and 5 mM DTT containing the following volumes and ammonium sulfate concentrations: two bed volumes of 1.0 M, six bed volumes each of 0.8 M and 0.7 M ammonium sulfate, two bed volumes of each of 0.6, 0.5, 0.4, 0.3, 0.2 and 0 M ammonium sulfate. UMSBP was eluted from the column with 0.4 M ammonium sulfate. A 1.0 mg amount of purified recombinant UMSBP was obtained from a 1.0 l cell culture.
One unit of UMSBP activity is defined as the amount of protein required for the binding of 1 fmol of the UMS DNA (5′-GGGGTTGGTGTA-3′) ligand, under the standard binding assay conditions (5,6).
Electrophoretic Mobility Shift Analysis (EMSA)
Analyses were carried out as described previously (5–7). The 20 µl standard binding reaction mixture contained 25 mM Tris–Cl pH 7.5, 2 mM MgCl2, 20% (vol/vol) glycerol, 1 mg/ml bovine serum albumin, 25 µg/ml poly(dI-dC)·poly(dI-dC) and 12.5 fmol of 5′-32P-labeled UMS (5′-GGGGTTGGTGTA-3′) DNA. Reactions were started by the addition of the indicated concentrations of recombinant UMSBP, prepared as described above, incubated at 30°C for 30 min and electrophoresed in an 8% native polyacrylamide gel (1:29, bisacrylamide/acrylamide) in TAE buffer (6.7 mM Tris–acetate, 3.3 mM sodium acetate, 1 mM EDTA, pH 7.5). Electrophoresis was conducted at 0–2°C at 250 V, for 1.5 h. Protein–DNA complexes were quantified by exposing the dried gels to an imaging plate and analyzing by phosphoimager.
Oxidation of free and DNA-bound UMSBP by H2O2
A 0.9 pmol of recombinant UMSBP was oxidized by the indicated H2O2 concentrations in 125 µl assay containing 25 mM Tris–Cl pH 7.5, 2 mM MgCl2 and 1 mg/ml BSA. Following 15 min incubation at 30°C, H2O2 was removed by passing the reaction mixture through ZEBA Desalt Spin Columns (PIERCE), pre-equilibrated with 25 mM Tris–Cl pH 7.5 and 2 mM MgCl2 at 4°C. The samples were then placed on ice before further analysis. To study the effect of oxidation on pre-bound UMSBP, ∼58 fmol UMSBP of the H2O2-untreated sample was added to each of five test tubes which contained 1.66 fold concentrated standard binding reaction mixture, depleted of DTT, glycerol and poly(dI-dC)·poly(dI-dC). Binding was performed at 30°C for 5 min, then H2O2 was added to each tube to the indicated final concentrations, and the binding reaction was continued for an additional 15 min at 30°C. To study the binding of pre-oxidized UMSBP, ∼58 fmol UMSBP from each of the indicated H2O2 pre-oxidized samples was added to standard binding reaction mixture and incubated for 15 min at 30°C. Glycerol was added to 20% final concentration to allow gel loading and the samples were analyzed by gel electrophoresis, as described above. We found that various recombinant UMSBP preparations differ in their degree of sensitivity to the inhibitory effect of hydrogen peroxide.
Surface plasmon resonance (SPR) analysis
Analyses were conducted, using BIAcore 3000, at the BIAcore unit, the Hebrew University of Jerusalem. 3′-biotinylated UMS DNA, immobilized to a streptavidin-coated SA sensor chip (BIAcore), was used as a DNA ligand. The immobilized DNA yielded a signal of 250 resonance units (RU) (representing 66.3 fmol mm−2 of bound UMS DNA). DNA-binding activity of UMSBP was measured as described previously (17), with the following modifications: poly(dI-dC)·poly(dI-dC) was added to both association and dissociation buffer (150 mM NaCl, 2 mM MgCl2, 10 mM Hepes–Cl pH 8.0) at 25 µg/ml. UMSBP was pretreated and diluted in the above buffer supplemented with either 20 mM H2O2 or 5 mM DTT. Qualitative analysis was performed by injection of 62.5 nM UMSBP. Quantitative analyses were performed by injection of 3.125–50 nM UMSBP, under reducing conditions and 62.5–1000 nM under oxidizing conditions.
Oxidation of UMSBP by diamide
UMSBP-binding assay was conducted in a 20 µl standard binding reaction mixture, containing 0.6 mM DTT, 36.5 fmol UMSBP and the indicated diamide [diazenedicarboxylic acid bis(N,N-dimethylamide), (Sigma)] concentrations. For the oligomerization assay, diamide was added, at the indicated concentrations, to a 50 µl reaction mixture, containing either 0.91 or 1.83 pmol UMSBP in 25 mM Tris–Cl pH 7.5, 2 mM MgCl2, 0.6 mM DTT and 20% (v/v) glycerol. The reactions were incubated at 30°C for 30 min and their products electrophoresed on non-reducing 16.5% Tris–Tricine SDS–PAGE, followed by western blot analysis, using anti UMSBP antibodies. The ECL reaction was developed in a LAS-3000 (Fuji) and quantified by TINA software.
