Abstract
Circadian mRNA oscillations are the main feature of core clock genes. Among them, period 2 is a key component in negative-feedback regulation, showing robust diurnal oscillations. Moreover, period 2 has been found to have a physiological role in the cell cycle or the tumor suppression. The present study reports that 3′-untranslated region (UTR)-dependent mRNA decay is involved in the regulation of circadian oscillation of period 2 mRNA. Within the mper2 3′UTR, both the CU-rich region and polypyrimidine tract-binding protein (PTB) are more responsible for mRNA stability and degradation kinetics than are other factors. Depletion of PTB with RNAi results in mper2 mRNA stabilization. During the circadian oscillations of mper2, cytoplasmic PTB showed a reciprocal expression profile compared with mper2 mRNA and its peak amplitude was increased when PTB was depleted. This report on the regulation of mper2 proposes that post-transcriptional mRNA decay mediated by PTB is a fine-tuned regulatory mechanism that includes dampening-down effects during circadian mRNA oscillations.
INTRODUCTION
The suprachiasmatic nucleus (SCN) in the hypothalamus governs circadian clock oscillation in mammals (1,2). Circadian oscillations in response to various stimuli have been demonstrated not only at the SCN in the brain but also in various peripheral tissues, and even in cultured cell lines (3). These oscillation mechanisms are regulated by a transcriptional negative feedback loop involving binding of the BMAL1/CLOCK heterodimer to the E/E′ box in the promoter region of clock genes and feedback inhibition by accumulated Per-Cry protein heterodimer, and by post-translational modification of protein products by cellular kinases and degradation by proteasomes (4). Among the core clock gene family, period 2 and period 1 are responsive to photic entrainment from the retina to the SCN (1,2,5,6). Robust circadian oscillations of per2 mRNA and Per2 protein are not only evident in the brain, but are also present in various peripheral tissues (7,8). period 2 is also known to have a physiological role in tumor suppression (9,10) and feeding/energy metabolism (11). For these reasons, period 2 regulation has been studied more than any other circadian molecule at the translational or post-translational level; however, no reports have demonstrated regulation at the level of mRNA degradation.
In general, mRNA degradation is controlled by two major components, the mRNA binding motif (cis-acting element, CAE) and its binding partners (trans-acting factors, TAFs) (12); thus mRNAs containing a CAE in their 3′-UTR may be regulated by various binding factors. The AU-rich element (ARE) in the 3′-UTR is well characterized and is the most commonly used CAE in post-transcriptional regulation. The fate of the mRNA is therefore largely determined by associations between the ARE and its binding factors (13).
The polypyrimidine tract-binding protein (PTB), also known as heterogeneous nuclear ribonucleoprotein (hnRNP) I, is a cellular RNA-binding protein that is abundant in many tissues. PTB binds to oligopyrimidine tracts in introns and regulates negative or positive exon definition in the differential splicing of various cellular mRNAs (14–17). More recently, PTB was shown to be involved in other aspects of RNA metabolism, such as modulation of polyadenylation efficiency (18) and the stability of nitric oxide synthase mRNA during inflammation (19). Moreover, PTB represents the prototype of a heterogeneous group of cellular RNA-binding proteins that are directly involved in the regulation of translation, a function that was discovered during investigation of translation of picornaviruses (20,21). Activated cytoplasmic PTB appears to affect the nutrient-dependent stabilization of mRNAs involved in insulin synthesis and secretion (22,23). In addition, regulation of protein kinase A by various hormone signaling pathways promotes nucleocytoplasmic relocalization of PTB (24), suggesting a direct role of PTB in the regulation of mRNA stability.
