Abstract
We present a new random array format together with a decoding scheme for targeted multiplex digital molecular analyses. DNA samples are analyzed using multiplex sets of padlock or selector probes that create circular DNA molecules upon target recognition. The circularized DNA molecules are amplified through rolling-circle amplification (RCA) to generate amplified single molecules (ASMs). A random array is generated by immobilizing all ASMs on a microscopy glass slide. The ASMs are identified and counted through serial hybridizations of small sets of tag probes, according to a combinatorial decoding scheme. We show that random array format permits at least 10 iterations of hybridization, imaging and dehybridization, a process required for the combinatorial decoding scheme. We further investigated the quantitative dynamic range and precision of the random array format. Finally, as a demonstration, the decoding scheme was applied for multiplex quantitative analysis of genomic loci in samples having verified copy-number variations. Of 31 analyzed loci, all but one were correctly identified and responded according to the known copy-number variations. The decoding strategy is generic in that the target can be any biomolecule which has been encoded into a DNA circle via a molecular probing reaction.
INTRODUCTION
For almost two decades now, microarrays have assumed a central role in global analyses of gene expression (1), genotyping (2) and genomic copy-number variations (array CGH) (3,4). Microarrays have also been applied as a platform for resequencing (5) and for protein analyses (6). The multiplexing capacity makes microarrays one of the leading tools for parallel analyses. However, the sensitivity, dynamic range and quantitative precision of DNA microarrays are limited, due to cross-hybridization and because the data are based on average measurements of fluorescence from a number of target molecules. These limitations can in principle be avoided by applying a noise-free single-molecule detection and quantification approach, counting molecules in a digital way instead of integrating fluorescence over populations of molecules. A number of single-molecule approaches have been described. Assays that depend on detection of single fluorescent probes, however, are often associated with considerable detection noise. Therefore, approaches based on compartmentalized amplification of target molecules have been developed to achieve robust amplified single molecule (ASM) detection. ASMs have been prepared through PCR from individual template molecules (7) by compartmentalized amplification of molecules in miniature reactions volumes (8), or using primers anchored to gels, to planar surfaces, or to individual beads in water-in-oil emulsions (9–11). The last two approaches are used in current next-generation sequencing instruments (12–14), which use random arrays of ASMs to achieve sufficient signals from the sequencing reactions.
Rolling-circle amplification (RCA) generates another type of ASMs by replicating circularized DNA strands (15). The ASMs produced by RCA are true single molecules, each composed of tandem repeats of typically around 1000 complements of a 100-mer DNA circle after one hour of polymerization (16). Hybridization with fluorescence-labeled oligonucleotide probes enriches the fluorophores locally in the ASMs so that these become readily visible as discrete bright micron-sized objects by fluorescence microscopy (17–22). DNA circles can be formed by general or target-directed circularization reaction, catalyzed by a DNA ligase. General circularization can be achieved by equipping randomly fragmented genomes with adaptor sequences and then template circularization using a general sequence hybridizing to the adaptor sequences. This strategy was used by Pihlak et al.as sample preparation for shotgun sequencing of ASMs (17). Target-directed DNA circularization reactions can be achieved using any of a number of probing techniques. For example, padlock probes are linear oligonucleotides that become circularized in a strictly target-dependent ligation reaction (23). Padlock probes consist of target-specific end sequences, linked together by a sequence that can contain sequence elements for amplification and tag sequences for identification of the amplification product (24,25). Padlock probes have been used for genotyping (25), gene copy number (26) and expression analysis (22,27), as well as detection of infectious pathogens (19). The selector probe is another category of reagents for probe-directed DNA circularization (28). Selector probes are similar to padlock probes and have target-specific ends for target recognition, flanking a DNA sequence with elements for amplification. In contrast to padlock probes, however, selector probes are designed to hybridize to the end-sequences of restriction digested genomic DNA fragments and thus template DNA ligase assisted circularization of specific genomic DNA sequences. Selector probes have been used for gene copy-number analysis (29,30) and for preparation of large sets of exons for parallel sequencing (31). A third category of target-directed DNA circularization reagents is the proximity ligation probes. These are pairs of protein-binding reagents equipped with DNA strands that can form reporter molecules through DNA ligation upon coordinated binding of both probes in a probe pair (32). The reporter molecule can be circularized through a number of reaction mechanisms (19,33). The proximity ligation assay has been used to detect proteins, protein modifications and interactions in serum samples and in situ (32–35). Rolling-circle ASMs have been used for readout in several genotyping assays (20,21,36), for detection of protein and protein complexes in situ using proximity ligation (33), and for detecting microbes with padlock probes followed by counting individual rolling-circle ASMs pumped through a microfluidic channel (19).
We present a method for targeted quantitative multiplex analysis of biomolecules based on a combination of molecular probing and decoding reactions. The biomolecules are first probed with techniques that generate DNA circles upon recognition. ASMs are generated through RCA, and then attached to glass slides in a random pattern. The rolling-circle ASMs include sets of tags that are used for identification following a combinatorial decoding scheme, similar to that used to identify hundreds of thousand bead species in random bead arrays (37). Our approach is generic and can be applied for multiplex quantification of ASMs created from any assay that results in circular DNA molecules. We demonstrate our approach for quantitative multiplex analysis by using it to measure relative copy numbers of 31 autosomal and sex chromosome loci, targeted by selector probes (28).