In vivo oxidation by H2O2
Total 7.5 × 107 logarithmic C. fasciculata cells were grown in 5 ml brain heart infusion medium (Difco Laboratories, Inc., Detroit, MI), containing the indicated H2O2 concentrations, for 3 h. Cleared cell lysates (Fraction I) were prepared by the gentle disruption of the cell membrane using a nonionic detergent in hypotonic solution, as described previously (24), except that 0.2% (wt/vol) Brij-58 was used (5) and DTT was omitted from the lysis solution. Protein concentration was measured using Bradford protein assay (BIO-RAD). 86.4 µg protein was loaded on non-reducing 16.5% Tris–Tricine SDS–PAGE, followed by western blot analysis, using anti UMSBP antibodies. Membrane was developed in a LAS 3000 instrument and protein bands were quantified by TINA software. DNA-binding assay was preformed as described above, except that MgCl2 and DTT were omitted. Three hundred and fifty nanograms of whole cell lysate [Fraction I (5)] were assayed in the reaction conducted under non-reducing conditions.
Analysis of methionine sulfoxide residues
Seventy-three micromolar His-tagged UMSBP in 20 mM Hepes–Cl pH 7.4 and 0.25 mM DTT, was diluted 1:10 in a 50 µl solution containing 20 mM HCl and 20 mM H2O2. The oxidation reaction was incubated for 30 min at 0°C. The reaction mixtures were dialyzed overnight at 4°C against 1 l of 20 mM HCl, followed by additional 1 h dialysis against a fresh 1 l 20 mM HCl. The solution was neutralized with 2 M Tris–base and stored in liquid nitrogen until its use in a cyanogen bromide (CNBr) cleavage reaction. For CNBr cleavage assay, formic acid was added to the samples to final concentration of 70%. Nitrogen was flown into the tubes, followed by addition of 4.4 µmol CNBr. The solution was incubated in darkness overnight. Nitrogen was flown again into the tubes, which were then left open in the chemical hood to evaporate for several hours, followed by speed-vac evaporation. The samples were dissolved in 25 mM Tris–Cl pH 7.5 and electrophoresed by 16.5% Tris–Tricine SDS–PAGE, followed by western blot analysis, using anti UMSBP antibodies. For mass spectrometry (MS) analysis, samples were reduced in 50 mM Tris–Cl pH 8.0 containing 10 mM DTT and 6 M guanidinium chloride, for 60 min, at 56°C and were alkylated by 22 mM iodoacetamide, in the same solution, in the dark, for 30 min at room temperature. The reduced and alkylated samples were acidified with formic acid to final concentration of 5% and subjected to solid phase extraction on ZipTip C4 resin-filled tips (Millipore). The extracted samples were dried and resuspended in a 25 μl trypsin solution (0.15 μg per sample) in 25 mM NH4HCO3, pH 8.0 solution. The samples were incubated at 37°C for 16 h and the reactions were stopped by addition of formic acid to final concentration of 28%. The samples, containing tryptic digests were solid-phase extracted with C18 resin-filled tip and nanosprayed into the Qtof MS system in 50% acetonitryl, 1% formic acid solution. Data analysis was performed using the Biolynx package (Micromass, England) and database searches were performed using the Mascot package (Matrix Science, England). MS analysis was carried out by the Bletterman Laboratory of the Interdepartmental Equipment Unit of the Faculty of Medicine, The Hebrew University of Jerusalem.
Mutagenesis of UMSBP Met16
Substitution of UMSBP methionine16 (Met16) into alanine (Ala) or leucine (Leu) was conducted by PCR in two steps. First, two fragments, containing the sequences upstream and downstream to the Met16 residue, were amplified, using primers containing the desired mutations. Second, the two amplified fragments were used as templates for a second PCR reaction, in which only a primer for the 5′ end of the first fragment and a 3′ end primer for the second fragment were used, resulting in a full length Met16 mutated UMSBP. Upper fragment primers for Met16-L substitution (amino acids 1–16): Met16-A-up-F (5′-CTCGATCCCG CGAAATTAATACGACTCACT–3′), Met16-L-up-R (5′-CGGCTCAGGTGGCCAGCCTC-3′). Downstream fragment primers for Met16-L substitution (amino acids 17–124): Met16-L-down-F (5′-GAGGCTGGCCACCTGAGCCG-3′), Met16-A-down-R (5′-AGCCGGATCTCAGTGGTGGTGG-3′). Upper fragment primers for Met16-A substitution (amino acids 1–16): Met16-A-up-F, Met16-A-up-R: (5′-CGGCTAGCGTGGCCAGCCTC-3′). Downstream fragment primers for Met16-A substitution (amino acids 17–124): Met16-A-down-F (5′-GAGGCTGGCCA CGCTAGCCG-3′), Met16-A-down-R. Primers used for the preparation of full length UMSBP were Met16-A-up-F and Met16-A-down-R. PCR was conducted as followed: 1 min 98°C, followed by 30 rounds of 1 min 95°C, 1 min 65°C, 1 min 72°C, followed by 5 min 72°C. UMSBP Met16A/L mutated sequences were cloned into pet22b + expression vector and transformed into E. coli BL21-DE3 Origami strain. Overexpression and purification on nickel beads, followed by phenyl Sepharose chromatography, was conducted following (17), as described above.
