Abstract
Riboswitches regulate gene expression through direct, small molecule–mRNA interactions. The creation of new synthetic riboswitches from in vitro selected aptamers benefits from rapid, high-throughput methods for identifying switches capable of triggering dramatic changes in gene expression in the presence of a desired ligand. Here we present a flow cytometry-based screen for identifying synthetic riboswitches that induce robust increases in gene expression in the presence of theophylline. The performance characteristics of our newly identified riboswitches exceed those of previously described natural and synthetic riboswitches. Sequencing data and structure probing experiments reveal the ribosome binding site to be an important determinant of how well a switch performs and may provide insights into the design of new synthetic riboswitches.
INTRODUCTION
Riboswitches are genetic control elements that regulate gene expression via small molecule–RNA interactions (1). Riboswitches are typically comprised of an aptamer domain, which recognizes the ligand, and an expression platform that couples ligand binding to a change in gene expression. Over the past several years, a number of riboswitches that control gene expression in a variety of organisms have been reported (2). In addition to these natural riboswitches, several groups have developed synthetic riboswitches, which operate through similar principles to those of natural riboswitches, but are created from aptamers that have been selected in the laboratory (3–7). Because methods to select RNA aptamers are well established (8,9), it is, in principle, possible to develop synthetic riboswitches that respond to a great variety of ligands. Such riboswitches could be useful for controlling gene expression in a variety of organisms that lack orthogonal inducible expression systems, and would open the door to reprogramming cell metabolism and behavior (10). A key to these efforts is the ability to rapidly and inexpensively create new synthetic riboswitches from new or existing aptamers.
Ideally, a method for discovering riboswitches should be rapid, inexpensive, and very high-throughput. Additionally, the method should have the ability to quantitatively distinguish the switches with the best performance characteristics (i.e. for an ‘on’ switch, this would typically mean low background expression in the absence of ligand and robust increases in gene expression in the presence of the ligand) from those switches that are leaky, or only weakly activate (or repress) gene expression in the presence of the ligand. Recently, our lab (11,12), and others (13,14) have reported a variety of methods to screen RNA libraries for robust-performing synthetic riboswitches. Using a yeast-based screening method, Suess and coworkers screened libraries of up to 50 000 members and isolated riboswitches that reduced gene expression 7.5-fold in the presence of the antibiotic neomycin (13). Working in Escherichia coli, Wieland and Hartig screened a 64-member library of theophylline-dependent aptazymes and discovered constructs that activated gene expression 10-fold (14). Also working in E. coli, our lab has developed both a high-throughput screen (11) and a high-throughput selection (12) for theophylline-dependent riboswitches. In our screening method, the theophylline aptamer was cloned into the 5′-untranslated region of the β-galactosidase reporter gene (lacZ), and the sequence of the region between the aptamer and the ribosome binding site (RBS) was randomized to create libraries of up to 65 000 members. These libraries were used to transform E. coli, which were then grown on media containing X-gal and lacking theophylline. The whitest clones (i.e. those clones with the lowest levels of β-galactosidase expression) were picked and assayed for β-galactosidase activity in the presence and absence of theophylline. Functioning riboswitches were identified by increased β-galactosidase expression in the presence of theophylline. The switches identified using this screen displayed improved characteristics over our previously reported synthetic riboswitch (3), including lower levels of gene expression in the absence of theophylline and robust increases (up to 36-fold) in its presence (1 mM). Although sequence data allowed us to propose and test a mechanism for how these new riboswitches functioned, and illuminated some principles that may ultimately enable the de novo design of new synthetic riboswitches from existing RNA aptamers, we believe that at present, genetic screens or selections remain the best strategy for discovering new synthetic riboswitches.
Toward that end, we developed a motility-based genetic selection for synthetic riboswitches (12). In this method, a library of potential riboswitches is cloned upstream of the cheZ gene that enables cells to transition between a tumbling and smooth-swimming phenotype. In the absence of cheZ, cells tumble in place and do not migrate on semi-solid agar. When cheZ is expressed, the swimming motion is restored. Using these principles, we were able to screen libraries for theophylline-dependent riboswitches by plating cells onto semi-solid media in the absence of theophylline and selecting the cells that did not move. This process was repeated, and the remaining cells were then grown on semi-solid media containing theophylline. Picking the cells that moved in the presence of theophylline, but not in its absence, allowed us to discover riboswitches with low background levels of gene expression and robust (24-fold) activation ratios. This assay has several advantages over our lacZ-based screen in that it is relatively simple to perform, extremely cost-effective, requires no special equipment, and can easily be adapted to screen libraries with >106 members. However, one disadvantage of motility screens based on cheZ is that over expression of cheZ leads to a decrease in cell motility as cells become embedded in the media. Thus, discovering riboswitches that strongly activate gene expression using the cheZ assay requires careful, empirical optimization of the promoter strength to be certain that excellent candidates are not eliminated.