MATERIAL AND METHODS
Preparation of DNA circles
Padlock probes (Table S6) were designed using the ProbeMaker oligonucleotide design software (38). Stock solutions of DNA circles were prepared by ligating 20 nM of the different phosphorylated padlock probes, using 60 nM of their respective synthetic ligation targets (Table S6). The ligation reaction was performed in a volume of 50 μl containing 4 mU/μl of T4 DNA ligase (Fermentas, Lithuania), 1 mM ATP, 0.2 μg/μl BSA (New England Biolabs, USA), in 1× phi29 buffer (34 mM Tris–acetate, 10 mM magnesium acetate, 66 mM potassium acetate, 0.1% Tween 20 and 1 mM DTT with a pH of 7.9 at 37°C; Fermentas, Lithuania). Dilution series of DNA circles were carried out in 1× PBS containing 0.2 μg/μl BSA. The ligation reactions were incubated for 15 min at 37°C. The oligonucleotides used for preparing synthetic DNA circles were purchased from DNA Technology (Denmark).
RCA
DNA circles were replicated by adding 5 μl of circles to a 45 μl replication mix containing 100 μM dNTP, 0.2 μg/μl BSA, 1× phi29 buffer and 2 U phi29 DNA polymerase (Fermentas). Polymerization was carried out for 1 h at 37°C and terminated by incubation at 65°C for 5 min. Unless stated otherwise, the ligation template also served as a primer for the RCA.
Selector probe ligation
The selector probe assay was designed using the PieceMaker and ProbeMaker software (38,39). Restriction digestion was performed with 8 U restriction enzyme Mnl I (Fermentas), 800 ng genomic DNA sample, 50 mM potassium acetate, 20 mM Tris–acetate, 10 mM magnesium acetate, 1 mM DTT (pH 7.9 at 25°C) and 2 µg BSA in a total volume of 20 µl. The reaction was carried out at 37°C for 1 h, followed by 20 min at 65°C to inactivate Mnl I. The circularization reaction contained 200 ng digested DNA, 22 nM vector oligo (P-CTCGACCGTTAGCAAAGCTTTCTACCGTTATCGT) and 100 pM of each selector probe (Table S2). The circularization reaction was performed in 0.5× PCR buffer (Invitrogen) supplemented with 9.7 mM MgCl2, 0.8 mM NAD and 0.2 U Ampligase (Epicentre Biotechnologies, USA) in a total volume of 15 µl. The reaction was incubated at 95°C for 5 min, followed by 60°C for 16 h. Genomic DNA samples from subjects diagnosed with Down syndrome were collected and isolated from blood (Flexigene, Qiagen, Germany) at the Department of Clinical Genetics, Uppsala University, with the subjects’ permissions. Pooled female and pooled male reference samples were purchased from Promega (cat.#G147A 20745 001, cat. #G152 20215 001). The selector probe oligonucleotides were purchased from Integrated DNA Technology (USA).
Amplification of DNA circles
Amplification of DNA circles was performed using the circle-to-circle (C2CA) procedure essentially as described in Dahl et al. (16). In this procedure a 100-mer DNA circle produces about 1000 new circles after a 1 h RCA. In brief, 10 µl of the circularized selector probes (∼133 ng genomic DNA) were replicated using RCA primed by 25 nM replication oligonucleotide (ACGATAACGGTAGAAAGCTTTGCTAACGGTCGAG) in a total volume of 20 μl. The RCA reaction was incubated at 37°C for 1 h and 45 min, followed by 5 min at 65°C to inactivate the phi29 polymerase. Subsequent reaction steps were initiated by 5 μl additions of mixes containing the appropriate reagents, enzymes and oligonucleotides in 1× phi29 buffer. The monomerization step was carried out by addition of 7.5 U Hind III (Fermentas) and 600 nM restriction oligonucleotide (CTCGACCGTTAGCAAAGCTTTCTACCGTTATCGT) and incubating the sample at 37°C for 10 min, followed by inactivation at 80°C for 10 min. Monomerized RCA products were re-circularized by adding 0.1 U T4 DNA ligase together with 4 mM ATP and incubating the sample at 37°C for 10 min. The ASMs were then generated in a second RCA by addition of 2 U of phi29 DNA polymerase and 1.25 mM dNTP. The polymerization reaction was performed at 37°C for 1 h, followed by 5 min at 65°C. The oligonucleotides used in the C2CA procedure were purchased from DNA Technology (Denmark).
Hybridization of ASMs
Ten microlitres of the ASMs were deposited on a poly-l-lysine slide (Sigma-Aldrich) and dried at 55°C for approximately 15 min. A silicone mask was applied to create a hybridization chamber (17 mm in diameter) on the slide. A blocking solution containing 10 ng/μl sonicated salmon sperm DNA (Invitrogen), 2× SSC buffer and 0.05% Tween-20 was added and the slide was incubated shaking for 15 min at 37°C. Washing was performed at 20°C with 1 ml 1× PBS per hybridization chamber. For hybridization of the ASMs generated from ligated padlock probes, 0.5 ml of a hybridization mix containing 10 nM fluorescence-labeled oligonucleotides (Table S6), 0.05% Tween-20, 2× SSC buffer and 5% dextran sulfate was added and incubated on shake at 55°C for 1 h. For hybridization of the ASMs generated from selector probe-guided ligation reactions, 0.5 ml of a hybridization mix was added that contained 10 nM of Cy3 and Texas Red tag probes and 100 nM of Cy5 tag probes (Table S5), 10 nM general FITC-labeled tag probe (Table S5), 10 nM sandwich probe-mixture (340 nM in total, Table S3), 0.05% Tween-20, 2× SSC buffer and 5% dextran sulphate, and the reactions were incubated on shake at 55°C for 1 h. All arrays were washed in 1× PBS at room temperature, followed by 1 min incubation in 70% ethanol. The slides were spun dry in a bench centrifuge, and mounted with approximately 10 μl VectaShield (Immunkemi, Sweden) and a 24× 55 mm2 cover slip (Menzel-Gläser, Germany). Sandwich probes were purchased from Integrated DNA Technology (USA), whereas the fluorescent tag probes were purchased from Biomers (Germany).
