Abstract
Whole genome shotgun sequence analysis has become the standard method for beginning to determine a genome sequence. The preparation of the shotgun sequence clones is, in fact, a biological experiment. It determines which segments of the genome can be cloned into Escherichia coli and which cannot. By analyzing the complete set of sequences from such an experiment, it is possible to identify genes lethal to E. coli. Among this set are genes encoding restriction enzymes which, when active in E. coli, lead to cell death by cleaving the E. coli genome at the restriction enzyme recognition sites. By analyzing shotgun sequence data sets we show that this is a reliable method to detect active restriction enzyme genes in newly sequenced genomes, thereby facilitating functional annotation. Active restriction enzyme genes have been identified, and their activity demonstrated biochemically, in the sequenced genomes of Methanocaldococcus jannaschii, Bacillus cereus ATCC 10987 and Methylococcus capsulatus.
INTRODUCTION
In 1981, Messing and colleagues (1) introduced a radical concept into the field of DNA sequencing. They took a small phage genome and prepared random clones in an M13 vector that permitted the determination of terminal sequences from each of these clones, thereby permitting a large amount of sequence data to be obtained in an essentially random fashion. Over the subsequent years this basic technique of shotgun sequencing has been applied to larger and larger genomes, greatly facilitating the sequencing of many phage genomes, among the largest of which, was that of cytomegalovirus (2). In 1995, Craig Venter applied the same approach to the sequencing of the first bacterial genome, that of Haemophilus influenzae Rd (3). Since then the whole genome shotgun approach has provided the initial sequence data, not just for bacterial genome sequencing projects, but also for the human genome sequence (4).
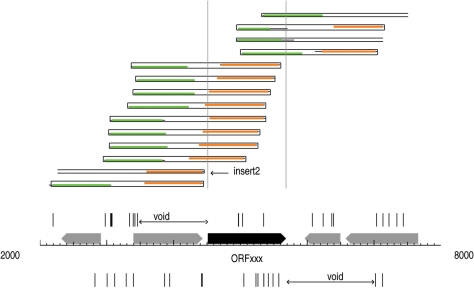
When preparing the libraries for microbial genome sequencing projects, the sizes of the fragments cloned are typically 2–3 kb in length. Since this size is large enough to encode most typical bacterial genes, the preparation of this shotgun library really constitutes a biological experiment. That experiment tests as to which segments of a given genome can be cloned successfully into Escherichia coli. Imagine that within the 2–3 kb fragment that is being cloned, there is a gene that is lethal to E. coli. Clones containing such fragments should be missing from the shotgun genome sequence data set and so should provide a visible feature, namely a gap in coverage, when analyzing the sequence reads. Typical of such lethal genes, are those that encode restriction enzymes (5). Such genes require the presence of a companion DNA methyltransferase gene to provide protection against the deleterious action of the encoded restriction enzyme as is found in the host organism (6). However, during the preparation of shotgun sequence clones, restriction enzyme genes become separated from their DNA methyltransferase gene and so if expressed in E. coli they would cleave the unmethylated E. coli genome unless a protective methylation is present, such as that provided by the dam methyltransferase gene used by E. coli as part of the mismatch repair machinery (7). This means that clones that would be expected to be present in the shotgun genome sequence data will be missing from the set and their absence can be detected by an absence of sequence reads beginning in the region either immediately upstream or downstream of the restriction enzyme gene. That is to say that clones that would normally begin immediately upstream of the gene and hence would contain the gene intact, should be missing from the data set. Similarly, clones that begin downstream of the gene and would provide sequence reads going back into the gene, would similarly be missing. This is illustrated schematically in Figure 1.
Figure 1.
Distribution of shotgun read starts around an uncloneable gene. A 6-kb long sequence is represented on the horizontal axis. Vertical marks above (and below) the axis show read starts mapping on the forward (and reverse) strand. The regions marked as ‘voids’ indicates that no reads were present beginning within the regions flanking the ORFxxx gene. Thick arrows represent ORFs. The rectangles above the marks show how the reads align to the sequence, green shows the read from the forward strand and orange shows the read from the complementary strand. Thinner black extension of the colored lines show parts of the reads that cannot be aligned to the final sequence. Paired end reads are shown by closed rectangles to display the full extent of the cloned insert. For unpaired reads, estimated sizes of the corresponding inserts are shown with open-ended boxes (e.g. insert 13). The absence of marks, labeled ‘voids’, on opposite strands before and after ORFxxx, and the specific staggered arrangement of boxes indicate that the gene was not cloned intact during the sequencing project.
We have written software that presents a visualization of the shotgun sequence coverage permitting the data to be analyzed very easily. In the case of H. influenzae, the very first genome studied, two of the known restriction enzymes had previously been cloned and characterized (8,9). Similarly, in the two sequenced strains of Helicobacter pylori, all of the potential Type II restriction-modification systems (RM systems) had been cloned out independently and assayed for activity (10,11). Thus, the shotgun sequence data for these genomes provides a test of the efficacy of the method. We now report the results of such tests as well as the analysis of several new genome shotgun sequence data sets where there had been no previous report of active restriction systems.
We would note that a recent paper has appeared, while this manuscript was in preparation that also addresses the issue of missing open reading frames (ORFs) in shotgun sequence data (12). That paper provided a genomewide view of ‘uncloneable genes’ in the context of horizontal gene transfer. Surprisingly, they did not mention any genes from RM systems as falling into the uncloneable category.
MATERIALS AND METHODS
Shotgun sequence data sets
Shotgun sequence data from three early genome sequencing projects (H. influenzae, H. pylori ATCC 26695 and Methanocaldococcus jannaschii) were provided by TIGR, Rockville, MD, USA. Additional shotgun sequence sets were downloaded from NCBI's TraceDB (www.ncbi.nlm.nih.gov/Traces/). Additional information was imported from TraceDB auxiliary data files, when these were available. This included the source of the DNA sequence information, whether or not reads had paired mates and whether the data came from shotguns, finishing primer walks, 454 experiments or from large insert clones where internal stops may interfere with strong expression of internal genes. We have used this information to identify matched paired ends (‘clone mates’) to estimate insert sizes, to exclude sequences from 454 experiments (which do not involve cloning), and to distinguish classes of inserts (initial shotgun, finishing primer walk, etc). In the absence of such explicit data, identification of reads from the same inserts (from opposite ends, or from internal positions), and computation of insert sizes was attempted by parsing the names of the reads. In particular, for the trace data obtained for H. pylori we calculated the average size of the reads, based on unequivocal paired ends, as 1550 with an SD of plus or minus 249. The key features of the shotgun data set used for the M. jannaschii analysis are summarized in Table 1.