Faced with these challenges, we sought to develop a screen for synthetic riboswitches that would offer the throughput of a genetic selection and also provide the tunable nature of a genetic screen. In the past several years, advances in fluorescence activated cell sorting (FACS) and the development of new genetically encoded fluorophores (15) have led to remarkable increases in throughput and reductions in cost. Moreover, while FACS has most often been used to sort populations of eukaryotic cells, technological advances have opened the door to sorting large (∼108 member) libraries of small cells, such as bacteria (16), and even cell-like compartments, such as oil–water emulsions (17,18). Because FACS can readily distinguish small differences in fluorescence emission intensity, it has become a powerful screening technique in directed evolution experiments (19), where it is critical to quantitate small changes in gene expression or enzyme activity. We thus anticipated that FACS would be useful for discovering synthetic riboswitches that display low background levels of gene expression in the absence of a ligand and robust increases in gene expression in its presence. [While this manuscript was under review, Fowler and coworkers reported a FACS-based approach to screening libraries of artificial riboswitches able to modulate gene expression at a transcriptional level. Although their approach identified functional switches, the activation ratios of the switches are modest (∼8-fold) compared to those reported here (20).]
Here we show that FACS can rapidly enrich a population of rare, dynamic riboswitches from a large library. The enriched population contained a high percentage of functioning riboswitches that displayed improved characteristics compared to those isolated using our and others’ previous screens and selections. Sequence data from these switches also sheds new light on the importance of the RBS, or Shine–Dalgarno sequence, on the function of synthetic riboswitches. Taken together, these results may provide new insights into the design of new synthetic riboswitches using in vitro selected aptamers.
MATERIALS AND METHODS
General considerations
All plasmid manipulations utilized standard cloning techniques and all constructs were verified by DNA sequencing. Purifications of plasmid DNA, PCR products and enzyme digestions were performed using kits from Qiagen. Theophylline, o-nitrophenyl-β-d-galactopyranoside (ONPG) and ampicillin were purchased from Sigma. X-gal was purchased from US Biological. Synthetic oligonucleotides were purchased from IDT. All PAGE supplies were purchased from Bio-Rad Laboratories. All experiments were performed in E. coli TOP10 F’ cells (Invitrogen) cultured in media obtained from EMD Bioscience.
Plasmid SAL006.1 (Ptac1 – mTCT4-8 aptamer – TATAAAAG – AGACAACAAG – IS10-DsRedExpress)
A cassette mutagenesis strategy was used to generate pSAL006. A PCR product (A) was generated by using pSKD445.1 as a template with forward primer SKD-178 which anneals to pSKD445.1 upstream of the mTCT4-8 aptamer and the reverse primer SKD-163 which anneals to the IS10 region of our reporter gene. A separate PCR product (B) was generated using forward primer SAL-002 and reverse primer SAL-003 using pDsRedExpress as a template. PCR products A and B contain an overlapping region and were mixed and amplified using the forward primer SKD-178 and reverse primer SAL-003, to give PCR product C. PCR product C was digested with NdeI and AflIII and cloned into those sites in pSKD850.2 (Supplementary Data) to yield plasmid pSAL006.1.
N8 Library construction—expression platforms with 8-base randomized region (Ptac1 – mTCT4-8 aptamer – NNNNNNNN – AGACAACAAG – IS10-DsRedExpress)
Cassette A for the 8-base library (N = 8) was created using forward primer SKD-178, which anneals to pSAL006.1 upstream of the mTCT4-8 aptamer, and reverse primer SKD-148. SKD-148 contains a randomized region of eight bases that is flanked by constant sequences that anneal to the region including the RBS and the ATG start codon (5′) and the mTCT4-8 aptamer (3′). Cassette B was created using forward primer SKD-147 and reverse primer SAL61 which anneals 3′ to the NcoI site of our IS10-DsRed-Express reporter gene. SKD-147 also contains a randomized region of 8 bases flanked by constant sequences that anneal to the mTCT4-8 aptamer (5′) and the region from the RBS and the ATG start codon (3′) PCR fragments were amplified and gel purified before assembly. Constant sequences from SKD-147 and SKD-148 overlap and allowed for cassette pieces A and B to be assembled and amplified using primers SKD-178 and SAL-061. The assembled PCR product was gel purified, digested with NdeI and NcoI and again gel purified. Digested insert was then ligated into vector pSAL006.1 digested with the same restriction enzymes, dephosphorylated and gel purified.
N12 library construction—expression platforms with 12-base randomized region (Ptac1 – mTCT4-8 aptamer – NNNNNNNNNNNN – CAACAAG-IS10-DsRedExpress)
Library was created as described earlier with the following exceptions. For cassette 1, reverse primer SAL-045 was used and for cassette 2, forward primer SAL-044 was used. Primers SAL-044 and SAL-045 contain regions of 12 randomized bases with overlapping constant sequences that anneal to the mTCT4-8 aptamer and the region between and including RBS and start codon of the IS10-DsRedExpress reporter gene.
Flow cytometry
To determine the number of transformed bacteria, library transformations were first plated on large (241 mm × 241 mm) bioassay trays from Nalgene containing 300 ml of LB/agar, supplemented with ampicillin (50 μg/ml). Plates were scraped using 2 ml of liquid media with 500 μl of the cultivated bacteria being used to inoculate a 50 ml culture of LB supplemented with ampicillin. This culture was incubated for 14 h at 37°C with shaking (250 rpm). The following day, 50 μl of the overnight culture was used to inoculate 5 ml of LB supplemented with ampicillin and incubated at 37°C with shaking for approximately 3 h to an OD600 between 0.3 and 0.5. At this time, 750 μl of the culture was centrifuged at 5000 rcf for 10 min, the media was removed, and the cell pellet was resuspended in 1.5 ml of PBS buffer (177 mM NaCl, 2.7 mM KCl, 5.3 mM Na2HPO4·7H2O, 1.8 mM KH2PO4, pH = 7.4). Cultures were immediately analyzed on a Becton Dickinson FACSVantage SE flow cytometer using an Innova70 spectrum laser tuned to 568 nm for excitation. Fluorescence was detected through a 630/22 bandpass filter.