RESULTS
Redox affects UMSBP binding to DNA but not its dissociation from the nucleoprotein complex
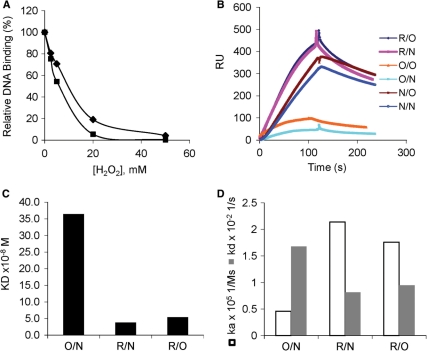
Regulation of UMSBP interactions with the minicircle replication origin may include either the control of the protein loading onto this site, prior to replication initiation, or the release of the bound protein from the nucleoprotein complex, or both. To determine whether redox affects the origin binding activity of the free unbound UMSBP or its DNA-bound form, binding of UMSBP to UMS was monitored under reducing and oxidizing conditions, using both EMSA (Figure 1A) and SPR (Figure 1B–D) analyses. EMSA analyses revealed that addition of hydrogen peroxide to the binding reaction resulted in the inhibition of UMSBP binding to DNA (Figure 1A). When free UMSBP was pretreated with hydrogen peroxide and then, following removal of the oxidizing agent, interacted with DNA, UMSBP binding to DNA was inhibited to approximately the same extent (Figure 1A). These observations suggested that oxidation of the free protein per se, was sufficient to prevent the generation of the UMSBP–UMS nucleoprotein complexes. However, the EMSA data, has not distinguished between the effects of redox on the association of UMSBP with the origin sequence, versus its dissociation from the nucleoprotein complex. This question was further addressed by an SPR approach using BIAcore, with biotinylated UMS DNA ligand attached to a streptavidin coated sensor chip (Figure 1B–D). The BIAcore analysis enabled the monitoring of the effect of UMSBP treatment with either reducing (R) or oxidizing (O) agents and in the absence of any redox affecting agents (N), during either UMSBP association with the DNA, or dissociation from the nucleoprotein complex. Treatment of UMSBP with H2O2 in its unbound form and during its association reaction, followed by dissociation of the complex under either oxidizing conditions (O/O), or in the absence of any redox-affecting agent (O/N) resulted in a dramatic decrease in its DNA-binding capacity, while its DNA-binding activity was enhanced upon association under reducing conditions and dissociation under either oxidizing conditions (R/O) or in the absence of a redox-affecting agent (R/N). Quantitative measurement were obtained by injection of 3.125–50 nM UMSBP, which was pre-incubated under reducing conditions and 62.5–1000 nM UMSBP, which was pre-incubated under oxidizing conditions (Figure 1C and D). The equilibrium binding constant (KD) measured for the interaction between pre-oxidized UMSBP and UMS DNA was an order of magnitude higher than the value measured for this interaction with pre-reduced UMSBP (Figure 1C, compare R/O, R/N to O/N). The apparent decrease measured in the affinity of UMSBP to the UMS DNA, has resulted from a decrease in the association constant and increase in the dissociation constant of the reaction (Figure 1D, compare R/O, R/N to O/N). No difference could be measured in the dissociation rates of the reaction, when UMSBP was pre-bound to the DNA under reducing conditions (or in the absence of a redox-affecting agent) and then allowed to dissociate under either oxidizing conditions or in the absence of a redox affecting-agent (Figure 1B, compare R/N to R/O, or N/N to N/O; Figure 1D, compare the dissociation constants in R/O and R/N).
Figure 1.
Oxidation affects the binding of UMSBP onto UMS DNA, but not its dissociation from the nucleoprotein complex. Free unbound, or DNA-bound UMSBP (2.9 nM), was incubated under various redox conditions and the effect on its binding to UMS was monitored. In (A) UMSBP-binding activity analyzed by EMSA. Squares, H2O2 was added directly into the binding assay; diamonds, UMSBP was preincubated with H2O2 to oxidize unbound UMSBP. Values represent UMS-binding activity relative to the maximal activity measured. (B–D) UMSBP-binding activity analyzed by BIAcore. Binding was measured under reduced (R), oxidized (O) or non-reduced (N) association and/or dissociation conditions. In (B), binding curves of 62.5 nM UMSBP incubated either before and during the association phase, or throughout the dissociation phase, under different redox conditions. Redox conditions are indicated [e.g. R/O, association under reducing (R) conditions and dissociation under oxidizing (O) conditions]. RU, in the sensogram ordinate, denotes Resonance Units. In (C) and (D), quantitative analyses of the redox effect on the equilibrium binding constant [KD, (C)] and the reaction kinetic constants (D) of the interactions of UMSBP with the UMS ligand, were conducted for three representative redox conditions. For R/N and R/O analysis, UMSBP, which was pre-incubated under reducing conditions, was injected at 3.125–50 nM UMSBP and for O/N analysis pre-oxidized UMSBP was injected at 62.5–1000 nM.
Overall these observations suggest that oxidation of the free UMSBP affects both the association and dissociation constants measured in its interactions with UMS DNA but has no significant effect on the dissociation of the pre-bound UMSBP from the UMSBP–UMS DNA complex.
Redox effect on UMSBP–DNA interactions is independent of UMSBP oligomerization
In a previous study we have demonstrated that UMSBP oxidation results in the inhibition of its binding to the origin sequence (17). This study has also revealed that oxidation of UMSBP results in its oligomerization. As UMSBP oxidation results in the protein oligomerization and loss of its capacity to bind DNA, while its reduction leads to UMSBP monomerization and resumption of its DNA-binding activity, it has been presumed that binding of UMSBP to UMS may be regulated via redox-mediated reversible interconversions of the active UMSBP monomers and its inactive oligomers (17).