Table 1.
Analysis of M. jannaschii shotgun sequence data
| Complete sequence | |
| Chromosome | 1 664 970 nt |
| Extra-chromosomal element 1 | 58 407 nt |
| Extra-chromosomal element 2 | 16 550 nt |
| Total shotgun sequence data | 32 564 505 nt |
| Total no. of clones | 21 109 |
| Total no. of reads | 39 519 |
| No. of reads from the ends of inserts | 36 015 |
| No. of reads internal to the insert | 3504 |
| Total no. of matched pair ends | 15 716 |
| Average read length | 824 |
| Fold coverage | 19.6 |
Complete genome sequences and annotations were retrieved from GenBank (13). In this study, we collected and analyzed 39 prokaryotic genomes (complete list available in Supplementary Table S1) for which REBASE (6) designations of RM systems were available. Most importantly, REBASE contained annotations for putative DNA methyltransferase genes. Since restriction enzyme and methyltransferase genes usually lie very close to one another we focused our search for potential gaps in shotgun sequence to regions of the genome close to those putative methyltransferase genes. We easily identified 32 potential Type II restriction enzymes with genes short enough (around 900 bp in size) to fit entirely into single inserts and a number of these were selected for biochemical analysis. These are the inserts that we do not expect to see in the shotgun set, because they possess genes for proteins that are likely to be lethal to the cloning host. A dozen of the selected genes encoded known restriction enzymes that had been characterized in other studies, while the rest are designated as putative restriction enzymes. When identifying proteins with similar functions, we used the COG system (14) and its annotations.
Trace data analysis programs
Our analysis first maps all shotgun sequence reads onto the finished genome, using the BLAST program. Strict criteria have been set to record hits (minimum score of 300, minimum aligned segments of 300 bp, with at least 94% identity). Only top scoring hits are retained, while secondary, lower scoring hits (5% below the top scores), which result from matches to diverged repeats of a sequence, are discarded. Some reads map with almost equal ‘top’ score to different segments of the genome where almost perfectly repeated sequences are present (e.g. the six ribosomal RNA operons in H. influenzae). In such cases it is not possible to identify the true origin of the clones, especially without knowing the entire sequence of the insert. Since we are looking for genes that are not covered, we chose to overestimate gene coverage, so we included every mapped, almost perfect repeat match.
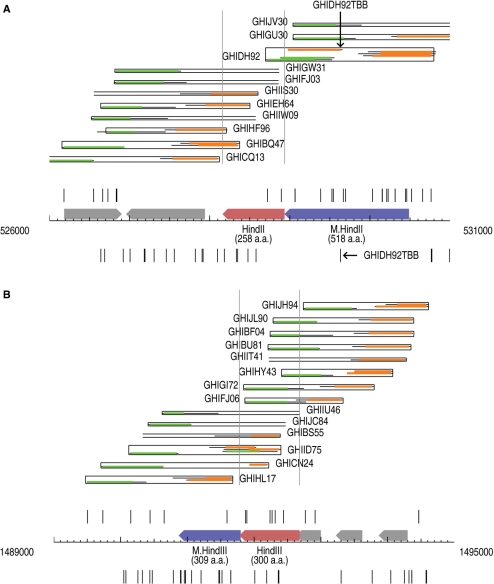
Based on the alignments, the starts of the mapped reads are recorded in a strand-specific manner along the genome. Random shotgun fragments would produce a random distribution of starts on the two strands, irrespective of gene locations, with a density typically around 10 per kilobase (number of reads/genome size). Because of the cloning step, however, starts corresponding to inserts for lethal genes will be missing from the plot, and we will not see reads that start close to the complete gene and would be likely to contain it intact. Thus, a matching pair of gaps in read starts is expected on the two strands surrounding the lethal gene. The data for the HindII and HindIII RM systems are shown in Figure 2.
Figure 2.
(A) Schematic representation of the shotgun reads in the vicinity of the HindII RM system. Extensive voids can be seen flanking the HindII R gene, but not the M gene. The shotgun clones do not contain the entire restriction enzyme gene. Note that the apparent exception indicated by the horizontal arrow at the bottom of the schematic does not represent a true clone start point, but is an internal read from the middle of insert GHIDH92, highlighted above by the vertical arrow. (B) Schematic representation of the shotgun reads in the vicinity of the HindIII RM system. See the legend for Figure 1 for an explanation of the symbols used.
When both ends of an insert are sequenced and mapped, we have calculated the exact length of the inserts. For clones where only one end is known we assume that the length is the average of the known lengths. We can also identify and discard chimeric, recombinant inserts, where presumably matching ends map to distant locations, where opposite ends map onto the same strand or where the expected ‘end’ reads lie internal to the insert. For unpaired reads, the full reach of each insert is guessed by successive invocation of parameters in the following order: annotated insert size, published insert sizes for classes, calculated average size of paired inserts for the same class, default values. We associated gene coverage numbers (number of inserts that contain the full length of a protein encoding gene) with the genes. Since insert lengths are typically around 2000 bp, longer genes tend to have low coverage numbers. For shorter genes, however, low coverage numbers may indicate toxicity. The targets of our specific interest, Type II restriction enzymes, typically have short genes (average length 900 bp). Therefore, our analysis looked for short genes with low coverage numbers. We overlaid insert coverage with read start distribution and with ORF data (Figure 2) around suspected restriction enzyme genes. These panels were inspected visually.