For all negative selections, at least 2 × 105 clones were collected. For all positive selections, at least 2 × 104 clones were collected at greater than 90% purity. Sorted bacteria were cultured overnight at 37°C in LB supplemented with ampicillin. Plasmids were isolated from the overnight culture of sorted bacteria and used to clone the enriched pool of riboswitches in front of an IS10-LacZ reporter gene. PCR reactions were set up using the isolated plasmids as a template with forward primer SKD-178 and reverse primer SKD-56. The resulting PCR product was gel purified, digested with KpnI and HindIII and again gel purified. The digested insert was then ligated into vector pSKD445.1 digested with the same restriction enzymes, dephosphorylated and gel purified. Bacteria were transformed with the resulting ligation and screened enzymatically with Miller units determined as previously described (11,21).
Structure probing
Synthetic oligonucleotides SAL-073 and its reverse complement, SAL-074 were mixed, heated to 95°C for 2 h and cooled 1°C per minute to 4°C to yield a double-stranded DNA pool with a T7 RNA polymerase promoter fused directly 5′ to the mTCT4-8 aptamer and expression platform of switch SAL-12.1. Using 1 ng of the double-stranded DNA pool, a 20 μl in vitro transcription reaction was prepared using the AmpliScribe™ T7-Flash™ Transcription Kit from Epicentre Biotechnologies and incubated at 37°C for 1 h. Following transcription, 1 μl of DNaseI was added to the reaction mixture to remove the DNA template. Transcribed RNA was diluted to a volume of 100 μl, phenol:chloroform extracted and ethanol precipitated. The RNA was resuspended in 44 μl of nuclease-free water and dephosphorylated with calf intestinal alkaline phosphatase (New England Biolabs). Again, the reaction was diluted to 100 μl, phenol:chloroform extracted, ethanol precipitated and the RNA was resuspended in 43 μl of nuclease-free water. Resuspended RNA was 5′-end-labeled using T4 polynucleotide kinase (New England Biolabs) and 0.5 μl [γ-32P]ATP (7000 Ci/mmol, 150 mCi/ml, MP Biomedicals). Radiolabeled RNA was purified using denaturing gel electrophoresis. The RNA band was then excised from gel and eluted in 5 ml of nuclease-free water overnight at 37°C with shaking. The eluted RNA was ethanol precipitated and resuspended in 50 μl of nuclease-free water.
The secondary structure of the end-labeled RNA was probed using T1 nuclease (Ambion). Structure analysis reactions were performed under denaturing conditions either in the absence of theophylline, or the presence of theophylline (0.1 mM or 1 mM). To each structure analysis reaction mixture [9 μl: ∼1 pmol RNA, 1× structure buffer from Ambion, 6 mM MgCl2, and theophylline (0, 0.1 or 1 mM)] and sequencing reaction (9 μl: 1 pmol RNA, 1× sequencing buffer from Ambion) 1 μl of T1 RNase (1U) was added and incubated at room temperature for 15 min. Reactions were stopped by adding 10 μl stop buffer (8 M Urea:50 mM EDTA) and separated using denaturing gel electrophoresis [7 M urea; 10% (29:1) acrylamide: bisacrylamide], imaged using a phosphorimager and the data analyzed using ImageQuant software.
Analyses of expression platforms
To investigate the effects of the Shine–Dalgarno sequence on the expression levels of riboswitches, the theophylline aptamer was deleted from each construct. To accomplish this PCR products were made using forward primers SAL-085, 086 and 087 which contain KpnI restriction sites and anneal to the expression platforms of switches 8.1, 8.1* and 12.1, respectively, and reverse primer SKD 056. PCR products were digested with KpnI and HindIII, gel purified and cloned into vector pSKD1248.2 digested with the same restriction enzymes, dephosphorlyated and gel purified. pSKD1248.2 is nearly identical to pSKD445.1 with the following exceptions. Oligonucleotide mutagenesis was used to insert a multiple cloning site 5′ to the theophylline aptamer and replace the Ptac promoter with the weaker IS10 promoter.
RESULTS
As a proof-of-principle experiment to determine whether FACS can identify synthetic riboswitches from large libraries, we reexamined a library we had screened using our previously reported lacZ assay (11). This library consisted of the theophylline aptamer, followed by a sequence of eight randomized bases, the RBS, and the red-fluorescent reporter gene DsRed express (22). E. coli were transformed with the plasmid library (theoretically 65 536 members), plated onto selective LB-agar, and grown overnight. Approximately 65 000 transformants were scraped from the plate, grown in selective LB media in the absence of theophylline, and this culture was analyzed using flow cytometry (Figure 1a). Analysis of the population indicated that a large percentage of clones displayed levels of fluorescence emission within the first decade of the log-scale. This was somewhat surprising as blue/white screening of the same library using the lacZ reporter gene revealed nearly all (>99%) of the population was blue in the absence of theophylline (i.e. most cells appeared to have high levels of gene expression, rather than the apparently low levels observed here). These results highlight a limitation of our plate-based lacZ assay, which only allowed qualitative ranking of gene expression and may have led us to overlook clones that appeared pale blue in the absence of theophylline, but nonetheless had acceptably low levels of background expression.
Figure 1.