Circadian rhythms have been studied using transgenic or knock-out animal models to investigate the molecular mechanisms underlying the control of gene expression at the transcriptional level (25–28), whereas transcriptional feedback regulation is considered a representative model for understanding the phenomenum of circadian oscillation, little is known about the role of post-transcriptional regulation in circadian rhythm. Several reports have demonstrated oscillatory mechanisms mediated by post-transcriptional regulation in the fruit fly (29,30) and recent studies in mammalian systems revealed that regulation of mRNA decay is actively involved in circadian oscillation (31,32). Kojima et al. also reported post-transcriptional regulation of the core clock gene PER1; however, this occurred through modulation of the translational process. Regulatory mechanisms acting at many different stages from core clock gene transcription to protein activation allow finely-tuned rhythmic oscillation. With the ongoing addition of new molecular components to the core model, our understanding of the oscillation mechanism is becoming more comprehensive (33).
The present reports proposes that mRNA stability of the circadian core clock gene mper2 is modulated by the 3′UTR and cytoplasmic PTB, and that mRNA instability is mediated by cytoplasmic increases in PTB protein. Our study also suggests that cytoplasmic PTB-mediated mRNA decay plays an important role in the modulation of peak amplitude or in dampening down the mper2 mRNA circadian oscillation.
MATERIALS AND METHODS
Plasmid constructions
mper2 3′-UTRs were amplified from full-length mouse per2 cDNA using specific primers (GenBank accession number NM011066). Amplification of cDNA was performed with pfu polymerase (Solgent) and confirmed by sequencing. To generate pcNAT-3′-UTR (mper2 3′-UTR inserted into 3′-UTR of AANAT), pGL3-3′-UTR, and pSK′-3′-UTR, PCR products were digested with EcoRI/XbaI and inserted into the EcoRI/XbaI site of pcNAT control which expresses rat aanat without 3′-UTR (32), modified pGL3-control (Promega), and modified pSK′ (32) using EcoRI/XbaI site, respectively. To dissect the regions involved in post-transcriptional regulation, serially deleted and mutated 3′-UTR fragments were cloned into the 3′-UTR region of the reporter using the EcoRI/XbaI digestion site.
Cell culture and dexamethasone treatment
CHO-K1 cells were cultured in DMEM/F-12 (Welgene) with 10% bovine calf serum (Hyclone)/1% antibiotics (Welgene) and HEK 293T cells in DMEM (Welgene) with 10% fetal bovine serum (Welgene)/1% antibiotics. NIH-3T3 and HeLa cells were cultured in DMEM with 10% fetal bovine serum/1% antibiotics, and maintained in humidified 95% air/5% CO2 incubator.
Circadian oscillation of NIH 3T3 cells was synchronized by treatment with 100 nM dexamethasone for 2 h. Instead of the usual single dexamethasone treatment, dexamethasone shock was applied twice at −24 h and 0 h before the onset of the circadian period, and cells were monitored throughout the following 2 days.
Transient expression, reporter assay, and mRNA quantification
For expression of the reporter constructs, cells were counted and plated in a 24-well plate at a density of 105 cells/well. After 24 h, 0.7 μg DNA was introduced into each well by lipid-mediated transfection using Welfect transfection reagent (Welgene) following the manufacturer's manual. Post-transfected cells were treated with the transcription blocker actinomycin D (5 μg/ml) at time 0 h and harvested at indicated times thereafter. AANAT assay and luciferase assay for reporter activity were performed as previously reported (34) and following the manufacturer's manual, respectively. mRNA levels of reporters were detected by northern blot analysis with an [α-32P] dCTP(NEN)-labeled specific probe or by quantitative real-time PCR using a MyIQ single time real-time detector system (Bio-Rad) with SYBR Green PCR mixture (Bio-Rad, Takara) as described previously (31). Specific primer pairs for firefly luciferase, endogenous mper2, mgapdh, mrpl32, and mtbp were used (primer sequences are provided in supplementary Table2).