Image acquisition
An epifluorescence microscope (Axioplan II, Zeiss) equipped with a 20× objective NA 0.75 (Fluar, Zeiss) was used to image the ASMs using the AxioVision LE 4.3 software (Zeiss). The microscope was equipped with a 100 W mercury lamp and a charge-coupled device camera (C4742-95, Hamamatsu). Excitation and emission filters for visualization of FITC, Cy3, Texas Red and Cy5 were used. Between 12 and 14 areas were imaged per subject sample, corresponding to a total imaged area of 0.18–0.21 mm2 containing between 27 000 and 62 000 ASMs.
Dehybridization of ASMs
After each imaging, the slides were washed in 1× PBS at room temperature for about 10 min to remove the cover slip. The slides were then washed in a dehybridization buffer containing 50% formamide and 2× SSC buffer at 50°C for 1 min. A second wash in 1× PBS was followed by an ethanol series and the slides were spun dry with a bench centrifuge.
Image processing
The identity of an ASM species is defined by its unique combination of tags. This is recorded as image data after hybridization of fluorescence-labeled tag probes. The image data acquired from all cycles of hybridization was analyzed to decode ASM identities. Before ASM identities could be decoded, images were pre-processed. First, images from different hybridization steps showing the same imaged area were aligned, resulting in an image stack. Thereafter, a filter was applied to enhance image structures with ASM-like size and shape and at the same time reducing background variation. As a final step, the image contrast was normalized. All steps were fully automated and implemented as dedicated Matlab scripts (Wählby et al. manuscript in preparation).
Image decoding
After pre-processing, each image stack was reduced into a single image by maximum intensity projection. Every picture element (pixel) in this projection with intensity exceeding a general background threshold was identified as belonging to a potential ASM. The intensity information from the hybridization pattern of each such pixel was thereafter extracted from the image stack and represented as a vector. By also representing the patterns of the combinatorial decoding scheme as vectors, the patterns of the potential ASM-pixel could be compared to the expected patterns of the decoding scheme. Ratios of intensities produced more stable results than absolute values and the identities were therefore decoded by searching for the minimal angular deviation between a pixel vector and each vector of the decoding scheme (Wählby et al. manuscript in preparation). Groups of connected pixels of the same identity were identified and counted as true, decoded ASMs. This strategy for ASM identification minimizes the risk of detecting false ASMs, as background noise is usually random. Furthermore, the decoding scheme is error-tolerant because any misclassification requires both a loss and a gain of signal.
Normalization of ASM counts
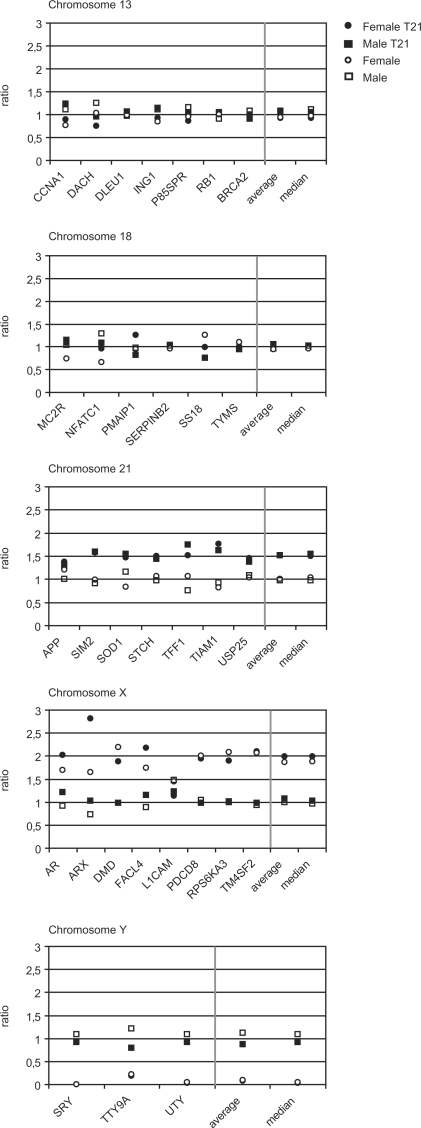
The number of counted ASMs may vary due to variation in ASM density between different imaged areas, but also due to inter-sample variation. For dilution series, counts were therefore normalized against counts of a second ASM type that was present in a known, constant concentration. For copy number variation analysis, counts were normalized against a pooled reference sample. The pooled sample was constructed to mimic a normal male sample by compensating female and trisomy 21 samples for known variations in chromosome. That is, the counts deriving from loci on chromosome 21 of trisomy subjects were compensated by dividing with 1.5 and counts from loci on chromosome X of female samples were divided by two. To compensate for variations in ASM density, all counts for a subject were divided by the total number of identified ASMs for that subject. The pooled reference sample was thereafter defined as the average of these relative ASM counts (for chromosome Y, only male samples were used). Finally, the relative ASM counts for each subject were compared to the pooled sample to generate the ratios plotted in Figure 4.
Figure 4.