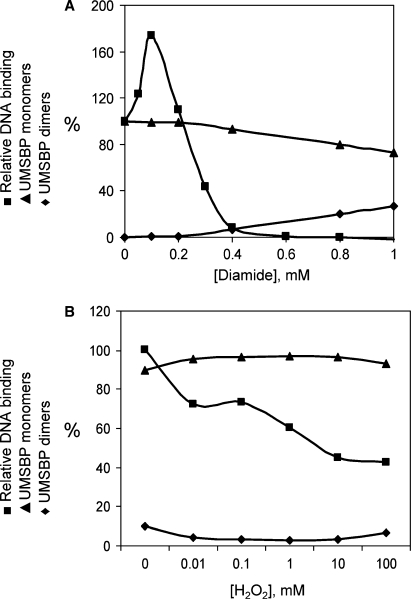
The hypothesis that redox affects the binding of UMSBP to DNA through cellular control of its oligomeric state, was challenged here by monitoring the correlation between the loss of UMSBP-binding activity and the protein oligomerization, in the presence of increasing concentrations of an oxidizing agent. For this purpose, UMSBP, which was pre-reduced by DTT, was oxidized gradually using increasing concentrations of the thiols-oxidizing agent diamide. The effect of oxidation on the binding of UMSBP was monitored by EMSA analysis and its oligomerization by SDS–PAGE analysis under non-reducing conditions. The protein capacity to generate UMSBP–UMS complexes and the relative abundance of its monomeric versus oligomeric forms were compared. These analyses demonstrate that while a complete loss of UMSBP DNA-binding activity could be observed in the presence of 0.6 mM diamide, only 10% of the protein molecules were converted into UMSBP dimers and oligomers under these oxidation conditions (Figure 2A). These results indicate that the loss of UMSBP-binding activity by oxidation is not the result of the protein oligomerization per se, as it occurs while 90% of the protein molecules retain their monomeric state. It is suggested that UMSBP inactivation under these conditions, could rather be the result of the generation of intra-molecular disulfide bonds by the oxidized cysteins’ thiol groups in UMSBP's zinc fingers, which may enhance the ejection of the zinc ions, thus causing a conformational change that impairs the protein binding to DNA (16,25). The reason for the initial increase observed in binding of UMSBP to the DNA at low diamide concentrations is not clear.
Figure 2.
Inhibition of UMSBP binding to DNA by oxidation is not mediated by the protein oligomerization. The effect of oxidation on UMSBP DNA-binding activity and its oligomerization was measured in vitro (A) and in vivo (B). In (A), UMSBP was incubated in the presence of the indicated diamide concentrations. DNA binding was measured using EMSA with UMS ligand and UMSBP oligomerization using SDS–PAGE under non-reducing conditions. Squares, DNA-binding activity; diamonds, UMSBP oligomers; triangles, UMSBP monomers. In (B), C. fasciculata cell culture was treated with the indicated concentrations of H2O2 and cell lysates were assayed for UMS-binding activity and UMSBP oligomerization. Squares, UMSBP DNA-binding activity; diamonds, UMSBP oligomers; triangles, UMSBP monomers. UMS-binding activity is presented relative to the activity of untreated UMSBP, used as a 100%. The sum of the monomer and oligomer fractions are used as a 100% in each oxidation point and their relative abundance (%) is presented.
Next we have examined the possible interrelations between the effects of oxidation of UMSBP on its DNA-binding activity and oligomerization in vivo. For this purpose C. fasciculata cells were incubated in the presence of increasing concentrations of hydrogen peroxide and its DNA-binding activity and oligomerization were assayed in cell extracts, as described above. The results revealed (Figure 2B) that while DNA-binding activity was significantly impaired in the presence of increasing concentrations of oxidizing agent, no detectable change could be observed in UMSBP's oligomeric state (Figure 2B). In accord with the in vitro observations described above, these results demonstrate that under in vivo oxidation conditions, inhibition of UMSBP's DNA-binding activity is independent of the protein oligomerization. Overall, these results imply that the redox-mediated regulation of UMSBP interactions with DNA is not mediated through the redox effect on the protein oligomeric state.
UMSBP oligomerizes through oxidation of cysteine residues in zinc-depleted zinc fingers
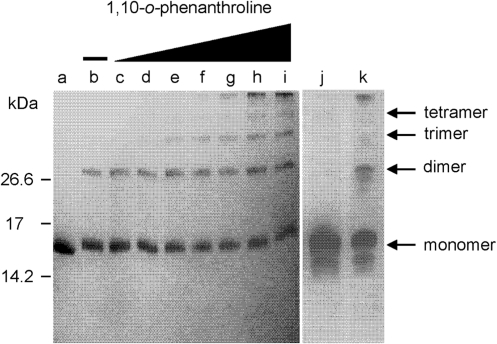
We have previously found that chelation of zinc ions by 1,10-o-phenanthroline, results in a conformational change in UMSBP and DNA-binding inhibition (17), implying that a zinc-containing zinc finger is the active conformation in UMSBP–DNA interactions. To explore the role of the zinc ions in the process of UMSBP oligomerization, UMSBP was oxidized with 5 mM diamide under conditions, which have been previously shown to yield UMSBP oligomers (17), in the presence of increasing concentration of the zinc chelator 1,10-o-phenanthroline. Products of the reaction were analyzed by non-reducing SDS–PAGE, revealing that chelation of the zinc ions has not prevented the dimerization and further oligomerization of UMSBP (Figure 3). These observations suggest that the oxidation-driven oligomerization of UMSBP is not mediated by the zinc-containing zinc finger conformation that was shown to be active in the binding of UMSBP to DNA, but rather via cysteine residues in the zinc-depleted, unfolded zinc finger domains. In accord with this notion is the observation that although chelation of the zinc ions per se has not resulted in UMSBP oligomerization, this process was enhanced with zinc exclusion, as demonstrated by the generation of higher oligomers with increased concentration of the zinc chelator (Figure 3 compare lanes d–i to j).
Figure 3.