Establishment of a stable cell line
A pP2PL-3′-UTR target vector was constructed by replacing the CMV promoter region with the full length mper2 promoter (GenBank accession no. AF491941) using BglII/BamHI and replacing the AANAT ORF with the firefly luciferase ORF using the BamHI/XhoI site within the pcNAT-per2 3′-UTR. NIH-3T3 cells were transfected with target vector by the calcium phosphate precipitation method and transfected cells were selected by incubation in selection medium containing G418 (700 μg/ml). Selected colonies were screened by reverse transcriptase–polymerase chain reaction (RT–PCR) to confirm stable expression of reporter mRNA.
In vitro binding/UV cross-linking/immunoprecipitation
To identify proteins specifically bound to mper2 3′-UTR, in vitro binding and UV cross-linking assays were performed as described previously (35). pSK′-3′-UTR constructs were constructed for T7 RNA polymerase-based in vitro transcription (Promega, Roche). [α-32P] UTP (NEN)-abeled RNAs and unlabeled competitor RNA were transcribed in vitro using T7 RNA polymerase and XbaI linearized pSK′-3′-UTR constructs. Equal molar amount of labeled RNAs were incubated with 15 µg nuclear extracts or 30 μg cytoplasmic extract of either HEK293T cell or mouse SCN for 20 min at 30°C. The RNA-protein binding reaction was carried out in a 30 µl reaction mixture containing 0.5 mM DTT, 5 mM HEPES (pH 7.6), 75 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 4% glycerol, 20 U RNasin (Promega), and 4 µg yeast tRNA. When indicated, unlabeled competitor transcripts were included in the reaction mixture. After incubation, the samples were UV-irradiated on ice for 10 min with a CL-1000 UV-crosslinker (UVP). Unbound RNAs were digested with 5 µl RNase cocktail (2.5 µl RNase A [10 mg/ml] and 2.5 µl RNase T1 [100 U/ml; Roche]) at 37°C for 1 h. The reaction mixtures were analysed by SDS-PAGE and autoradiography.
For immunoprecipitation, RNase digested lysates were incubated with specific antibodies or pre-immune serum for the negative control. After overnight incubation, Protein G agarose beads (Amersham bioscience) were added to the sample, which was further incubated for 3 h. Washed beads were analysed by SDS-PAGE and autoradiography.
In vitro decay assay
In vitro decay assay was performed as described previously (36). Briefly, in vitro transcribed [α-32P] UTP-labeled/capped mRNAs were incubated with cytoplasmic S100 lysate with or without purified recombinant PTB protein (a gift of Dr Jang) or competitor RNA for the indicated time. Reactions were terminated by addition of urea lysis buffer (7 M urea; 2%SDS; 350 mM NaCl;10 mMEDTA;10 mM Tris-Cl pH 7.5). Phenol-chloroform extracted probes were dissolved in 95% formamide RNA loading buffer. Samples were separated in 6% SDS-PAGE/7 M Urea, then analysed by autoradiography.
RNA interference
siRNA and DNA based shRNA were designed for endogenous PTB knock down. Two target sites were selected by the web-based siRNA design programs: http://www.dharmacon.com/DesignCenter/DesignCenterPage.aspx http://www.ambion.com/techlib/misc/siRNA_tools.html
UTR target siRNAs for PTB were synthesized as recommended (Samchully Pharm.). Selected siRNA sequences are given in supplementary Table 1. DNA-based shRNA was generated using the H1 promoter from a modified pShuttle vector transfection system, as described previously (35). PAGE purified oligonucleotides were annealed in 2 × SSC and subcloned into the BglII/HindIII site. The oligonucleotide sequences are given in supplementary Table 1. To express the shRNA, cells were plated in a 24-well plate at a density of 105 cells/well. After 24 h, 0.7 μg DNA was introduced into each well by lipid-mediated transfection using Welfect transfection reagent (Welgene) following the manufacturer's instructions. For siRNA transfection into NIH 3T3 cells, a microporator (Digital-Bio) was used as recommended by the manufacturer with conditions of 1300 V/20 ms/2 pulses 12 h prior to dexamethasone shock.