FACS histograms representing a population of bacteria possessing a library of mutant theophylline riboswitches. (a) Profile of the N8 library grown in the absence of theophylline. For the initial negative selection, a gate was set to collect bacteria falling in the ‘OFF’ region. (b) Profile of the N8 bacteria collected in the initial negative selection grown in the absence and presence of theophylline (1 mM, red). Bacteria collected from the culture grown with theophylline were those that exhibited an increase in fluorescence when compared to the population of bacteria grown without. These are represented by the ‘ON’ region and amount to approximately 2% of the culture. (c) Profile of the N8 bacteria following one round of negative selection and one round of positive selection. Riboswitches from this enriched population were subsequently cloned in front of a β-galactosidase reporter gene and screened enzymatically leading to the discovery of 8.1*. All histograms were generated with identical instrument settings and display data from 10 000 events.
Encouraged by the initial flow cytometry results, we positioned a gate to collect bacteria displaying levels of fluorescence below the first decade of the log-scale (Figure 1a). These bacteria were cultured overnight, and aliquots of the overnight culture were used to inoculate two separate cultures, one with theophylline (1 mM) and one without. Analysis of these cultures revealed that our initial attempt to isolate clones displaying low levels of fluorescence in the absence of theophylline was successful, and furthermore that a small percentage of this sorted population exhibited a theophylline-dependent increase in fluorescence (Figure 1b). These cells were collected, cultured overnight, and aliquots of the overnight culture were used to inoculate two separate cultures, one with theophylline (1 mM) and one without. These cultures were grown to an OD600 between 0.4 and 0.6 and were analyzed using flow cytometry, which revealed a dramatic theophylline-dependent shift in fluorescence (Figure 1c).
To quantify how individual riboswitches performed, the culture was grown to saturation, the plasmid DNA of the entire population was isolated, and the 5′-untranslated regions were subcloned upstream of a lacZ reporter gene. This library was used to transform E. coli, which were grown on selective LB-agar containing X-gal, but no theophylline to give approximately 3000 transformants. Using a robotic colony picker, we picked the 48 whitest colonies from this plate and performed our previously described enzymatic assay for riboswitch activity (11). To our satisfaction, greater than half of these clones were functioning riboswitches, the best of which, clone 8.1* (nomenclature signifies the number of randomized bases, in this case 8, followed by the switch's rank amongst other identified switches identified from the same library), displayed a 59-fold activation ratio in the presence of 1 mM theophylline (Figure 2). By way of comparison, the best riboswitch isolated from our previous screen had an activation ratio of only 36, and its isolation required 846 separate β-galactosidase assays, compared to the 48 performed here.
Figure 2.
FACS histograms representing a population of bacteria possessing a library of mutant theophylline riboswitches. (a) Profile of the N12 library grown in the absence of theophylline. For the initial negative selection a gate was again set to collect bacteria falling in the ‘OFF’ region. A two-step enrichment sort was required to collect only the weakly fluorescing clones. (b) Profile of the N12 bacteria collected in the initial negative selection grown following the two-step enrichment in the absence and presence of theophylline (1 mM). Clones displaying fluorescence levels falling within the indicated ‘ON’ region were collected and again amounted to approximately 2% of the culture. (c) Profile of the N12 bacteria following one round of negative selection and one round of positive selection. Riboswitches from this enriched population were subsequently cloned in front of a β-galactosidase reporter gene and screened enzymatically.
Sequence analysis revealed that clone 8.1* harbored an expression platform identical to our previously reported switch (clone 8.1; 36-fold activation ratio), but had a point mutation that changed the Shine–Dalgarno sequence from AAG to the stronger AGGA (23). It is interesting to note that this mutation occurred immediately 3′ to the randomized 8-base region, suggesting that it was introduced by either an impurity in the oligonucleotides used to create the library, or by a replication error in E. coli. As such, we anticipate that this sequence would be underrepresented in the initial library. [Sequencing of the PCR product used to create the library revealed that the vast majority of the population contained 8 nt between the aptamer and the ribosome binding site (AGA). While this low resolution assay cannot detect a rare GGA sequence, it does rule out the possibility that the majority of the library was comprised of GGA ribosome binding site sequences.] This proof-of-principle experiment suggested three things: First, the FACS-based assay can discover new synthetic riboswitches; second, the assay has the ability to find rare sequences; and third, the sequence of the RBS is critical to the performance of this family of synthetic riboswitches.
With these lessons in mind, we sought to optimize the entire expression platform, including the sequence of the RBS. To do this, we created a much larger ‘N12’ library, which theoretically contains 1.7 × 107 members, a number that would be virtually impossible to screen enzymatically. This N12 library was cloned into the 5′-untranslated region of DsRed express and the plasmid library was used to transform E. coli. This transformation yielded approximately 250 000 clones, which were subsequently sorted using the flow cytometer. We note, however, that the number of clones was limited by the transformation efficiency of this particular batch of electrocompetent cells. Although the cell-sorting experiments were performed using only a fraction of the theoretical library size, this does not represent a limitation of the throughput of flow cytometry, which can approach 109 cells/day (16). It is also important to note that our previously described lacZ-based assay proved only marginally successful in screening this library for functioning switches, as nearly all of the whitest clones selected displayed no enzymatic activity in the presence or absence of theophylline, presumably due to poor ribosome binding sites.