The first phase of a sequencing project (random fragmentation—cloning—insert end sequencing) is usually followed by a round of gap closing and repeat sequencing to resolve ambiguities. These experiments typically involve the creation of a small set of large insert (10–50 kb) fragments, and may use cloning vectors and sequencing strategies different from those of the first stage. The experiments may not involve cloning at all. When possible, we isolated data from these finishing stage experiments, and analyzed them separately.
When a gene is toxic to a new host, a homolog of the gene, presumably with the same function, is also expected to be toxic to the host. To test for such congruency, we compared the coverage of homologous genes, based on their COG assignments (14). For each non-covered gene, we checked the coverage of the orthologs. We also calculated the average coverage for all COGs.
Our batch analysis programs have been complemented with interactive visual analysis aids. We have developed computer software that works from precompiled data files and tables, and creates a graphic display of clone start distributions and insert reach figures by creating appropriate HTML pages dynamically. Selected genes can be highlighted to direct scrolling and zooming into regions of specific interest. At high resolution, vertical bars represent individual clone starts. These, and the gene-representing arrows are active; pop-up balloons appear on mouse-over to display relevant information (clone or gene names, sizes, locations, etc.). Clickable links bring up detailed sequence and alignment information, and additional visualization windows. Translation blocking and frameshifting mutations (identified by matching read translations to the gene products of genome CDS annotations), and chimeric clones can be optionally hidden from view. This tool that analyzes the trace data sets is available upon request from the authors. It should be noted that a certain amount of manual intervention is required to build the databases from which the visualizations are calculated. The results of the analyses described in this article can be found on the genome page of the REBASE server (http://tools.neb.com/~vincze/genomes/) (6). Genomes for which analyses are available are indicated by an ‘yes’ in the SG column and clicking on the ‘yes’ leads to a page with links to each RM system analysed. For each system the trace start diagrams are shown, but only the potential Type II R genes also have the trace map diagrams showing the detailed breakdown of the clones and the lengths of the individual reads.
Analysis of the HindV RM system of H. influenzae
The ORF HI1041 encoding the putative HindV methyltransferase was amplified from H. influenzae genomic DNA using primers H1 and H2 (Supplementary Table S2). The amplified DNA was cut with BamHI and SalI, ligated into the vector pACYC184 previously cut with BamHI and SalI and transformed into ER2566. Plasmid and genomic DNAs were isolated from cells carrying the HindV methyltransferase gene, and both DNAs were found to be resistant to digestion with BsaHI endonuclease, which recognizes the DNA sequence GRCGYC that HindV is predicted to recognize (R.D.M., unpublished results).
The ORF encoding the putative HindV endonuclease gene, HI1040, was amplified from H. influenzae genomic DNA using primers H3 and H4 (Supplementary Table S2). The amplified DNA was cut with NdeI and SalI and ligated into the T7 expression vector pAII17 (15), previously cut with NdeI and SalI, and transformed into competent ER2566 cells carrying the HindV methyltransferase expressing construct on the compatible plasmid pACYC184. Plasmid DNA was isolated from the transformed host, sequenced and clones containing the correct putative HindV endonuclease sequence were tested for endonuclease expression. Cells were grown to late log phase in 250 ml LB broth containing 0.1 mg/ml ampicillin and 0.025 mg/ml chloramphenicol, induced with 0.5 mM IPTG and grown for an additional 2 h. Cells were harvested by centrifugation. Cells (0.9 g) were suspended in 10 ml sonication buffer (20 mM Tris–HCL, 1 mM DTT, 0.1 mM EDTA, pH7.5 at 25°C), lysed by sonication and assayed for endonuclease activity using λDNA (40 sites for GRCGYC) and pBR322 DNA linearized with PstI (six sites for GRCGYC) in NEBuffer 4. No endonuclease activity was observed for any of the clones, even though the plasmids had the correct HindV putative gene sequence. A vector carrying the HindV putative endonuclease gene was transformed into ER2566 cells lacking the methyltransferase construct, but even when induced there was no deleterious effect on cell growth, nor was any endonuclease activity detected in cell extracts. Thus, even though on the basis of sequence similarity it seemed likely that this gene might encode an active restriction endonuclease we have been unable to detect any evidence for activity.
Analysis of ORFs MJ563, MJ1200, MJ1209 of M. jannaschii
MJ1209 was originally reported (GenBank: NC_000909) as one large ORF (485 AAs) containing two frameshifts and labeled as a DNA methyltransferase. This large ORF is really two smaller ORFs (1 152 607–1 153 263 and 1 153 254–1 154 044) that were expressed separately. Two sequence errors were found and corrected by resequencing (GenBank: EU363462). The methyltransferase ORF was expressed in pLITMUS38. This plasmid DNA was fully protected against in vitro cleavage by MspI when isolated from stationary phase cultures. A clone of the putative endonuclease gene was not obtained in E. coli cells expressing the methylase construct, suggesting that the endonuclease was toxic to the host, likely due to incomplete methyltransferase protection of the host genome during rapid growth conditions.
ORFs MJ563 and MJ1200 were cloned and crude extracts of cells were prepared. Attempts to detect methyltransferase activity using 3H-S-adenosylmethionine as methyl donor and bacteriophage λDNA as substrate were inconsistent. 3H counts incorporated into λDNA by MJ563 and MJ1200 varied from one to two times background, in contrast to MJ1209 extracts that produced counts of 400 times background.
In vitro transcription/translation analysis
A reconstituted in vitro transcription/translation (IVTT) system, the PURESYSTEM (Post Genome Institute, Tokyo, Japan), was used to assay the DNA cleavage activity of the products of the predicted ORFs. The template DNA for IVTT was generated by a two-step PCR process: the first PCR to amplify the complete ORF from the genomic DNA and the second PCR to attach a T7 promoter together with a ribosome binding site for driving in vitro protein synthesis. All PCRs were carried out with Phusion polymerase (Finnzymes, Espoo, Finland) with proofreading activities to minimize amplification errors (see Supplementary Table S2 for a list of primers used). All PCRs were purified using the spin-column procedure (Qiagen, Valencia, CA, USA). In vitro protein expression was carried out by combining 1 μl of purified template DNA with 8 μl of PURESYSTEM mix and incubating at 37°C for 1 h. IVTT reaction mix (2 μl) was then used to digest 1 μg of λDNA at 37°C for 1 h in a 50 μl reaction volume. The digestion reaction was supplemented with 2 μl of RNase A (0.02 μg/μl, Ambion, Austin, TX, USA) to remove any excess RNA species present in the PURESYSTEM mix. The reaction mix was then purified using a spin-column (Qiagen) and subjected to agarose gel electrophoresis.