Cytoplasmic/nuclear protein preparation and immunoblot analysis
Fractionation of CHO-K1 cells, NIH-3T3 cells, and mouse SCN into cytoplasmic and nuclear extracts was performed as described previously (35,37). Immunoblot analyses were performed with polyclonal anti-PTB, monoclonal anti-14-3-3ξ, and monoclonal anti-GAPDH (ICN) as primary antibodies. HRP-conjugated species-specific secondary antibodies (KPL) were visualized using a SUPEX ECL solution kit (Neuronex) and a LAS-4000 chemiluminescence detection system (FUJIFILM), and acquired images were analysed using Image Gauge (FUJIFILM) according to the manufacturer's instructions.
RESULTS
mper2 mRNA is degraded in a phase-dependent manner
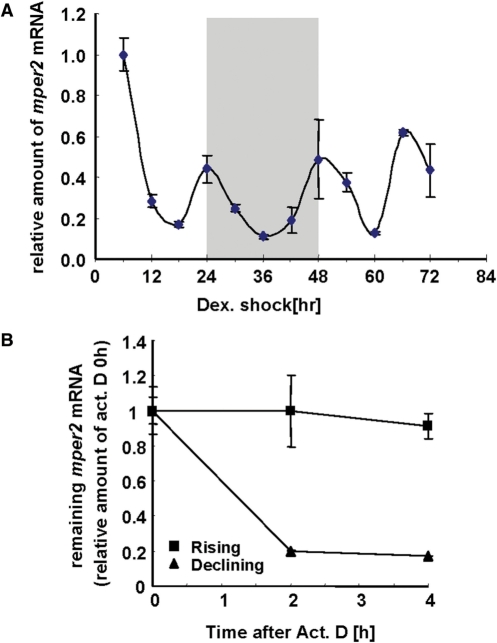
mRNA oscillation of mper2 is well established in various tissues including the brain and even in fibroblast cell lines (34,35). To elucidate the mechanism of phase-dependent mRNA decay the mper2 mRNA oscillation pattern was monitored in the NIH 3T3 fibroblast cell line after synchronization by dexamethasone treatment. Under our modified treatment conditions, robust mper2 mRNA oscillation could be observed (Figure 1A) and the mRNA profile was not significantly different from the pattern obtained after a classic single dexamethasone treatment (data not shown) (40). In this oscillatory condition, mRNA degradation kinetics were compared in the rising and declining phases during day 2 (shaded box in Figure 1A). To avoid any influence of media change (3), we monitored all profiles during the 2nd day after induction instead of the 1st day. As shown in Figure 1B, when actinomycin D was used to inhibit transcription, more dramatic decay was observed in the declining phase (closed triangles) than in the rising phase (closed squares). The role of transcriptional regulation in circadian mRNA oscillation is well established, and such oscillation is generally thought to be mainly governed by the action of transcription factors such as the BMAL1/CLOCK heterodimer. However, our results indicate that mRNA decay also plays a role in the circadian mRNA regulatory mechanism.
Figure 1.
mper2 mRNA is degraded in a phase dependent manner. (A) Oscillation of endogenous mper2 mRNA was measured by quantitative real-time PCR in NIH 3T3 cells. NIH 3T3 cells were treated with 100 nM dexamethasone twice for 2 h at −24 and 0 h to synchronize the circadian oscillation. Total RNA (1 μg) from each time point was subjected to quantitative real time RT–PCR with specific oligonucleotides for mper2 and mtbp (TATA box binding protein) as a normalizing control. All results are expressed as the mean ± SEM of three independent experiments. (B) mRNA degradation kinetics were measured during both declining (closed triangles) and rising (closed squares) phases in the 2nd day after dexamethasone treatment (shaded box in Figure 1A). Cells were treated with 5 μM actinomycin D at 32 h, during the rising phase, and at 40 h, during the declining phase, and then harvested at 2 h intervals. Total RNA (1 μg) was subjected to quantitative real time RT–PCR with specific primer pairs against mper2, or mrpl32 as a normalizing control. All results are expressed as the mean ± SEM of three independent experiments.