As with the N8 library, we sorted our N12 library by positioning a gate to collect cells that exhibited levels of fluorescence emission below the first decade of the log-scale in the absence of theophylline (Figure 2a). The collected cells were cultured overnight in the absence of theophylline and again analyzed with the cytometer. While clones with low fluorescence dominated the population, a portion of the population remained highly fluorescent, which required an additional sort to collect only the weakly fluorescing clones. The clones collected following the second round of sorting were cultured overnight, and aliquots of the cultures were used to inoculate two new cultures: one with theophylline (1 mM) and one without. Analysis of these cultures revealed a small percentage of clones displaying high levels of theophylline-induced fluorescence (Figure 2b). These highly fluorescent cells were collected and cultured overnight. Aliquots of this overnight culture were used to inoculate one culture containing theophylline (1 mM) and one without. Subsequent flow cytometry analysis of these cultures revealed the fluorescence of the population to be highly theophylline-dependent, with a significant portion of the population displaying levels of fluorescence in the presence of theophylline far greater than the same population grown in the absence (Figure 2c). These highly fluorescent cells were collected and again cultured overnight.
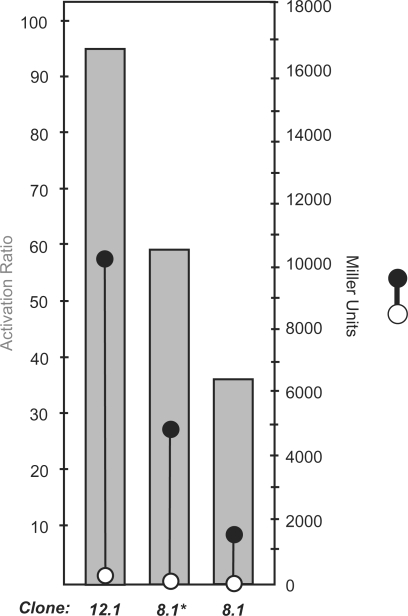
The plasmid DNA from the saturated overnight culture was isolated, and the 5′-untranslated regions were again subcloned upstream of a lacZ reporter gene. This library was used to transform E. coli, which were grown on selective LB-agar containing X-gal, but no theophylline to give approximately 12 000 transformants. Using a robotic colony picker, we picked the 288 whitest colonies from this plate and performed our previously described enzymatic assay for riboswitch activity. Over 80 of these clones were verified as functioning theophylline-dependent synthetic riboswitches. The most impressive switch, clone 12.1, activated gene expression 96-fold in the presence of theophylline (1 mM), and produced greater than 10 000 Miller units of β-galactosidase activity, compared to 4800 Miller units for clone 8.1*, and 1820 Miller units for clone 8.1 (Figure 3). To our knowledge, this represents the largest activation ratio of any synthetic riboswitch reported to date. Furthermore, the activation ratio of riboswitch 12.1 exceeds that of many natural riboswitches. For example, fusion of glycine and adenine sensing RNAs from Bacillus subtilis with β-galactosidase reporters yielded, in each case, approximately 10-fold increases in the presence of ligand (24,25).
Figure 3.
Measures of β-galactosidase activity (in Miller units) for switches identified using the high-throughput, FACS enrichment (12.1 and 8.1*) and best switch isolated using a previously reported β-galactosidase assay (8.1). Right axis: β-Galactosidase activities for switches in the absence (open circle) or presence of theophylline (1 mM, closed circle). Errors in the measurements (SEM.) are smaller than the diameter of the circles. Left axis: Activation ratio (gray bar) is determined by the ratio of activities in the presence and absence of theophylline.
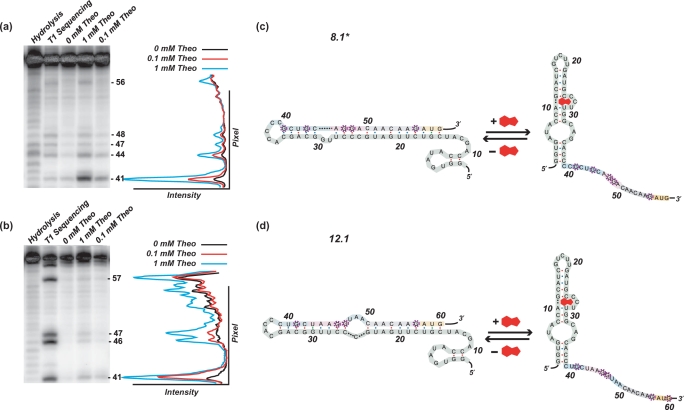
Given the substantial improvements in how these switches functioned relative to our previously reported synthetic riboswitches, we were interested in exploring their mechanisms of action. Sequencing data suggested that these new switches likely regulated translation in the same manner as our previously reported synthetic riboswitches, and secondary-structure predictions provided by mFold (26,27) suggested that these new switches adopted similar folds. We hypothesized that in the absence of theophylline, the switches adopt a conformation in which the RBS (Shine–Dalgarno sequence) is paired, and translation is inhibited. Upon binding of theophylline, the RBS becomes accessible, and translation occurs. Furthermore, because the RBS is stronger, translation is activated to a higher level than previously observed. To test these hypotheses, we performed in vitro T1 ribonuclease digestions of clone 8.1* and 12.1 both in the absence and in the presence of theophylline (0.1 and 1 mM). RNase T1 cleaves 3′ to solvent-accessible, single-stranded G residues, and our mFold model suggested that many of the G's near the RBS would become unpaired in the presence of theophylline. Results of the digestions are shown in Figure 4a and b. Switch 8.1* shows clear increases in the cleavage of residue G41 and minor increases at residues G44, G47, G48 and G56 near the RBS in the presence of theophylline, consistent with the model in Figure 4c. Similar results were achieved with switch 12.1 where there are clear increases in the cleavage of residue G41 and, to a lesser extent, residues G46, G47 and G57 near the RBS in the presence of theophylline, consistent with the model in Figure 4d.