RESULTS
Haemophilus influenzae Rd RM systems
The H. influenzae Rd genome is known to encode two fully active Type II restriction enzymes, HindII and HindIII (16,17). These are encoded by ORFs HI0512 and HI1393, respectively. We examined the sequence reads from the original shotgun sequence data set in the vicinity of these two genes as shown in Figure 2. It can be seen that in both cases there are substantial gaps in the sequence coverage immediately upstream and downstream of the restriction enzyme genes. It should be noted that one read (GHID92TBB), beginning 600 nt downstream of the HindII gene, is present in the data set and would normally be expected to contain the intact HindII gene. However, examination of the DNA sequence of this clone shows that it is actually an internal read from a larger insert, GHIDH92. This insert clearly does not cover the entire restriction enzyme gene.
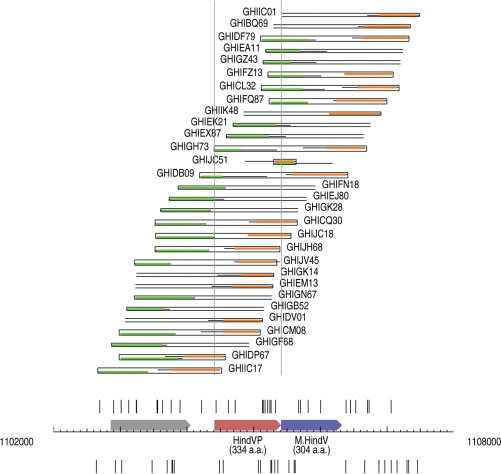
One interesting potential system, called the HindV system, is encoded by ORFs HI1040 and HI1041, which show strong similarity to a related system, HgiDI (recognition sequence: GRCGYC), from Herpetosiphon giganteus Hp2 (18). Prior to the current data analysis we had previously checked this system as a potential new RM system encoded by H. influenzae and had found that the M gene was active based on its ability to protect DNA against the action of the restriction enzyme BsaHI (an isoschizomer of HgiDI) in vitro. We also checked genomic DNA from H. influenzae and found that it too was resistant to BsaHI, indicating the gene was fully active in vivo also. However, the R gene was inactive both in vivo, where no activity has ever been detected, and in vitro when overexpressed from the T7 expression vector pAII17. The shotgun sequence data confirms the view that the restriction enzyme gene in this case is inactive as it can be cloned from a whole genome shotgun quite successfully into E. coli without any deleterious effects (Figure 3). Our analysis identifies three clones (GHIJC18, GHICQ30, GHIDB09) with certainty, and three more (GHIGK28, GHIEJ80, GHIFN18, with missing read mates) which potentially contain the entire gene. Table 2 contains a summary of our analysis of the H. influenzae Rd RM systems.
Figure 3.
Schematic representation of the shotgun reads in the vicinity of the HindV RM system. The read starts around the HindVP (HI1040) gene show that several shotgun clones cover the gene fully. The gene is cloneable and presumed to lead to an inactive R protein. Note that the strange-looking insert in GHIJC51 is a very small one and the trace sequence contains flanking regions from the cloning vector.
Table 2.
Summary of predicted and experimentally determined RE genes
| ORF # | Enzyme name | Predicted from shotgun data | Experimental results | Reference |
|---|---|---|---|---|
| Haemophilus influenzae Rd (fold coverage = 12.0) | ||||
| HI0512 | HindII | Active | Active | (14) |
| HI1393 | HindIII | Active | Active | (15) |
| HI1041 | HindVP | Inactive | Inactive | This work |
| Methanocaldococcus jannaschii (fold coverage = 19.6) | ||||
| MJ0984 | MjaI | Inactive | Active | This work + US patenta |
| MJ1449 | MjaII | Active | Active | This work |
| MJ0600 | MjaIII | Inactive | Active | RDM unpublished |
| MJ1327 | MjaIV | Active | Active | This work |
| MJ1500 | MjaV | Inactive | Active | RDM unpublished |
| MJ1209 | MjaVIP | Active | Unknown | This work |
| Helicobacter pylori ATCC 26695 (fold coverage = 12.0) | ||||
| HP0052 | HpyAVIP | Inactive | Inactive | (11) |
| HP0053 | HpyAV | Active | Active | (11) |
| HP0091 | HpyAIII | Inactive | Active | (11) |
| HP0503 | HpyAXII | Inactive | Active | (19) |
| HP0909 | HpyAIXP | Inactive | Inactive | (11) |
| HP1209 | HpyAIP | Inactive | Inactive | (11) |
| HP1351 | HpyAIV | Active | Active | (11) |
| HP1366 | HpyAII | Inactive | Active | (11) |
| Bacillus cereus ATCC 10987 (fold coverage = 11.2) | ||||
| BCE4604 | BceSII | Active | Active | This work |
| Methylococcus capsulatus (fold coverage = 13.8) | ||||
| MCA1617 | McaTI | Active | Active | This work |
aRoberts, R.J., Byrd, D.R., Morgan, R.D., Patti, J., Noren, C.J. US Patent Office (2004).
US 6689573 B Method for screening restriction endonucleases based on database homology searching of cognate DNA methylase sequences.
Methanocaldococcus jannaschii RM systems
Analysis of the shotgun data shows that MjaII, MjaIII and MjaV cannot be cloned, which confirms that they are active restriction enzymes. At first glance, MjaIV appears able to be cloned, in that one insert (GG79) covers the gene entirely. However, close inspection reveals that a frameshift occurs in the sequence of the only read (GG79TAA226B), which extends into the restriction enzyme gene. As a result, the insert would encode a truncated, probably inactive version of the otherwise toxic protein that is 71 amino acids shorter than the full-length gene product. The CTAG-specific enzyme, MjaI, is the only M. jannaschii restriction enzyme where shotgun data does not indicate cloning difficulties. Eight inserts contain the gene in full and show no sign of frameshifts or other problems. The coding region for MjaI contains an unusual start codon, GTG instead of ATG. While GTG can be acceptable as a start codon in E. coli, since about 17% of its own genes start with GTG, there may be other local sequence effects that prohibit expression at a high enough level to cause problems. It should be noted that an isoschizomer of MjaI has been successfully cloned in E. coli and it too contains a GTG start codon but shows no activity in that system despite being active in its original host (20).