Zinc is not required for UMSBP oligomerization. UMSBP, at final concentration of 7.2 µM in 20 mM Hepes pH 7.4, was treated with increasing concentrations of 1,10-o-phenanthroline, in the presence of 5 mM diamide and the reaction products were electrophoresed in a non-reducing 16.5% Tris–Tricine SDS–PAGE. The gel was stained using GelCode Blue Stain Reagent. In lane a, no diamide and no 1,10-o-phenanthroline are present. In lanes b–i: 5 mM diamide and 0, 50, 100, 200, 400, 600, 800 and 1000 µM of 1,10-o-phenanthroline, respectively. Lanes j and k represent a separate analysis: In lane j, UMSBP treated with 1000 µM of 1,10-o-phenanthroline in the absence of diamide and in lane k, UMSBP treated with 5 mM diamide in the absence of 1,10-o-phenanthroline.
UMSBP's unique methionine residue is not involved in the redox-mediated regulation of UMSBP–DNA interactions
Previous studies have shown the specific involvement of the N-terminal zinc finger of UMSBP in the protein oligomerization (17). Sequence alignment of the five CCHC-type zinc fingers motifs in UMSBP [(26); Figure 4] revealed that the N-terminal zinc finger also differs in its sequence, which contains four unique residues. This includes a methionine (Met) residue, located at position 16 in the UMSBP sequence, substituting a conserved leucine (Leu) residue in the other four zinc finger domains in this protein. This Met16 residue, located 10 residues downstream of the first Cys residue in the N-terminal CCHC motif, is conserved in the sequences of UMSBP orthologues from other Kinetoplastida species (Supplementary Table S1). One significant difference between Met and Leu residues is the presence in methionine of a sulfur atom, which renders this residue redox-sensitive. Considering the regulatory effect of redox on UMSBP activity (17,23), we have addressed the potential functional role of the conserved redox-sensitive Met residue in the regulation of UMSBP function.
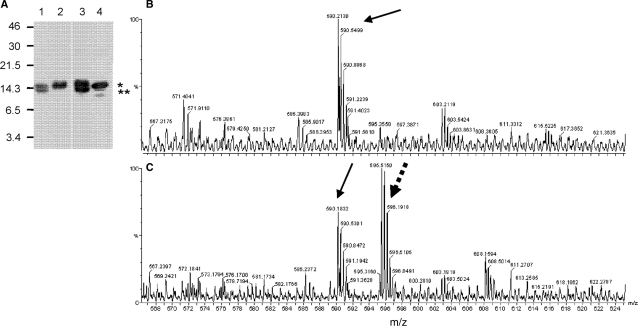
To examine capacity of the unique Met16 residue to be oxidized, UMSBP was treated with hydrogen peroxide, under acidic conditions. Under these conditions Met residues are being oxidized, while Cys residues are fully protonated and thus are not affected by the oxidant (27). As UMSBP oligomerization occurs through the generation of inter-molecular interactions of cysteine thiols (17), we have used the protein oligomerization under these conditions, as an indicator for the protonation of cysteine residues, hence the lack of UMSBP oligomerization. Met16 oxidation, yielding methionine sulfoxide residue was identified using the CNBr cleavage assay, based on CNBr capacity to cleave the polypeptide chain next to a Met, but not next to a Met-sulfoxide residue (28), as well as by mass spectrometry. CNBr cleaved untreated or acid-treated UMSBP (Figure 5A, lanes 1 and 3), but not UMSBP that was treated with the oxidant under acidic conditions (Figure 5A, lane 2), indicating the oxidation of Met16 under these conditions. In addition, MS analysis of UMSBP, either untreated (Figure 5B) or oxidant-treated (Figure 5C), under acidic conditions, revealed ions with a mass of 1783.5 Da (595.51 3+), which were present only in the oxidized form (Figure 5C, hatched arrow). Ions corresponding to a mass of 1767.6 Da (590.21 3+) were present in both samples (Figure 5B and C). Sequencing the peptide by MS/MS analysis identified a peptide of the sequence SAAVTCYKCGEAGHMSR, where Met in the oxidized sample has a mass of 147 Da, suggesting the presence of a methionine sulfoxide at position 16, as compared to a mass of 131 Da for the methionine residue at this position in the reduced sample.
Figure 5.
Met16 is oxidized by H2O2. (A) CNBr cleavage of UMSBP was carried out as described in the Materials and methods section. UMSBP was pretreated as follows: lane 1, UMSBP was incubated in the presence of 20 mM HCl; lane 2: With 20 mM H2O2 under acidic conditions (as in lane 1); lane 3: neutral conditions, with no oxidant; lane 4: untreated. CNBr uncleaved UMSBP (i.e. containing Met16-O) and cleaved UMSBP (i.e. containing Met16) reaction products are designated with * and **, respectively. (B) and (C), MS analysis of a tryptic digest of UMSBP, treated under acidic conditions as above (B), or treated with 80 mM H2O2 under acidic conditions (C). Arrows indicate ion corresponding to the SAAVTCYKCGEAGHMSR sequence in UMSBP, as revealed by MS/MS analysis. Hatched arrow, denotes ion with the above sequence, but carrying Met16-sulfoxide modification instead of Met16, as revealed by MS/MS analysis. Numbers represents mass/charge ratio.