Analysis of relative copy number of loci distributed along chromosomes 13, 18, 21, X and Y. The relative copy-number ratios were obtained by comparing the relative counts of the different ASMs with a normalized reference profile (y-axis). Gain of one copy of an autosomal chromosome should yield a ratio of about 1.5 and a ratio of about 2 would be expected for the X chromosomal loci in female samples. A total loss of a chromosome should yield a value of 0. The ratios are plotted in separate graphs for the different chromosomes. Squares denote male samples and circles female. Filled symbols represent samples from subjects with Down syndrome (trisomy 21) and open symbols represent pooled DNA samples. Median and average ratios from all loci of a chromosome are included to the right in the diagrams.
RESULTS
Evaluation of format properties
Any multiplex molecular analysis utilizing fluorescence for readout is limited by the number of fluorescence spectra that can be resolved (40). To enhance the number of distinguishable objects, serial targeting can be applied (37). We have developed a decoding scheme for identification of ASMs that uses a combination of spectral and serial targeting of sets of tags. By combining the information from several tags, more identities than there is tag probes can be decoded.
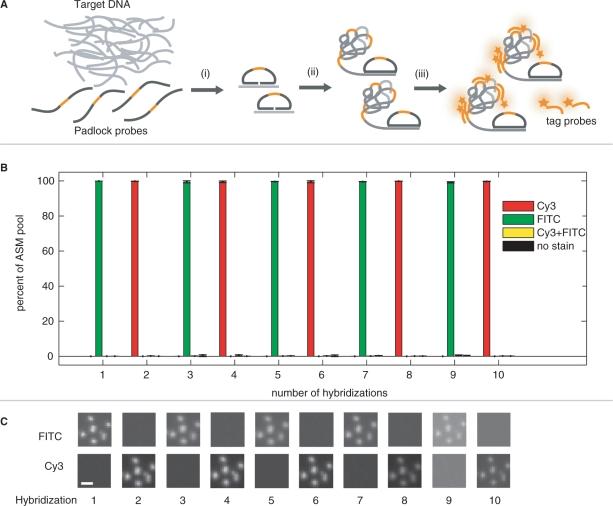
In our procedure, ASMs are first randomly attached to a glass surface in order to create an array. Since our approach to decode ASMs is based on serial hybridization reactions, we first investigated the ASM immobilization along with the efficiency of the hybridization and dehybridization reactions. ASMs were produced from circularized padlock probes, then immobilized on a poly-l-lysine modified glass surface and subjected to 10 cycles of hybridization and dehybridization. Two tag probes of the same sequence, but labeled with different fluorophores, were used in alternating hybridization cycles. In total, 20-images were collected with FITC- and Cy3-filters and subsequently processed and analyzed with dedicated Matlab software. More than 94% of the ∼23 000 detected ASMs responded with alternating FITC/Cy3 signals throughout all 10 hybridizations, with no tendency for loss of ASMs during the experimental series (Figure 1B).
Figure 1.
The stability of immobilized ASMs through iterations of hybridization, imaging and dehybridization. (A) Generation and decoding of random arrays of ASMs. Padlock probes are mixed with target DNA and (i) correct target recognition enables enzymatic joining of DNA circles. (ii) RCA of the circles generates concatemer products (ASMs) which are (iii) immobilized and labeled with fluorescent tag probes for visualization by fluorescence microscopy. (B) Iterative hybridizations to arrayed ASMs. A series of 10 cycles of hybridizations and dehybridization reactions were carried out to evaluate immobilization chemistry and the efficiency of the hybridization and dehybridization reactions. Two tag probes of the same sequence but labeled with different fluorescence dyes (Cy3 or FITC) were added in alternating cycles of hybridization and dehybridization reactions. The graph illustrates the proportion and fluorescence label of the detected ASMs after each step in the hybridization series. Six image areas were captured per cycle with an average of 7481 counted ASMs. Green label represents labeling with FITC in odd hybridization reactions and red implies labeling with Cy3 in even hybridization reactions. Yellow label represents ASMs that were both green and red, and black ASMs that were not detected in the respective hybridization step. (C) A small subset of raw ASM images illustrates the alternating hybridization and dehybridization cycles. The scale bar is 2 μm.
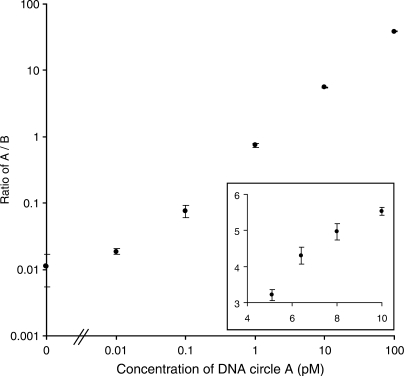
The linear range and quantitative precision are two key parameters for accurate relative quantifications. In order to investigate the quantitative precision and dynamic range of the ASM array readout, different concentrations of ligated padlock probes A were titrated in a fixed concentration of ligated padlock probes B and amplified by RCA. The ASMs were immobilized and analyzed using A and B tag probes labeled with different fluorophores. For each concentration of probe A, five areas of the corresponding random array were imaged. ASM concentration differences of 20% were discriminated and the linear dynamic range spanned at least 103 with an R2 value for the four data points in the linear range of 0.9986 (Figure 2). The average coefficient of variation between the imaged areas was 4% for the data points based on ASM counts higher than 200.
Figure 2.
Evaluation of dynamic range and quantitative precision. Ligated padlock probe A was titrated in steps of 10 in a fixed concentration of ligated padlock probe B (10 pM). The DNA circles were amplified by RCA, arrayed and detected by hybridizing tag probes to A and B specific tags. The counts from A probes ranged from 8 to 19 272 over the dilution series and the average B count was 464. In the graph, the A/B ratios of the two ASM types are plotted on the y-axis, with the corresponding concentrations of ligated padlock probe A on the x-axis. Inset shows a titration with 20% decrements. Axes of the inset are the same as those of the large graph. SDs are from A/B ratios in five imaged areas.