The mper2 3′-UTR plays a role in its own degradation
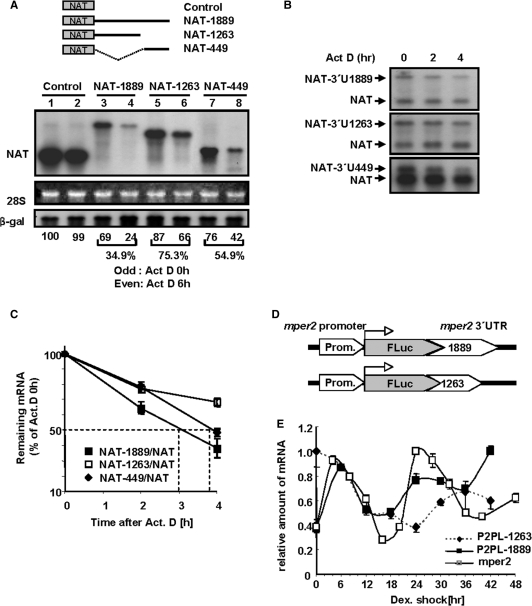
Because mRNA decay is typically mediated by the 3′-UTR, we introduced serially deleted NAT(serotonin-N-acetyltransferase) reporter constructs containing various lengths of the mper2 3′-UTR into CHO-K1 cells (32) to determine the critical cis-acting element. In cells treated with the transcription blocker actinomycin D for 6 h, (Figure 2A) the full-length (1889nts) and 3′-end (449nts) of mper2 3′-UTR triggered dramatic decreases in mRNA levels to 34.9% and 54.9% of the untreated level respectively, whereas the reporter gene alone and reporter fused to a construct with a 3′-end truncation (1263 nts) were relatively stable (99 and 75.3%, respectively). This difference in stability was also observed at the level of the reporter protein (supplementary Figure S1). These trends in stability were also observed in the NIH 3T3 cell line which is typically used as a circadian model system (data not shown).
Figure 2.
mper2 3′-UTR is responsible for its own mRNA degradation.(A) Northern blot analysis of NAT reporter: 3′-UTR constructs. CHO-K1 cells were transiently transfected by electroporation with NAT reporter alone or NAT reporter fused to serially deleted mper2 3′-UTR (shown in schematic diagrams, the numbers indicate nucleotide lengths of the 3′-UTR). β-gal was used as a transfection control. Transfected cells were incubated for 12 h before treatment with 5 μM actinomycin D. Total RNA (10 μg) was hybridized with a probe specific for AANAT coding sequence. Et-Br stained 28S rRNA provided a loading control. Odd numbered lanes, 0 h; even numbered lanes, after 6 h treatment with 5 μM actinomycin D. (B) Degradation kinetics and half-life of 3′-UTR:reporter constructs were analysed by actinomycin D pulse chasing and northern blot hybridization. Experimental schemes are the same as in panel A. Representative results of three independent experiments are shown. (C) The band intensities in panel B were quantified by phosphoimaging and normalized to the intensity of cotransfected AANAT control in each lane. The graph shows the mean ± SEM of three independent experiments. (D) Diagrams of recombined reporter constructs consisting of mper2 promoter (∼1.7 kb), firefly luciferase open reading frame, and either full-length mper2 3′UTR or 3′ region deleted mper2 UTR. (E) Firefly luciferase mRNA expression from stably recombined reporter vectors and endogenous mper2 mRNA were measured by quantitative real-time RT–PCR using cDNA harvested at 6 h (cell line) or 4 h (endogenous mper2) intervals from stably transfected NIH 3T3 cell lines that were synchronized by treatment with dexamethazone. All results are representative of three independent experiments from each selected line.