Figure 4.
Sequencing gels [7 M urea; 10% (29:1) acrylamide: bisacrylamide] indicating the cleavage pattern for 5′ radiolabeled RNA from 5′-untranslated region of switches 8.1* and 12.1 digested by RNase T1 in the presence and absence of theophylline and under denaturing conditions (a and b). A graph of band intensity versus pixel location is shown to the right indicating a significant increase in cleavage at circled G residues. Cleavage patterns indicate G residues surrounding and included in the SD sequence are solvent accessible and single-stranded in the presence of theophylline and base-paired in its absence. Predicted mechanisms of riboswitch 8.1* and 12.1 function (c and d). The 5′-UTR of switch. 1 adopts a highly folded structure with extensive base-pairing in the region including the RBS. This base-pairing in the absence of theophylline prevents translation. In the presence of theophylline, the transcript adopts a secondary structure in which the RBS is exposed. The secondary structure shown for the theophylline aptamer predicted by mFold is identical to the secondary structure determined by NMR (30–32).
While the nuclease digestion studies suggest a switching mechanism, they alone do not explain the differences in the activation ratios between the switches. Inspection of the expression platforms revealed that the switches with the highest levels of expression in the presence of theophylline all have at least four consecutive bases that are complementary to the anti-Shine–Dalgarno sequence of E. coli 16S RNA. Furthermore, our best switch, clone 12.1, not only possessed a longer Shine–Dalgarno sequence (UAAGG) than the other switches, but it also appeared to be spaced optimally, 6 nt upstream from the AUG initiation codon (28) (Figure 5). In this context, we refer to the aligned spacing as the number of bases separating the AUG start codon and the 5′-A of the anti-Shine–Dalgarno sequence following its alignment with the Shine–Dalgarno sequence. Optimal aligned spacing places the AUG start codon 4–6 bases from the 5′-A of the anti-Shine–Dalgarno sequence (28).
Figure 5.
Nucleotide sequence of the putative anti-Shine–Dalgarno sequence from the 3′-end of the E. coli 16s rRNA aligned with expression platforms from high-performing theophylline riboswitches. Expression platforms include the Shine–Dalgarno sequence (pink) and start codon (green). The putative Shine–Dalgarno sequence (UAAGG) of switch 12.1 (96-fold activation ratio) is longer than that of the other switches, and has the optimal 6-base aligned spacing between the 5′-A of the anti-Shine–Dalgarno and the start codon.
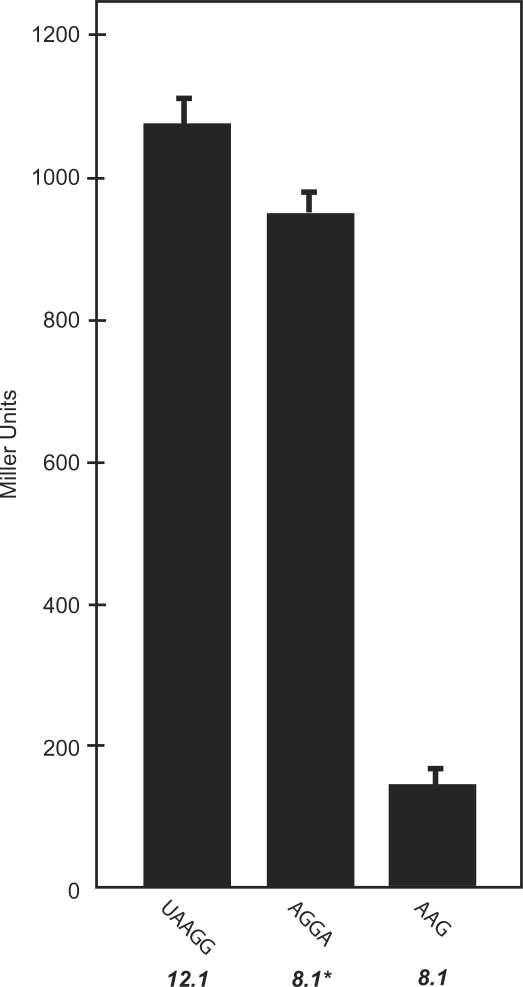
To test whether the Shine–Dalgarno sequences were responsible for the different behaviors of the switches, we removed the aptamer sequences from clones 8.1, 8.1* and 12.1 and determined the overall levels of gene expression. Because removing the aptamer drastically increases gene expression and leads to cell death due to the toxicity of β-galactosidase, we also replaced the Ptac promoter in all three constructs with a weaker IS10 promoter to reduce the overall level of gene expression. Figure 6 shows that the Shine–Dalgarno sequences from clones 8.1* and 12.1 both lead to higher expression levels than the sequence from clone 8.1. These results are consistent with the predicted strengths of the Shine–Dalgarno sequences and their positions relative to the initiation codon.
Figure 6.
Measures of β-galactosidase activity (in Miller units) for constructs with 5′-untranslated regions lacking aptamers, containing only the expression platforms of switches 12.1, 8.1* and 8.1. Previous work has shown that longer Shine–Dalgarno sequences typically give rise to higher expression levels (23) as is the case here.