The putative RM systems in M. jannaschii were examined for potential restriction enzyme activity by cloning individual systems into E. coli and using a traditional biochemical assay to test for activity. The systems MjaI (when the GTG native start codon was replaced by ATG), MjaII, MjaIII, MjaIV and MjaV all showed restriction enzyme activity when expressed from clones and could also be detected in M. jannaschii when large amounts of cells were used to prepare extracts.
The putative MjaVI restriction system, encoded by ORF MJ1209, was originally annotated as a single ORF with frameshifts. We recloned and sequenced this region of the M. jannaschii genome and discovered two sequence errors (G replaces T at position 1 153 482 and an extra T needed to be inserted after position 1 153 489) (GenBank: EU363462). This results in the annotated pseudogene being split into two separate ORFs: the first from position 1 153 254 to position 1 154 044 encoding a putative methyltransferase, while the second from 1 152 607 to 1 153 263 encoding a putative restriction enzyme. Cloning the first ORF in pLITMUS38 showed that it possessed methyltransferase activity in that it rendered the plasmid DNA completely resistant to MspI. Since MspI can cut when the inner cytosine in its recognition sequence, CCGG, is either 5-methylcytosine or N4-methylcytosine (6), this means that M.MjaVI must be methylating the outer cytosine. Examination of the conserved motifs found in this protein show that it is clearly an N4-cytosine methyltransferase that would thus form m4CCGG. The smaller ORF that follows the frameshift could not be successfully cloned in E. coli. We presume that this means there is an active endonuclease gene, although further work is required to confirm that. It should be noted that there is very high sequence similarity between both of these genes and a putative methyltransferase and restriction endonuclease in Thermosipho melanesiensis BI429 (GenBank: NC_009616; Copeland, A. et al., unpublished results).
There are two putative m5C methyltransferases encoded by ORFS MJ563 and MJ1200. These two ORFs have the sequence PCE rather than PCQ in the catalytic motif 4 found in confirmed m5C DNA methyltransferases (21). We cloned both of these ORFs and tried to detect DNA-specific methyltransferase activity by looking for incorporation of 3H-SAM into λDNA or other DNA. Unfortunately, the results were not definitive and it seems likely that neither of these genes encodes an m5C-DNA methyltransferase. While we think it unlikely these are DNA methyltransferases, we cannot rule out the possibility that they are protein or RNA methyltransferases. Table 2 contains a summary of our analysis of the M. jannaschii RM systems.
Helicobacter pylori RM systems
Previously the RM systems in H. pylori had been identified by sequence analysis and each of the candidate systems had been carefully tested for activity of both the DNA methyltransferase genes and their associated putative restriction enzyme genes (10,11). Four active Type II restriction enzymes had been identified and experimentally verified in H. pylori strain ATCC 26695: HpyAII, HpyAIII, HpyAIV and HpyAV (11). The analysis of shotgun sequence data also indicates that HpyAII is active, since no insert fully contains the gene. No insert with paired ends covers fully the HpyAV gene either, consistent with toxicity of the restriction enzyme gene. Two unpaired reads, however, spoil the otherwise significant gaps in read starts around this gene. A calculation of the length of inserts with paired ends shows that the shotgun clones are considerably shorter (closer to 1500 bp), than the expected 2000 bp average based on earlier whole genome shotguns (3) which would suggest that the two inserts probably do not cover the HpyAV gene entirely. Several inserts fully cover the HpyAIII gene, which means that the host tolerates this restriction enzyme gene. This is expected since the specificity of HpyAIII is GATC, so the dam+ host, in which the shotgun was prepared, would be protected from HpyAIII restriction.
Shotgun sequence coverage of HpyAIV suggests the restriction enzyme is inactive, since three inserts (two with paired ends) extend over the entire gene. A detailed analysis of the sequences offers several possible explanations for this unexpected result. In all four sequenced Helicobacter orthologs of HpyAIV, the endonuclease genes are downstream from, and overlapping with their methylase partners. The production of restriction enzyme protein may require polycistronic mRNA of the entire RM system, which cannot be transcribed from the observed shotgun inserts. Another possibility is that the published sequence does not encode an active enzyme. The almost identical ortholog Hpy99IXP from the J99 strain has been proven inactive (11). The genes contain homopolymer runs, and are susceptible to phase variation (22). It is conceivable, therefore, that the previous work showing active HpyAIV picked up a non-canonical, active variant of the gene (11). Table 2 contains a summary of our analysis of the H. pylori RM systems.
Bacillus cereus ATCC10987
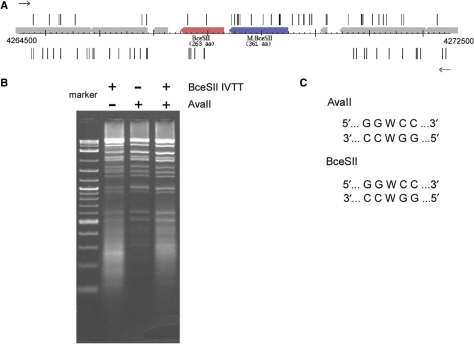
Recently, the complete genome sequence for B. cereus ATCC10987 has been reported (23). One putative RM system encoded by genes BCE4604 and BCE4605 could be predicted to recognize GGWCC on the basis of similarity to the M gene of the previously characterized AvaII RM system and the R gene of the HgiCII RM system (both recognize GGWCC) (6). The M gene was 46.7% identical to residues 73–425 of M.AvaII, while the R gene showed limited similarity (e−7) to HgiCII. Analysis of the first stage (short insert) shotgun sequence data for this genome showed that the putative restriction enzyme gene, BCE4604, had large gaps both upstream and downstream of the coding sequence (Figure 4A and Supplementary Figure S1) indicating that it was likely an active restriction endonuclease. The large insert library panel shows the same gaps around the ORF, suggesting that the B. cereus promoter driving this system works well in E. coli. PCR primers were prepared and the gene was amplified from genomic DNA and subjected to an in vitro transcription/translation reaction. A portion of this reaction was then used to digest bacteriophage λDNA. Analysis of the digest by agarose gel electrophoresis is shown in Figure 4B. It can be seen that it gives rise to the characteristic banding pattern produced by AvaII and the double digest between the new enzyme called BceSII and AvaII showed no increase in the number of bands, indicating that they both had the same specificity. While the position of cleavage has not been determined for BceSII, based on previous experience (6) it is likely that it is identical to the cleavage specificity of AvaII and HgiCII as shown in Figure 4C. Table 2 contains a summary of our analysis of the B. cereus Rd RM systems.