Multiplex targeted copy-number variation analysis
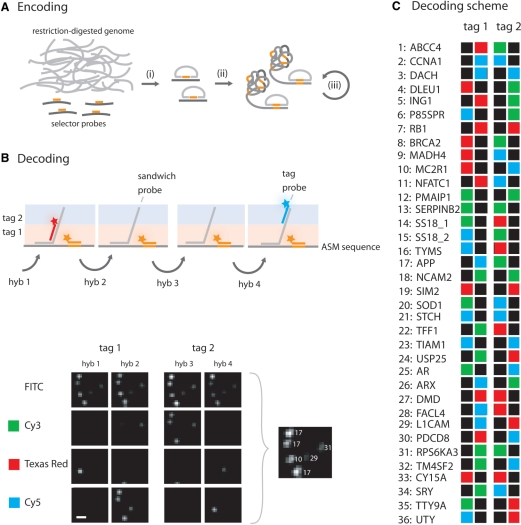
As a demonstration, we applied our decoding procedure for multiplex targeted copy-number variation analysis, using selector probes to generate DNA circles from genomic fragments in a targeted way (Figure 3A) (28). In this experiment, we chose to use four fluorescence channels, three for tag probes labeled with different fluorescence dyes and one for a general tag probe labeling all ASMs. The arrayed ASMs were exposed to cycles of hybridization, imaging and dehybridization using different sets of tag probes (Figure 3B). The ASMs were identified according to a combinatorial decoding scheme and counted (Figure 3C).
Figure 3.
Multiplex encoding and decoding of genomic loci targeted by selector probes. (A) A genomic sample is prepared by restriction digestion of the sample genome with an appropriate restriction enzyme. (i) Selector probes designed to target specific genomic sequences are added and genomic DNA circles are formed through the action of a thermostable DNA ligase. The circles are either (ii) directly amplified by RCA, or (iii) enriched for by a process which includes restriction digestion of the RCA products into monomers that can be ligated into new circles. The array is created by random immobilization of the ASMs to a poly-l-lysine slide. (B) Arrayed ASMs are decoded by sequential hybridizations of sandwich probes (grey), tag probes (red or blue) and a general tag probe (orange). The sandwich probes contain two regions, one complementary to a specific ASM and one region contains the two decoding tags. These decoding tags, denoted tag 1 and tag 2, are targeted by the tag probes during the hybridization series and their positions in the sandwich probe are illustrated as a pink or blue shade. A small 20 × 20 pixles image subset from one experiment is included, illustrating labeled ASMs after the different hybridization reactions along with an image showing the identified ASMs (scale bar is 2 μm). (C) The decoding scheme used for multiplex decoding of genomic fragments. The names of the gene loci and their corresponding number are listed vertically and the labels from the two tags are illustrated horizontally. Green labels are for Cy3, red labels represent Texas Red and blue correspond to Cy5 labeling. Black means no labeling, i.e. absence of a detectable signal (tag probe).
The performance of the procedure was evaluated by using genomic DNA samples from subjects with previously characterized chromosomal copy-number differences (trisomy 21 and sex chromosomes in male and female samples). In total, 36 selector probes targeting loci distributed along chromosomes 13, 18, 21, X and Y were designed using PieceMaker and ProbeMaker software (38,39). The target fragments for the selector probes were generated through digestion of total genomic DNA with the restriction enzyme Mnl I. Then selector probes along with a thermostable DNA ligase (Ampligase) were added to the fragmented DNA samples. The resulting DNA circles were of different size, which allowed us to verify probe performance using the recently described multiplex ligation-dependent genome amplification technique, via quantitative analysis of electropherograms (29). This analysis together with initial ASM experiments revealed that two of the selector probes targeted repeated regions (data not shown). These two selector probes were therefore excluded from the probe pool. The 34 remaining selector probes were applied to two samples of genomic DNA from trisomy 21 subjects (male, female), while two samples represented pooled male and female samples, respectively. To obtain a suitable density of ASMs on the slides, the ligated DNA circles were amplified with an initial RCA reaction (see Materials and methods section for detailed description of this procedure), under conditions that should generate between about 1200 to 2000 new ASMs per DNA circle, depending on the size of the circle (16). The resulting ASMs were randomly immobilized on a poly-l-lysine modified microscope slide. The ASM arrays were decoded by four cycles of hybridization and dehybridization. Since the ASM targets consisted of genomic DNA, the decoding tags were introduced via an intermediate ‘sandwich probe’. The sandwich probe is divided into two sections: a 5′-region which is complementary to a specific ASM target and a 3′-tail that is composed of a series of tags. In this study we used two tags (Figure 3B). Each hybridization reaction contained (i) a set of 34 sandwich probes specific for each of the targeted genomic loci, (ii) one of four sets of three tag probes (labeled with Cy3, Texas Red or Cy5) and (iii) the general tag probe (labeled with FITC) targeting a motif introduced in the circularized fragments via the selector probes (Figure 3A and Tables S3–S5). The general tag probe served to identify true ASMs. Only objects with the general tag probe qualify for further analysis, as described in Materials and methods section.