DISCUSSION
Our results demonstrate that FACS is capable of isolating excellent synthetic riboswitches from large libraries. Using FACS, we were able to discover what is, to our knowledge, the best-performing synthetic riboswitch to date. Clone 12.1 displays a low level of gene expression in the absence of ligand, and a robust (96-fold) increase in the presence of theophylline (1 mM). The key to these improvements involved optimizing the expression platform, including the RBS. In a previous study, the randomized expression platforms were limited to a sequence length of eight bases while sequence of the putative, non-specific RBS (AGA) was held constant, thus restricting the number of accessible Shine–Dalgarno sequences. This was necessary because randomizing the RBS would lead to libraries that would be too large to screen fully using an enzymatic assay. While our recently reported motility-based selection method was capable of screening such large libraries (>106 members), it was challenged to discover the switches that most strongly activated gene expression. The FACS-based assay described here offers the best of both worlds, in that its throughput is competitive with a genetic selection, but it also has the ability to semi-quantitatively determine gene expression to reveal the best synthetic riboswitches.
To perform well as an ‘on’ switch, a synthetic riboswitch must carefully balance the ability to repress gene expression in the absence of the ligand, while strongly activating gene expression in its presence. For synthetic riboswitches that regulate gene expression post-transcriptionally, this typically involves base-pairing between the expression platform (which often includes the Shine–Dalgarno sequence) and the aptamer. If these interactions are too strong (or if ligand binding is weak), the switch will not turn on; if they are too weak, the switch will be leaky in the absence of the ligand. Furthermore, the activation ratio will depend on other factors, including, but not limited to the strength of the Shine–Dalgarno sequence, and the relative stabilities of the mRNA in the presence and absence of the ligand. Because so many of these parameters are variable, some in not entirely intuitive ways, it should come as no surprise that there are several different ways to create a synthetic riboswitch from a known aptamer. Furthermore, aptamer selection experiments typically isolate multiple aptamers that bind the target ligand. The creation of new synthetic riboswitches will likely require the screening of pools of aptamers enriched for ligand binding, along with screening randomized expression platforms, which makes the need for reliable high-throughput screening methods even greater. While it is possible to design switches de novo, the new riboswitches reported here perform better than any rationally designed synthetic riboswitch.
While the results achieved using this flow cytometry-based screen are promising, we must emphasize that flow cytometry is unlikely to serve as a standalone method for discovering synthetic riboswitches. It is well-known that individual cells from bacterial cultures comprised of a single genotype may display a range of phenotypes (29) attributable to the stochastic nature of gene expression. As one considers that the selection criteria in flow cytometry experiments are determined by the phenotype of an individual cell, the inherent population heterogeneity of a bacterial culture, especially one containing a library of genotypes, makes the task of counter-selecting for one phenotype against another extremely challenging. For example, isolating a single bacterium displaying a low level of fluorescence does not guarantee that a culture grown from that single bacterium will possess, as a whole, those same fluorescence characteristics. With this in mind, the need for reinforcing a single-cell enrichment strategy with traditional assays, measuring average expression levels for an entire population, should be clear. The ability of a FACS-based screen employed in tandem with a β-galactosidase reporter gene to assay large libraries (>108 members theoretically) allows one to optimize the expression platform to discover outstanding synthetic riboswitches.
CONCLUSIONS
We have reported a screen to discover synthetic riboswitches using fluorescence activated cell sorting. The screen is very high throughput, and can, in principle, examine libraries of >108 members. Using the screen, we have discovered a new theophylline-dependent synthetic riboswitch, clone 12.1, that displays low levels of background gene expression in the absence of theophylline, and a 96-fold increase in gene expression in its presence. To our knowledge, this is the largest activation ratio of any synthetic riboswitch reported to date. Using nuclease mapping, we have provided evidence for the likely mechanism of action of the switch, which involves a ligand-dependent rearrangement of the RNA secondary structure to reveal the RBS. We also confirmed that the strength of the RBS, which is dictated by its length and distance from the initiation codon, plays a critical role in how the best switches function. Because FACS can sort through large libraries and reveal outstanding synthetic riboswitches, we anticipate that it will be a useful screening tool in the field.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: National Institutes of Health (GM074070 to J.P.G.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Robert Karaffa of the Emory University School of Medicine Flow Cytometry Facility for assistance with the cell sorting. This work was supported by the National Institutes of Health (GM070740 to J.P.G.). S.A.L. was supported by a National Science Foundation Problems and Research to Integrate Science and Mathematics (PRISM) Fellowship. J.P.G. is a Beckman Young Investigator, a Camille Dreyfus Teacher-Scholar, and an Alfred P. Sloan Research Fellow.