Figure 4.
In vitro transcription/translation of the BceSII RM system. (A) Map of the sequencing trace starts around the genomic region of the BceSII RM system. The R.BceSII gene is in red and the M gene is in blue. Trace starts are shown in the forward direction (upper) and the reverse direction (lower). (B) Lane 1, DNA size markers; Lane 2, digestion of λDNA using in vitro translated BceSII; Lane 3, digestion of λDNA using purified AvaII (NEB); Lane 4, double-digestion of λDNA using in vitro translated BceSII and AvaII. (C) DNA recognition sequences of BceSII and AvaII.
Methylococcus capsulatus
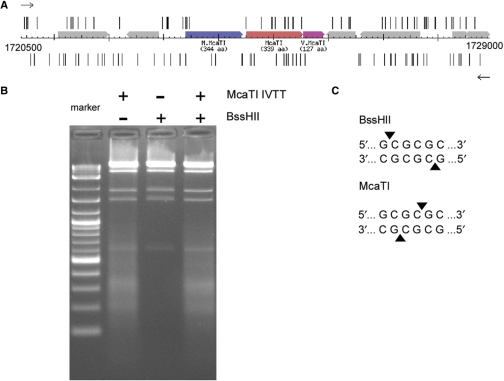
A second recently sequenced genome is that of M. capsulatus (24). In this case, the gene MCA1616 appears to encode a typical C5 DNA methyltransferase. Adjacent to it is an ORF, MCA1617, which shows no similarity to any other gene in GenBank, while the next gene downstream, MCA1618, shows strong similarity to the Vsr endonuclease of E. coli K (25,26). It is quite common for RM systems that use 5-methylcytosine as the protecting agent to have such a Vsr endonuclease gene associated with it and so the intermediate ORF, MCA1617, was a likely candidate for the restriction enzyme gene of this system. Analysis of the first stage shotgun sequence data set shows that the gene cannot be cloned into E. coli in either direction (Figure 5A and Supplementary Figure S2). Although several long inserts contain the ORF, the large insert panel still shows two flanking gaps (Supplementary Figure S3). The spacing of the two gaps is such that in all inserts, the start of the restriction enzyme gene is well downstream, at least 6 kb away from the upstream insertion site. The lethal gene may be tolerated because it is not expressed when long upstream sequences are present; the Methylococcus (63% G + C) regulatory elements are probably not recognized by the E. coli (50% G + C) machinery. Testing the gene's activity by in vitro transcription/translation shows that it does indeed encode an active restriction enzyme (Figure 5B). The specificity was determined to be GCGCGC, just like that of BssHII using run-off sequencing (27). However, when the cleavage site was determined, it was found to cleave at a different position within the recognition sequence, and in contrast to BssHII that leaves a 5′ tetranucleotide extension (G↓CGCGC), McaTI leaves a two nucleotide 3′ extension (GCGC↓GC) (Figure 5C). This is a new and unique specificity. Table 2 contains a summary of our analysis of the M. capsulatus RM systems.
Figure 5.
In vitro transcription/translation of the McaTI RM system. (A) Map of the sequencing trace starts around the genomic region of the McaTI RM system. The R.McaTI gene is in red, the M gene is in blue and the V.McaTI gene is in pink. Trace starts are shown in the forward direction (upper) and the reverse direction (lower). (B) Lane 1, DNA size markers; Lane 2, digestion of λDNA using in vitro translated McaTI; Lane 3, digestion of λDNA using purified BssHII (NEB); Lane 4, double-digestion of λDNA using in vitro translated McaTI and BssHII. (C) DNA cleavage specificities of the neoschizomers BssHII and McaTI.
The information analyzed by this method for many other potential restriction enzyme genes can be found in REBASE (6) and is accessible from the REBASE Genomes icon (tools.neb.com/∼vincze/genomes/).
DISCUSSION
In this article, we have described a new method by which restriction endonuclease genes can be identified in newly sequenced bacterial DNAs by careful examination of shotgun sequence data. Because such genes are lethal when expressed in E. coli in the absence of protective methylation, sequence reads corresponding to the 5′ ends of such clones that would normally contain these genes are missing from the sequence data set. This leads to gaps in the coverage that can be identified readily. Analysis of a number of such genome shotgun sequence data sets, summarized in Table 2, has shown that this is a useful general method for the identification of restriction enzyme genes and has already led to the identification of a new specificity present in M. capsulatus.
It should be noted that despite the fact that the gene for McaTI has two copies of its recognition site in the gene, this did not apparently interfere with our ability to make enough enzyme in vitro to test an external substrate. In general, most restriction enzyme genes lack the recognition sequence of the restriction enzyme they encode (6) and so it is unlikely that destruction of the template would be a general problem in detecting restriction enzyme activity using this in vitro technique. Also, with the exception of many Type IIS restriction enzymes, the high specific activity of most restriction enzymes means that sufficient quantities can easily be made in vitro to permit their detection and preliminary characterization.
Because many other genes are lethal when expressed in E. coli, the gaps in the sequence coverage are not limited to restriction endonuclease genes and so, one key feature of the method when applied to the identification of restriction enzyme genes is that in addition to a gap in the sequence, there must also be a nearby DNA methyltransferase gene. Fortunately, these genes are easy to spot because of the presence of characteristic motifs that enable their identification (21,28).