In each cycle, one image was captured for each of the fluorophores (FITC, Cy3, Texas Red and Cy5), resulting in a total of 16 images after completion of four cycles. Twelve to fourteen areas of each random ASM array were examined, containing 27 000 to 62 000 decoded ASMs. Images were pre-processed, detected ASM signal patterns were compared to the combinatorial decoding scheme and the number of ASMs of each species was counted. Species of ASMs that resulted in fewer than a median of seven counts in an imaged area were excluded. Data generated with this low number of counts were severely affected by background noise, estimated to between one and three counts per imaged area and ASM (data not shown). The counts per sample of the 31 remaining ASM species were normalized against a reference profile as described in the Materials and methods section. The observed ratios of ASMs from chromosome X were approximately 2-fold higher for female samples compared to male samples, with the exception of one locus that failed completely (L1CAM) and one sample in the ARX locus (T21 female sample). The loci distributed on the Y chromosome were detected only in the male samples and with a ratio of around 1. All loci distributed on chromosome 21 yielded ratios around 1.5-fold higher in the trisomy 21 samples compared to the normal samples, while all loci on chromosomes 13 and 18 yielded a ratio of about 1 (Figure 4). The results thus correspond well with the expected ratios.
DISCUSSION
We present a new random array format together with a decoding scheme for multiplexed readout of ASMs. By simply drying the ASM-containing solution onto the slide surface, a random array of all molecules in a sample is created that can be targeted by a series of hybridization reactions to decode the identity of the molecules. Therefore, potentially all molecules that have triggered the formation of a specific circular DNA molecule can be detected by scanning the complete random array area. We chose in this study to collect data from a subset of the total area, since the imaging was performed manually. The linear quantitative dynamic range and the detection sensitivity would be expected to increase linearly with the extended imaged area, while the quantitative precision improves with the number of detected objects, governed by Poisson sampling statistics.
The digital decoding technique was applied to estimate chromosome copy numbers using selector probes, targeting regions on three autosomal chromosomes and the sex chromosomes. ASMs were obtained from circularized probes and the products were identified using our decoding scheme (Figure 3C). We measured 31 loci in four genomic DNA samples from individuals of both sexes, including two trisomy 21 subjects. All but one of the interpreted loci responded as expected (Figure 4). In most cases, deviating data points can be explained by random variation due to low numbers of counted ASMs.
The selector probe generated genomic DNA circles were prepared to differ in size (76–133 nt in length), to allow verification of probe performance using the size-separation based multiplex ligation-dependent genome amplification technique (29). The length difference has a very predictable impact on the number of copies of a genomic sequence that is produced in the RCA. Since the selector probe generated circles were amplified twice in this study, first to generate more DNA circles and then to generate ASMs from these circles, both the number of ASMs and the number of monomers in each concatemer ASM will be affected by the size of the amplified circle. However, this can be expected to have little effect on the analysis since the amplification of DNA circles is a linear process and therefore the relative amount of ASMs should not be affected by the size difference. The number of monomers per ASM determines how many tag probes that can be hybridized and is therefore related to the fluorescence intensity of the ASMs. The absolute fluorescence intensity of the ASMs should have little effect on results, since our image analysis is based on fluorescence ratios. Even though the procedure is robust to difference in length, it is probably optimal to design an assay to generate DNA circles of the same size.
Our method serves to quantitatively decode multiplex populations of arrayed rolling circle ASMs that have been selected and prepared in solution. Recently, a new high-through-put sequencing approach was described, based on sequencing by hybridization using randomly arrayed rolling circle ASMs as sequencing substrates (17). The sequencing targets were unspecifically generated by ligating short randomly generated genome sequences flanked by general adaptors. Formed DNA circles were thereafter hybridized to an array of adaptor-complementary oligonucleotides and amplified with RCA from the immobilized primers. The products were sequenced by serial hybridizations of 582 Cy3-labeled pentamer probes (17). This strategy qualitatively assays an unspecifically generated pool of rolling circle ASMs that are grown directly on the array after hybridization of preformed DNA circles.
The ASM targets used in the copy-number analysis were generated from circularized genomic DNA sequences using selector probes. They can also be used to generate substrates for parallel high-throughput sequencing chemistries, many of which should be compatible with our ASM format. However, any biomolecule that can be represented as a DNA circle can be converted to an easily identifiable ASM. Padlock probes can be applied for gene-copy number analysis, as well as analysis of infectious pathogens and for mRNA expression. Also proteins or interacting pairs of proteins can be digitally monitored in this manner via the proximity ligation assay (33,35). Both padlock and selector probes have proven to work well in highly multiplexed reactions (25,28,31,41) and protein analyses so far have been scaled to 7-plex reactions using the proximity ligation assay, but with the potential to reach far higher numbers (34,42).
Padlock and proximity ligation probes, as well as other types of synthetic DNA, can be designed to include one or more tags in the probe sequence. This makes them directly suited for the decoding scheme presented here. As demonstrated in this study, multiple tags can be introduced to ASMs derived from genomic DNA by hybridization of sandwich probes. Regardless of how the tags are introduced, three variables affect the number of identities that can be decoded using the proposed strategy: the number of fluorophores that can be resolved in individual decoding reactions, the numbers of tags used and the number of serial hybridization reactions to the ASMs. As an example, we used two tags, each decoded by six different tag probes, requiring four hybridization reactions with sets of three tag probes. This design enabled identification of 36 (62) different genomic loci represented as ASMs. We have shown that the number of serial hybridization reactions can be extended to at least 10 without noticeable loss of signal (Figure 1B). The number of tags can practically be increased to at least three with the present design, and even more if overlapping or shorter tag probes are used. By extending the number of tags to three, each decoded by 12 different tag probes, (requiring nine hybridization reactions with sets of four tag probes) will enable simultaneous identification and quantification of 1728 (123) targets. The decoding potential would increase further by using more distinguishable fluorescence dyes, for instance quantum dots (43), and more tags.