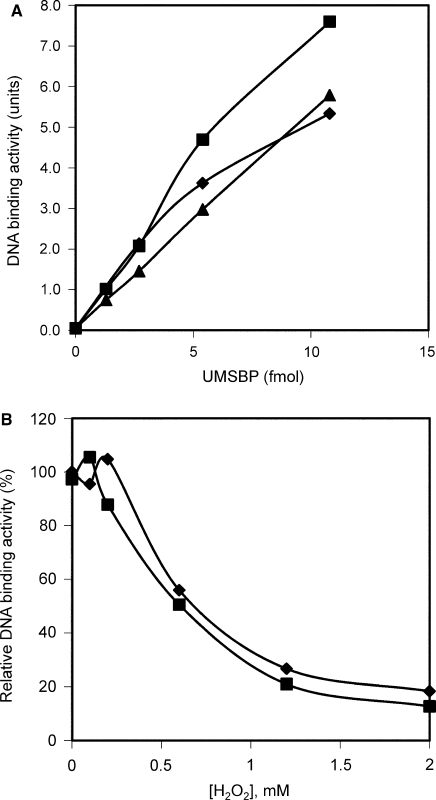
To study the role of the unique methionine residue in UMSBP activities, Met16 was substituted, using site-directed mutagenesis, by either a leucine (Leu) (which is the residue presents at this position in the other four zinc finger domains in UMSBP), or an alanine (Ala). The mutated proteins were expressed in E. coli, purified under reducing conditions and assayed for their capacity to bind the UMS DNA, in comparison to wt UMSBP. The observations described in Figure 6A reveal that substitution of the Met16 residue with either a Leu or an Ala residue had little effect on the binding of the protein to DNA (Figure 6A). These results suggest that Met16 is not a vital component of the DNA-binding motif in UMSBP, in accord with previous observations showing that the other four zinc finger domains in UMSBP, which contain a Leu residue at this position, are active in DNA binding, while the N-terminal zinc finger is involved in the protein oligomerization (17).
Figure 6.
Met16 has no detectable effect on the resistance of UMSBP to oxidation. In (A) UMS-binding activity of wt UMSBP (diamonds) or UMSBP, mutated in its Met16 residue, by substitution with either an alanine (Met16A) (squares) or a leucine (Met16L) (triangles) residue. 1.3, 2.7, 5.4 and 10.8 fmol of wild-type and mutated UMSBP were subjected to EMSA analysis under the standard binding assay conditions, except that glycerol, poly(dI-dC)·poly(dI-dC) and DTT were omitted from the reaction mixture. In (B), binding of 5.1 fmol wild-type UMSBP (diamonds), or 7.3 fmol Met16A mutated UMSBP (squares) (0.51 and 0.73 nM UMSBP, respectively) to UMS DNA was measured in the presence of increasing concentrations of H2O2 and quantified by phosphoimaging. EMSA was conducted under the standard binding assay conditions, except that a 10 μl reaction was used and DTT was omitted from the reaction mixture. H2O2 concentrations used were: 0, 0.1, 0.2, 0.6, 1.2 and 2 mM.
It has been previously suggested that methionine residues may play a role in the protection of proteins from critical oxidative damage, by serving as an endogenous antioxidants (29,30). Considering the observations presented here (Figure 5), demonstrating the capacity of UMSBP's Met16 to be oxidized into Met sufoxide, we have addressed the possibility that this unique residue may play a role in conferring an enhanced oxidation resistance to UMSBP. For this purpose, we have measured the binding to UMS DNA of either wt UMSBP or mutated protein, in which Met16 had been substituted by an Ala residue, in the presence of increasing concentrations of hydrogen peroxide. This analysis revealed (Figure 6B) no measurable difference in the sensitivity of the Ala16containing UMSBP mutant and the wild-type protein to hydrogen peroxide, implying that, under the in vitro oxidizing assay conditions, Met16 residue had apparently no contribution to the resistance of UMSBP to oxidative stress.
DISCUSSION
UMSBP binding to the replication origin, as well as its oligomerization have previously been shown to be affected by redox (17). These observations raised the possibility that the zinc finger motif could function as a redox switch in the regulation of UMSBP's functions in the trypanosomatid cell. This was further supported by recent observations showing that binding of UMSBP to DNA was tightly correlated to its redox state in vivo, in a cell cycle dependent manner (23). The observations described here, showing that the oxidation of UMSBP by hydrogen peroxide affects the free UMSBP but not the DNA-bound protein (Figure 1), implies that redox may control UMSBP loading onto the origin sequence, but has no significant effect on its dissociation from this site. A possible explanation for the limited effect of redox on the release of UMSBP from the nucleoprotein complex may be that the redox-responsive zinc fingers, which are available for interaction in the unbound state of UMSBP, are occupied by the bound DNA in the nucleoprotein complex and thus become significantly less accessible to redox changes. The mechanism which controls UMSBP unloading from the DNA remains to be clarified.
A previously proposed model (17) for the redox regulation of UMSBP, has coupled UMSBP's DNA-binding activity to the redox-mediated interconversions of active UMSBP monomers and its inactive oligomers. The current study provides several lines of evidence, indicating that loss of UMSBP's DNA-binding activity by oxidation is a consequence of intra-molecular generation of disulfide bonds, rather than of the intermolecular disulfide bonding involved in UMSBP oligomerization. First, binding of UMSBP was found to be significantly inhibited under oxidant concentrations, which yielded no measurable oligomerization of the protein (Figure 2A). Second, loss of UMSBP DNA-binding activity does not limit the extent of the protein oligomerization, which occurs under conditions in which DNA-binding activity had been completely abolished (Figure 3). Third, while incubation of cells in the presence of increasing concentrations of hydrogen peroxide affects the DNA-binding capacity of UMSBP in vivo, it has no detectable effect on the protein oligomerization (Figure 2B). These latter observations also raise questions regarding the capacity of UMSBP to oligomerize in the cell, in response to elevated intracellular levels of oxidation.