One important consideration in the analysis we present is that it is only those restriction enzyme genes that are lethal to the E. coli strain used for the shotgun sequence preparation that are detected by this methodology. In particular, it is common practice among the sequencing centers to use strains of E. coli that contain the Dam methylase. This methylase recognizes the sequence GATC and modifies the adenine residue to form N6-methyladenine (7). During the analysis of the shotgun sequence data from H. pylori we noticed that HpyAIII (recognition sequence: GATC) was easily cloned into the E. coli that was used to prepare the shotgun data set. This enzyme is part of an RM system in which the cognate methylase is also an N6A-methyltransferase, just like Dam (11). Also in M. jannaschii, we noticed that one restriction enzyme gene, encoding the enzyme MjaIII (recognition sequence GATC) could be cloned perfectly satisfactorily into E. coli. Again the reason is that the E. coli strain used to prepare the shotgun was dam+ and that protects against the action of the MjaIII restriction enzyme. The corresponding DNA methyltransferase, M.MjaIII, must be an N6A-methyltransferase since M. jannaschii DNA can be cleaved by Sau3AI, which is known to be blocked by both N4-methylcytosine and 5-methylcytosine in its recognition sequence (6).
There are several other important caveats about the analysis we present. First, only a positive signal—i.e. a pair of gaps on both side of the gene, combined with a proximal DNA methyltransferase gene—can be read as a strong indication of an active restriction enzyme gene. Some genes such as that encoding MjaI, which we know is an active restriction enzyme, can be cloned under the shotgun protocol. Presumably, there are sequence factors that preclude its strong expression in E. coli and do not lead to sufficient levels of the enzyme to induce lethality. This behavior of certain restriction enzymes has been noted before (29). Indeed, MthZI, an isoschizomer of MjaI from Methanococcus thermoformicicum that uses a GTG start codon is inactive when cloned into E. coli (20). Second, the data available from the initial stage of shotgun sequence analysis is more likely to give reliable gaps than that from later stages, where longer inserts are used. In this case, transcription of the potential restriction enzyme gene may be driven by the cognate promoter, rather than from a flanking promoter on the vector, and the strength may be insufficient to produce a lethal phenotype. It is already known that some restriction enzyme genes can be cloned alone if their activity is weak (R.D.M., unpublished results). Presumably in these cases the normal E. coli DNA repair machinery can absorb the damage. Finally, sometimes the data analysis can be misleading when apparent gaps are interrupted by one or more clones that either are short and do not extend through the gene or are chimeras that contain only a fraction of the gene. Thus, while positive results are very strong, negative results may be worth more careful interpretation.
One surprise from our analysis is that specificity subunits of Type I RM systems show up repeatedly on the non-cloneable list. In their original role, these proteins form complexes with restriction and methyltransferase components, and provide the recognition specificity for methylation and for restriction. By themselves, when produced incidentally in shotgun experiments, they may interfere with the host cell's normal functions by simply binding to specific sites on the chromosome. The S proteins could turn lethal also by complementing R and M subunits of a native Type I system. If the new specificity is different from the native one, the unmethylated chromosome of the host would be digested. However, such genes are usually not present in the strains used for shotgun sequencing. In any event, this induced lethality may be interesting and point to unknown biochemical problems that may occur in the presence of orphan S subunits.
While our analysis to date has focused on genes that form part of potential restriction modification systems, we have noticed that a number of other genes are detected by our shotgun data analysis. These include many other, non-cloneable genes and gene families, including DNA binding regulators, ribosome-related genes, transcription initiation and transcription elongation factors, cell division trigger factors, some kinases and several membrane proteins. Of the ∼10 000 short COGs (average gene length <1200 bp), 35 have multiple (between 4 and 8) non-covered members. No orthologous group will have all its members consistently implicated. For instance, some genes may be inactive or dead. In the case of membrane proteins we anticipate that the expression and insertion of a foreign protein into the membrane of E. coli may cause a severe disruption of its function and effectively kill the cells. Alternatively, a well-expressed gene may overwhelm the membrane synthesis machinery. There are likely many other genes that can be identified using this method that will provide the functional evidence that can be useful in annotating genomes. A recent article that appeared while this manuscript was being prepared has focused much more on these other ORFs that are missing from shotgun sequence data sets (12).
This analysis shows clearly that the experimental data which has proven so useful in assembling genome sequences can be of great use in discerning the function of the genes encoded by that genome. While many groups have deposited their data into the Trace archive run by NCBI, more groups have not. We would encourage these groups that have provided the final DNA sequence of their genomes into the DNA sequence databases to also deposit this trace data. It is an extremely valuable resource for the community and should be subject to the usual ethical principals of data deposition.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: New England Biolabs, Inc.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are extremely grateful to Dr Claire Fraser and Dr Steven Salzberg for recovering the original trace data for H. influenzae, M. jannaschii and H. pylori ATCC 26695 from the basement of TIGR. We also thank those other investigators who have deposited their trace data with NCBI. We thank Karen Otto for help in preparing the article.