Our method is easy to set up and does not require advanced equipment. In our approach we generate a rolling circle ASM library, which is significantly less time-consuming and labor intensive compared to the production of water-in-oil bead PCR ASM libraries. The array format is easy to set up and allows targeted analyses of for instance gene copy-numbers, as well as expression of mRNA and protein in highly multiplex assays with wide linear dynamic range and high quantitative precision.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
FUNDING
This work was supported by the Swedish Defence Nanotechnology Program, Uppsala BioX, the Swedish Research Council, the Knut and Alice Wallenberg Foundation, Göran Gustafsson Foundation and the European Union FP-7 research project READNA. Funding for open access charge: Swedish Research Council.
Conflict of interest statement. J.J. and M.N. have licensed the commercial rights to the technology to Olink AB (Uppsala, Sweden), a company in which J.J and M.N. also hold stock. J.G, C.W., M.I., and W.M.H declare no conflict of interest.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Ulf Landegren for critical review of the article.
REFERENCES
- 1.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 2.Syvanen AC. Accessing genetic variation: genotyping single nucleotide polymorphisms. Nat. Rev. Genet. 2001;2:930–942. doi: 10.1038/35103535. [DOI] [PubMed] [Google Scholar]
- 3.Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo WL, Chen C, Zhai Y, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat. Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 4.Solinas-Toldo S, Lampel S, Stilgenbauer S, Nickolenko J, Benner A, Dohner H, Cremer T, Lichter P. Matrix-based comparative genomic hybridization: biochips to screen for genomic imbalances. Genes Chromosomes Cancer. 1997;20:399–407. [PubMed] [Google Scholar]
- 5.Cantor CR, Mirzabekov A, Southern E. Report on the sequencing by hybridization workshop. Genomics. 1992;13:1378–1383. doi: 10.1016/0888-7543(92)90079-8. [DOI] [PubMed] [Google Scholar]
- 6.MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 7.Vogelstein B, Kinzler KW. Digital PCR. Proc. Natl Acad. Sci. USA. 1999;96:9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warren L, Bryder D, Weissman IL, Quake SR. Transcription factor profiling in individual hematopoietic progenitors by digital RT-PCR. Proc. Natl Acad. Sci. USA. 2006;103:17807–17812. doi: 10.1073/pnas.0608512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc. Natl Acad. Sci. USA. 2003;100:8817–8822. doi: 10.1073/pnas.1133470100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adessi C, Matton G, Ayala G, Turcatti G, Mermod JJ, Mayer P, Kawashima E. Solid phase DNA amplification: characterisation of primer attachment and amplification mechanisms. Nucleic Acids Res. 2000;28:E87. doi: 10.1093/nar/28.20.e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chetverina HV, Chetverin AB. Cloning of RNA molecules in vitro. Nucleic Acids Res. 1993;21:2349–2353. doi: 10.1093/nar/21.10.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 13.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, Church GM. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 15.Fire A, Xu SQ. Rolling replication of short DNA circles. Proc. Natl Acad. Sci. USA. 1995;92:4641–4645. doi: 10.1073/pnas.92.10.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dahl F, Baner J, Gullberg M, Mendel-Hartvig M, Landegren U, Nilsson M. Circle-to-circle amplification for precise and sensitive DNA analysis. Proc. Natl Acad. Sci. USA. 2004;101:4548–4553. doi: 10.1073/pnas.0400834101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pihlak A, Bauren G, Hersoug E, Lonnerberg P, Metsis A, Linnarsson S. Rapid genome sequencing with short universal tiling probes. Nat. Biotechnol. 2008;26:676–684. doi: 10.1038/nbt1405. [DOI] [PubMed] [Google Scholar]
- 18.Blab GA, Schmidt T, Nilsson M. Homogeneous detection of single rolling circle replication products. Anal. Chem. 2004;76:495–498. doi: 10.1021/ac034987+. [DOI] [PubMed] [Google Scholar]
- 19.Jarvius J, Melin J, Goransson J, Stenberg J, Fredriksson S, Gonzalez-Rey C, Bertilsson S, Nilsson M. Digital quantification using amplified single-molecule detection. Nat. Methods. 2006;3:725–727. doi: 10.1038/nmeth916. [DOI] [PubMed] [Google Scholar]
- 20.Larsson C, Koch J, Nygren A, Janssen G, Raap AK, Landegren U, Nilsson M. In situ genotyping individual DNA molecules by target-primed rolling-circle amplification of padlock probes. Nat. Methods. 2004;1:227–232. doi: 10.1038/nmeth723. [DOI] [PubMed] [Google Scholar]
- 21.Lizardi PM, Huang X, Zhu Z, Bray-Ward P, Thomas DC, Ward DC. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat. Genet. 1998;19:225–232. doi: 10.1038/898. [DOI] [PubMed] [Google Scholar]
- 22.Ericsson O, Jarvius J, Schallmeiner E, Howell M, Nong RY, Reuter H, Hahn M, Stenberg J, Nilsson M, Landegren U. A dual-tag microarray platform for high-performance nucleic acid and protein analyses. Nucleic Acids Res. 2008;36:e45. doi: 10.1093/nar/gkn106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nilsson M, Malmgren H, Samiotaki M, Kwiatkowski M, Chowdhary BP, Landegren U. Padlock probes: circularizing oligonucleotides for localized DNA detection. Science. 1994;265:2085–2088. doi: 10.1126/science.7522346. [DOI] [PubMed] [Google Scholar]