The lack of inhibition of the oxidation-driven oligomerization of UMSBP in the presence of 1,10-o-phenanthroline (Figure 3), indicates that zinc is not required for oligomerization. These observations suggest that these intermolecular interactions are not mediated by intact zinc fingers, but rather via disulfide bonding of Cys residues in zinc-depleted, unfolded zinc fingers. The enhancement of oxidation-dependent UMSBP oligomerization, observed in the presence of increasing concentrations of 1,10-o-phenanthroline, may indicate that UMSBP oligomerization is dependent on zinc exclusion. Thus, the relatively high levels of UMSBP oxidation, required for the protein oligomerization (Figure 2A), may correlate with the extensive depletion of its zinc ions (Figure 3) while DNA-binding activity is inhibited under relatively lower levels of oxidation. The stoichiometry of UMSBP-associated zinc ions, in correlation with the relatively low levels of oxidation required for the inhibition of UMSBP binding to the DNA and for the more extensive oxidation that supports its oligomerization has yet to be determined.
Previous studies have shown that methionine residues have the potential of being oxidized (27) and involved in oxidative stress scavenging, in several proteins (20–22). A unique methionine residue has been conserved in UMSBP orthologues of Kinetoplastida species (Supplementary Table S1), at the same location within the zinc finger motif, indicating a possible conserved functional role in the activity of UMSBP in the cell. Analysis of CfUMSBP, in which the unique methionine residue had been substituted for by alanine or leucine residues showed no significant difference in the DNA-binding capacity of the wild type versus the mutated proteins, implying that Met16 is not involved in UMSBP–DNA interactions, under reducing conditions. Moreover, despite its capacity of being oxidized into methionine sulfoxide, a Met16-containing UMSBP has not displayed higher resistance to increasing concentrations of the oxidant than an Ala16-containing UMSBP mutant, failing to support a potential function of this residue in oxygen radical scavenging. The possibility that this residue may play such a protective role under oxidative stress in vivo has yet to be determined.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This study was supported, in parts, by the Unites States-Israel Binational Science Foundation, Jerusalem Israel (grant no 2 005 023); and the Israel Science Foundation (grant no 54/06). Funding for open access charge: Israel Science Foundation and the United States-Israel Binational Science Foundation, Jerusalem, Israel.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Hagit Zer from the Institute of Life Sciences, The Hebrew University of Jerusalem, for her help and advice with the BIAcore analysis and Dr Ariel Gaathon and Dr Ofra Moshel from the Faculty of Medicine, The Hebrew University of Jerusalem, for their help and advice with the CNBr and MS analyses.
REFERENCES
- 1.Liu B, Liu Y, Motyka SA, Agbo EE, Englund PT. Fellowship of the rings: the replication of kinetoplast DNA. Trends Parasitol. 2005;21:363–369. doi: 10.1016/j.pt.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 2.Lukes J, Hashimi H, Zikova A. Unexplained complexity of the mitochondrial genome and transcriptome in kinetoplastid flagellates. Curr. Genet. 2005;48:277–299. doi: 10.1007/s00294-005-0027-0. [DOI] [PubMed] [Google Scholar]
- 3.Roy J, Faktorova D, Lukes J, Burger G. Unusual mitochondrial genome structures throughout the Euglenozoa. Protist. 2007;158:385–396. doi: 10.1016/j.protis.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Shlomai J. The structure and replication of kinetoplast DNA. Curr. Mol. Med. 2004;4:623–647. doi: 10.2174/1566524043360096. [DOI] [PubMed] [Google Scholar]
- 5.Tzfati Y, Abeliovich H, Avrahami D, Shlomai J. Universal minicircle sequence binding protein, a CCHC-type zinc finger protein that binds the universal minicircle sequence of trypanosomatids. Purification and characterization. J. Biol. Chem. 1995;270:21339–21345. doi: 10.1074/jbc.270.36.21339. [DOI] [PubMed] [Google Scholar]
- 6.Tzfati Y, Abeliovich H, Kapeller I, Shlomai J. A single-stranded DNA-binding protein from Crithidia fasciculata recognizes the nucleotide sequence at the origin of replication of kinetoplast DNA minicircles. Proc. Natl Acad. Sci. USA. 1992;89:6891–6895. doi: 10.1073/pnas.89.15.6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abu-Elneel K, Kapeller I, Shlomai J. Universal minicircle sequence-binding protein, a sequence-specific DNA-binding protein that recognizes the two replication origins of the kinetoplast DNA minicircle. J. Biol. Chem. 1999;274:13419–13426. doi: 10.1074/jbc.274.19.13419. [DOI] [PubMed] [Google Scholar]
- 8.Abu-Elneel K, Robinson DR, Drew ME, Englund PT, Shlomai J. Intramitochondrial localization of universal minicircle sequence-binding protein, a trypanosomatid protein that binds kinetoplast minicircle replication origins. J. Cell. Biol. 2001;153:725–734. doi: 10.1083/jcb.153.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drew ME, Englund PT. Intramitochondrial location and dynamics of Crithidia fasciculata kinetoplast minicircle replication intermediates. J. Cell. Biol. 2001;153:735–744. doi: 10.1083/jcb.153.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Onn I, Kapeller I, Abu-Elneel K, Shlomai J. Binding of the universal minicircle sequence binding protein at the kinetoplast DNA replication origin. J. Biol. Chem. 2006;281:37468–37476. doi: 10.1074/jbc.M606374200. [DOI] [PubMed] [Google Scholar]