REFERENCES
- 1.Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 2.Barrick J, Breaker R. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007;8:R239. doi: 10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Desai SK, Gallivan JP. Genetic screens and selections for small molecules based on a synthetic riboswitch that activates protein translation. J. Am. Chem. Soc. 2004;126:13247–13254. doi: 10.1021/ja048634j. [DOI] [PubMed] [Google Scholar]
- 4.Werstuck G, Green MR. Controlling gene expression in living cells through small molecule-RNA interactions. Science. 1998;282:296–298. doi: 10.1126/science.282.5387.296. [DOI] [PubMed] [Google Scholar]
- 5.Grate D, Wilson C. Inducible regulation of the S. cerevisiae cell cycle mediated by an RNA aptamer-ligand complex. Bioorg. Med. Chem. 2001;9:2565. doi: 10.1016/s0968-0896(01)00031-1. [DOI] [PubMed] [Google Scholar]
- 6.Harvey I, Garneau P, Pelletier J. Inhibition of translation by RNA-small molecule interactions. RNA. 2002;8:452–463. doi: 10.1017/s135583820202633x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suess B, Hanson S, Berens C, Fink B, Schroeder R, Hillen W. Conditional gene expression by controlling translation with tetracycline-binding aptamers. Nucleic Acids Res. 2003;31:1853–1858. doi: 10.1093/nar/gkg285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 9.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 10.Gallivan JP. Toward reprogramming bacteria with small molecules and RNA. Curr. Opin. Chem. Biol. 2007;11:612–619. doi: 10.1016/j.cbpa.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lynch SA, Desai SK, Sajja HK, Gallivan JP. A high-throughput screen for synthetic riboswitches reveals mechanistic insights into their function. Chem. Biol. 2007;14:173–184. doi: 10.1016/j.chembiol.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Topp S, Gallivan JP. Random walks to synthetic riboswitches - a high-throughput selection based on cell motility. ChemBioChem. 2008;9:210–213. doi: 10.1002/cbic.200700546. [DOI] [PubMed] [Google Scholar]
- 13.Weigand JE, Sanchez M, Gunnesch EB, Zeiher S, Schroeder R, Suess B. Screening for engineered neomycin riboswitches that control translation initiation. RNA. 2008;14:89–97. doi: 10.1261/rna.772408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wieland M, Hartig JS. Improved aptazyme design and in vivo screening enable riboswitching in bacteria. Angew. Chem. Int. Ed. Engl. 2008;47:2604–2607. doi: 10.1002/anie.200703700. [DOI] [PubMed] [Google Scholar]
- 15.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat. Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 16.Link AJ, Jeong KJ, Georgiou G. Beyond toothpicks: new methods for isolating mutant bacteria. Nat. Rev. Microbiol. 2007;5:680–688. doi: 10.1038/nrmicro1715. [DOI] [PubMed] [Google Scholar]
- 17.Hai M, Bernath K, Tawfik D, Magdassi S. Flow cytometry: a new method to investigate the properties of water-in-oil-in-water emulsions. Langmuir. 2004;20:2081–2085. doi: 10.1021/la035402+. [DOI] [PubMed] [Google Scholar]
- 18.Aharoni A, Amitai G, Bernath K, Magdassi S, Tawfik DS. High-throughput screening of enzyme libraries: Thiolactonases evolved by fluorescence-activated sorting of single cells in emulsion compartments. Chem. Biol. 2005;12:1281–1289. doi: 10.1016/j.chembiol.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 19.Aharoni A, Thieme K, Chiu CPC, Buchini S, Lairson LL, Chen H, Strynadka NCJ, Wakarchuk WW, Withers SG. High-throughput screening methodology for the directed evolution of glycosyltransferases. Nat. Methods. 2006;3:609–614. doi: 10.1038/nmeth899. [DOI] [PubMed] [Google Scholar]
- 20.Fowler CC, Brown ED, Li Y. A FACS-based approach to engineering artificial riboswitches. Chem. BioChem. 2008;9:1906–1911. doi: 10.1002/cbic.200700713. [DOI] [PubMed] [Google Scholar]
- 21.Jain C, Belasco JC. Rapid genetic analysis of RNA-protein interactions by translational repression in Escherichia coli. Meth. Enzymol. 2000;318:309–332. doi: 10.1016/s0076-6879(00)18060-7. [DOI] [PubMed] [Google Scholar]
- 22.Bevis BJ, Glick BS. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed) Nat. Biotechnol. 2002;20:83–87. doi: 10.1038/nbt0102-83. [DOI] [PubMed] [Google Scholar]
- 23.Barrick D, Villanueba K, Childs J, Kalil R, Schneider TD, Lawrence CE, Gold L, Stormo GD. Quantitative analysis of ribosome binding sites in E.coli. Nucleic Acids Res. 1994;22:1287–1295. doi: 10.1093/nar/22.7.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- 25.Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat. Struct. Mol. Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 26.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 28.Chen H, Bjerknes M, Kumar R, Jay E. Determination of the optimal aligned spacing between the Shine - Dalgarno sequence and the translation initiation codon of Escherichia coli m RNAs. Nucleic Acids Res. 1994;22:4953–4957. doi: 10.1093/nar/22.23.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 30.Zimmermann GR, Jenison RD, Wick CL, Simorre JP, Pardi A. Interlocking structural motifs mediate molecular discrimination by a theophylline-binding RNA. Nat. Struct. Biol. 1997;4:644–649. doi: 10.1038/nsb0897-644. [DOI] [PubMed] [Google Scholar]
- 31.Zimmermann GR, Shields TP, Jenison RD, Wick CL, Pardi A. A semiconserved residue inhibits complex formation by stabilizing interactions in the free state of a theophylline-binding RNA. Biochemistry. 1998;37:9186–9192. doi: 10.1021/bi980082s. [DOI] [PubMed] [Google Scholar]
- 32.Zimmermann GR, Wick CL, Shields TP, Jenison RD, Pardi A. Molecular interactions and metal binding in the theophylline-binding core of an RNA aptamer. RNA. 2000;6:659–667. doi: 10.1017/s1355838200000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.