- 24.Baner J, Isaksson A, Waldenstrom E, Jarvius J, Landegren U, Nilsson M. Parallel gene analysis with allele-specific padlock probes and tag microarrays. Nucleic Acids Res. 2003;31:e103. doi: 10.1093/nar/gng104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hardenbol P, Baner J, Jain M, Nilsson M, Namsaraev EA, Karlin-Neumann GA, Fakhrai-Rad H, Ronaghi M, Willis TD, Landegren U, et al. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat. Biotechnol. 2003;21:673–678. doi: 10.1038/nbt821. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Moorhead M, Karlin-Neumann G, Falkowski M, Chen C, Siddiqui F, Davis RW, Willis TD, Faham M. Allele quantification using molecular inversion probes (MIP) Nucleic Acids Res. 2005;33:e183. doi: 10.1093/nar/gni177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baner J, Marits P, Nilsson M, Winqvist O, Landegren U. Analysis of T-cell receptor V beta gene repertoires after immune stimulation and in malignancy by use of padlock probes and microarrays. Clin. Chem. 2005;51:768–775. doi: 10.1373/clinchem.2004.047266. [DOI] [PubMed] [Google Scholar]
- 28.Dahl F, Gullberg M, Stenberg J, Landegren U, Nilsson M. Multiplex amplification enabled by selective circularization of large sets of genomic DNA fragments. Nucleic Acids Res. 2005;33:e71. doi: 10.1093/nar/gni070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Isaksson M, Stenberg J, Dahl F, Thuresson AC, Bondeson ML, Nilsson M. MLGA – a rapid and cost-efficient assay for gene copy-number analysis. Nucleic Acids Res. 2007;35:e115. doi: 10.1093/nar/gkm651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salmon Hillbertz NH, Isaksson M, Karlsson EK, Hellmen E, Pielberg GR, Savolainen P, Wade CM, von Euler H, Gustafson U, Hedhammar A, et al. Duplication of FGF3, FGF4, FGF19 and ORAOV1 causes hair ridge and predisposition to dermoid sinus in Ridgeback dogs. Nat. Genet. 2007;39:1318–1320. doi: 10.1038/ng.2007.4. [DOI] [PubMed] [Google Scholar]
- 31.Dahl F, Stenberg J, Fredriksson S, Welch K, Zhang M, Nilsson M, Bicknell D, Bodmer WF, Davis RW, Ji H. Multigene amplification and massively parallel sequencing for cancer mutation discovery. Proc. Natl Acad. Sci. USA. 2007;104:9387–9392. doi: 10.1073/pnas.0702165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gustafsdottir SM, Ostman A, Landegren U. Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 2002;20:473–477. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- 33.Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 34.Fredriksson S, Dixon W, Ji H, Koong AC, Mindrinos M, Davis RW. Multiplexed protein detection by proximity ligation for cancer biomarker validation. Nat. Methods. 2007;4:327–329. doi: 10.1038/nmeth1020. [DOI] [PubMed] [Google Scholar]
- 35.Jarvius M, Paulsson J, Weibrecht I, Leuchowius KJ, Andersson AC, Wahlby C, Gullberg M, Botling J, Sjoblom T, Markova B, et al. In situ detection of phosphorylated platelet-derived growth factor receptor beta using a generalized proximity ligation method. Mol. Cell. Proteomics. 2007;6:1500–1509. doi: 10.1074/mcp.M700166-MCP200. [DOI] [PubMed] [Google Scholar]
- 36.Nie B, Shortreed MR, Smith LM. Scoring single-nucleotide polymorphisms at the single-molecule level by counting individual DNA cleavage events on surfaces. Anal. Chem. 2005;77:6594–6600. doi: 10.1021/ac051025p. [DOI] [PubMed] [Google Scholar]
- 37.Gunderson KL, Kruglyak S, Graige MS, Garcia F, Kermani BG, Zhao C, Che D, Dickinson T, Wickham E, Bierle J, et al. Decoding randomly ordered DNA arrays. Genome Res. 2004;14:870–877. doi: 10.1101/gr.2255804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stenberg J, Nilsson M, Landegren U. ProbeMaker: an extensible framework for design of sets of oligonucleotide probes. BMC Bioinformatics. 2005;6:229. doi: 10.1186/1471-2105-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stenberg J, Dahl F, Landegren U, Nilsson M. PieceMaker: selection of DNA fragments for selector-guided multiplex amplification. Nucleic Acids Res. 2005;33:e72. doi: 10.1093/nar/gni071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiegant J, Bezrookove V, Rosenberg C, Tanke HJ, Raap AK, Zhang H, Bittner M, Trent JM, Meltzer P. Differentially painting human chromosome arms with combined binary ratio-labeling fluorescence in situ hybridization. Genome Res. 2000;10:861–865. doi: 10.1101/gr.10.6.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hardenbol P, Yu F, Belmont J, Mackenzie J, Bruckner C, Brundage T, Boudreau A, Chow S, Eberle J, Erbilgin A, et al. Highly multiplexed molecular inversion probe genotyping: over 10,000 targeted SNPs genotyped in a single tube assay. Genome Res. 2005;15:269–275. doi: 10.1101/gr.3185605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fredriksson S, Horecka J, Brustugun OT, Schlingemann J, Koong AC, Tibshirani R, Davis RW. Multiplexed proximity ligation assays to profile putative plasma biomarkers relevant to pancreatic and ovarian cancer. Clin. Chem. 2008;54:582–589. doi: 10.1373/clinchem.2007.093195. [DOI] [PubMed] [Google Scholar]
- 43.Chan WC, Nie S. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.