- 11.Milman N, Motyka SA, Englund PT, Robinson D, Shlomai J. Mitochondrial origin-binding protein UMSBP mediates DNA replication and segregation in trypanosomes. Proc. Natl Acad. Sci. USA. 2007;104:19250–19255. doi: 10.1073/pnas.0706858104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katz RA, Jentoft JE. What is the role of the cys-his motif in retroviral nucleocapsid (NC) proteins? Bioessays. 1989;11:176–181. doi: 10.1002/bies.950110605. [DOI] [PubMed] [Google Scholar]
- 13.Rajavashisth TB, Taylor AK, Andalibi A, Svenson KL, Lusis AJ. Identification of a zinc finger protein that binds to the sterol regulatory element. Science. 1989;245:640–643. doi: 10.1126/science.2562787. [DOI] [PubMed] [Google Scholar]
- 14.Urbaneja MA, Kane BP, Johnson DG, Gorelick RJ, Henderson LE, Casas-Finet JR. Binding properties of the human immunodeficiency virus type 1 nucleocapsid protein p7 to a model RNA: elucidation of the structural determinants for function. J. Mol. Biol. 1999;287:59–75. doi: 10.1006/jmbi.1998.2521. [DOI] [PubMed] [Google Scholar]
- 15.South TL, Summers MF. Zinc- and sequence-dependent binding to nucleic acids by the N-terminal zinc finger of the HIV-1 nucleocapsid protein: NMR structure of the complex with the Psi-site analog, dACGCC. Protein Sci. 1993;2:3–19. doi: 10.1002/pro.5560020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baldwin MA, Benz CC. Redox control of zinc finger proteins. Methods Enzymol. 2002;353:54–69. doi: 10.1016/s0076-6879(02)53036-6. [DOI] [PubMed] [Google Scholar]
- 17.Onn I, Milman-Shtepel N, Shlomai J. Redox potential regulates binding of universal minicircle sequence binding protein at the kinetoplast DNA replication origin. Eukaryot. Cell. 2004;3:277–287. doi: 10.1128/EC.3.2.277-287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nadeau PJ, Charette SJ, Toledano MB, Landry J. Disulfide Bond-mediated multimerization of Ask1 and its reduction by thioredoxin-1 regulate H(2)O(2)-induced c-Jun NH(2)-terminal kinase activation and apoptosis. Mol. Biol. Cell. 2007;18:3903–3913. doi: 10.1091/mbc.E07-05-0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akhtar MW, Srinivas V, Raman B, Ramakrishna T, Inobe T, Maki K, Arai M, Kuwajima K, Rao Ch M. Oligomeric Hsp33 with enhanced chaperone activity: gel filtration, cross-linking, and small angle x-ray scattering (SAXS) analysis. J. Biol. Chem. 2004;279:55760–55769. doi: 10.1074/jbc.M406333200. [DOI] [PubMed] [Google Scholar]
- 20.Graumann J, Lilie H, Tang X, Tucker KA, Hoffmann JH, Vijayalakshmi J, Saper M, Bardwell JC, Jakob U. Activation of the redox-regulated molecular chaperone Hsp33–a two-step mechanism. Structure. 2001;9:377–387. doi: 10.1016/s0969-2126(01)00599-8. [DOI] [PubMed] [Google Scholar]
- 21.Ilbert M, Horst J, Ahrens S, Winter J, Graf PC, Lilie H, Jakob U. The redox-switch domain of Hsp33 functions as dual stress sensor. Nat. Struct. Mol. Biol. 2007;14:556–563. doi: 10.1038/nsmb1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanders CM, Sizov D, Seavers PR, Ortiz-Lombardia M, Antson AA. Transcription activator structure reveals redox control of a replication initiation reaction. Nucleic Acids Res. 2007;35:3504–3515. doi: 10.1093/nar/gkm166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sela D, Yaffe N, Shlomai J. Enzymatic mechanism controls redox-mediated protein-DNA interactions at the replication origin of kinetoplast DNA minicircles. J. Biol. Chem. 2008;283:32034–32044. doi: 10.1074/jbc.M804417200. [DOI] [PubMed] [Google Scholar]
- 24.Shlomai J, Linial M. A nicking enzyme from trypanosomatids which specifically affects the topological linking of duplex DNA circles. Purification and characterization. J. Biol. Chem. 1986;261:16219–16225. [PubMed] [Google Scholar]
- 25.Webster KA, Prentice H, Bishopric NH. Oxidation of zinc finger transcription factors: physiological consequences. Antioxid. Redox. Signal. 2001;3:535–548. doi: 10.1089/15230860152542916. [DOI] [PubMed] [Google Scholar]
- 26.Abeliovich H, Tzfati Y, Shlomai J. A trypanosomal CCHC-type zinc finger protein which binds the conserved universal sequence of kinetoplast DNA minicircles: isolation and analysis of the complete cDNA from Crithidia fasciculata. Mol. Cell. Biol. 1993;13:7766–7773. doi: 10.1128/mcb.13.12.7766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brot N, Weissbach H. Biochemistry and physiological role of methioninekaiser sulfoxide residues in proteins. Arch. Biochem. Biophys. 1983;223:271–281. doi: 10.1016/0003-9861(83)90592-1. [DOI] [PubMed] [Google Scholar]
- 28.Kaiser R, Metzka L. Enhancement of cyanogen bromide cleavage yields for methionyl-serine and methionyl-threonine peptide bonds. Anal. Biochem. 1999;266:1–8. doi: 10.1006/abio.1998.2945. [DOI] [PubMed] [Google Scholar]
- 29.Levine RL, Berlett BS, Moskovitz J, Mosoni L, Stadtman ER. Methionine residues may protect proteins from critical oxidative damage. Mech. Ageing Dev. 1999;107:323–332. doi: 10.1016/s0047-6374(98)00152-3. [DOI] [PubMed] [Google Scholar]
- 30.Stadtman ER, Moskovitz J, Levine RL. Oxidation of methionine residues of proteins: biological consequences. Antioxid. Redox Signal. 2003;5:577–582. doi: 10.1089/152308603770310239. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.