REFERENCES
- 1.Messing J, Crea R, Seeburg PH. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981;9:309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baer R, Bankier AT, Biggin MD, Deininger PL, Farrell PJ, Gibson TJ, Hatfull G, Hudson GS, Satchwell, S.C. Segui C, et al. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 3.Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb J.-F, Dougherty BA, Merrick JM, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 4.Venter JC, Adams MD, Myers G, Li P, Mural RJ, Sutton G, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 5.Howard KA, Card C, Benner JS, Callahan HL, Maunus R, Silber K, Wilson G, Brooks JE. Cloning the DdeI restriction-modification system using a two-step method. Nucleic Acids Res. 1986;14:7939–7951. doi: 10.1093/nar/14.20.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE–enzymes and genes for DNA restriction and modification. Nucleic Acids Res. 2007;35:D269–D270. doi: 10.1093/nar/gkl891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marinus MG, Morris NR. Isolation of deoxyribonucleic acid methylase mutants of Escherichia coli K-12. J. Bacteriol. 1973;114:1143–1150. doi: 10.1128/jb.114.3.1143-1150.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito H, Sadaoka A, Kotani H, Hiraoka N, Nakamura T. Cloning, nucleotide sequence, and expression of the HincII restriction-modification system. Nucleic Acids Res. 1990;18:3903–3911. doi: 10.1093/nar/18.13.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nwankwo DO, Moran LS, Slatko BE, Waite-Rees PA, Dorner LF, Benner JS, Wilson GG. Cloning, analysis and expression of the HindIII R-M-encoding genes. Gene. 1994;150:75–80. doi: 10.1016/0378-1119(94)90861-3. [DOI] [PubMed] [Google Scholar]
- 10.Kong H, Lin L.-F, Porter N, Stickel S, Byrd D, Posfai J, Roberts RJ. Functional analysis of putative restriction-modification system genes in the Helicobacter pylori J99 genome. Nucleic Acids Res. 2000;28:3216–3223. doi: 10.1093/nar/28.17.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin LF, Posfai J, Roberts RJ, Kong MH. Comparative genomics of the restriction-modification systems in Helicobacter pylori. Proc. Natl Acad. Sci. USA. 2001;98:2740–2745. doi: 10.1073/pnas.051612298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sorek R, Zhu Y, Creevey CJ, Francino MP, Bork P, Rubin EM. Genome-wide experimental determination of barriers to horizontal gene transfer. Science. 2007;318:1449–1452. doi: 10.1126/science.1147112. [DOI] [PubMed] [Google Scholar]
- 13.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2007;35:D21–D25. doi: 10.1093/nar/gkl986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong H, Kucera RB, Jack WE. Characterization of a DNA polymerase from the hyperthermophile archaea Thermococcus litoralis. Vent DNA polymerase, steady state kinetics, thermal stability, processivity, strand displacement and exonuclease activities. J. Biol. Chem. 1993;268:1965–1975. [PubMed] [Google Scholar]
- 16.Smith HO, Wilcox KW. A restriction enzyme from Hemophilus influenzae. Purification and general properties. J. Mol. Biol. 1970;51:379–391. doi: 10.1016/0022-2836(70)90149-x. [DOI] [PubMed] [Google Scholar]
- 17.Old R, Murray K, Roizes G. Recognition sequence of restriction endonuclease III from Hemophilus influenzae. J. Mol. Biol. 1975;92:331–339. doi: 10.1016/0022-2836(75)90232-6. [DOI] [PubMed] [Google Scholar]
- 18.Dusterhoft A, Erdmann D, Kroger M. Stepwise cloning and molecular characterization of the HgiDI restriction-modification system from Herpetosiphon giganteus Hpa2. Nucleic Acids Res. 1991;19:1049–1056. doi: 10.1093/nar/19.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Humbert O, Salama NR. Functional characterization of HP0503: an adenine methyltransferase required for the virulence of Helicobacter pylori. Abstr. Gen. Meet. Am. Soc. Microbiol. 2007;107:39. [Google Scholar]
- 20.Nolling J, de Vos WM. Identification of the CTAG-recognizing restriction-modification systems MthZI and MthFI from Methanobacterium thermoformicicum and characterization of the plasmid-encoded mthZIM gene. Nucleic Acids Res. 1992;20:5047–5052. doi: 10.1093/nar/20.19.5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pósfai J, Bhagwat AS, Pósfai G, Roberts RJ. Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res. 1989;17:2421–2435. doi: 10.1093/nar/17.7.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skoglund A, Bjorkholm B, Nilsson C, Andersson AF, Jernberg C, Schirwitz K, Enroth C, Krabbe M, Engstrand L. Functional analysis of the M.HpyAIV DNA methyltransferase of Helicobacter pylori. J. Bacteriol. 2007;189:8914–8921. doi: 10.1128/JB.00108-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rasko DA, Ravel J, Økstad OA, Helgason E, Cer RZ, Jiang L, Shores KA, Fouts DE, Tourasse NJ, Angiuoli SV, et al. The genome sequence of Bacillus cereus ATCC 10987 reveals metabolic adaptations and a large plasmid related to Bacillus anthracis pXO1. Nucleic Acids Res. 2004;32:977–988. doi: 10.1093/nar/gkh258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ward N, Larsen Ø., Sakwa J, Bruseth L, Khouri H, Durkin AS, Dimitrov G, Jiang L, Scanlan D, Kang KH, et al. Genomic insights into methanotrophy: the complete genome sequence of Methylococcus capsulatus (Bath) PLOS Biol. 2004;2:1616–1628. doi: 10.1371/journal.pbio.0020303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sohail A, Lieb M, Dar M, Bhagwat AS. A gene required for very short patch repair in Escherichia coli is adjacent to the DNA cytosine methylase gene. J. Bacteriol. 1990;172:4214–4221. doi: 10.1128/jb.172.8.4214-4221.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hennecke F, Kolmar H, Brundl K, Fritz H.-J. The vsr gene product of E. coli K-12 is a strand- and sequence-specific DNA mismatch endonuclease. Nature. 1991;253:776–778. doi: 10.1038/353776a0. [DOI] [PubMed] [Google Scholar]
- 27.Zhu Z, Samuelson JC, Zhou J, Dore A, Xu S-Y. Engineering strand-specific DNA nicking enzymes from the Type IIS restriction endonucleases BsaI, BsmBI, and BsmAI. J. Mol. Biol. 2004;337:573–583. doi: 10.1016/j.jmb.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 28.Klimasauskas S, Timinskas A, Menkevicius S, Butkiene D, Butkus V, Janulaitis A. Sequence motifs characteristic of DNA[cytosine-N4]methylases: similarity to adenine and cytosine-C5 DNA-methylases. Nucleic Acids Res. 1989;17:9823–9832. doi: 10.1093/nar/17.23.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lunnen KD, Barsomian JM, Camp RR, Card CO, Chen S.-Z, Croft R, Looney MC, Meda MM, Moran LS, Nwankwo DO, et al. Cloning Type II restriction and modification genes. Gene. 1988;7:25–32. doi: 10.1016/0378-1119(88)90242-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.