Actinomycin D pulse-chasing was applied to analyse reporter mRNA decay kinetics following co-transfection with a control reporter lacking the 3′-UTR and a 3′-UTR fused reporter (Figure 2B). When quantified relative to expression of the control construct, reporter genes fused to full-length or 3′-end UTR (3′-U449) had half-lives of ∼3 h or ∼4 h, respectively, whereas the 3′-end truncated UTR (3′-UTR1263) had a much longer half-life (Figure 2C).
To examine the effect of the 3′-UTR on mRNA oscillation, we constructed stable NIH 3T3 cell lines that express vectors containing the endogenous mouse per2 promoter, firefly luciferase open reading frame (ORF), and either full-length or 3′-truncated mper2 3′-UTR. Although several groups have reported that canonical or non-canonical putative E/E' box or D box elements in the promoter of clock genes are sufficient to generate robust oscillation (27,33), we used the full length mper2 promoter (∼1.7 kb) cloned from genomic DNA to more accurately reflect the endogenous transcriptional activity of mper2 (Figure 2D). As shown in Figure 2E, cells expressing vector with full-length 3′-UTR displayed a similar oscillation pattern to that of endogenous mper2, although the phase was slightly advanced. In contrast, cells expressing the 3′-end-deleted UTR showed an unusual pattern of mRNA oscillation, indicating that the 3′-truncated UTR is not sufficient to generate an oscillation pattern similar to the endogenous rhythm of mper2. The unusual oscillation of the truncated mRNA may be the results of mRNA stabilization. When we tested the oscillation profile of a reporter lacking per2 promoter the mRNA did not show a natural oscillating profile(data not shown) indicating that transcriptional regulation is required for circadian mRNA oscillation. These results suggest that the 3′-UTR, in particular the 3′-end sequence, can modulate mper2 mRNA oscillation in association with transcriptional regulation.
PTB binds to pyrimidine tract in mper2 3′-end sequence
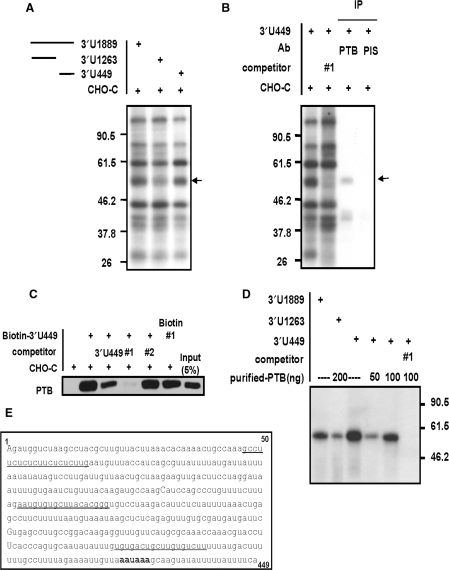
We next investigated mRNA binding proteins to identifie the protein(s) responsible for the changes in mRNA stability associated with truncation of mper2 3′-UTR. The mRNA binding profile was analyzed by an in vitro binding/UV cross-linking assay using in vitro transcribed mRNAs. Although the contribution of indirectly bound proteins could not be excluded, the direct binding profile of many proteins was examined. One protein of ∼57 kDa exhibited enhanced binding to the full-length 3′-UTR and 3′-U449, but not to the 3′-end truncated UTR (Figure 3A, supplementary Figure S2A). These results suggest that the 3′-end region of mper2 mRNA 3′-UTR functions as a critical region to destabilize mRNA and provides a binding site for the ∼57 kDa protein, which acts as a trans-acting factor. To identify the interacting protein, we used in vitro transcribed biotinylated mper2 3′-UTR to isolate RNA-bound proteins, which were separated by SDS-PAGE (data not shown). The 57 kDa protein (p57) was analysed by MALDI-TOF mass spectrometry as described previously (35) yielding a strong match to the polypyimidine tract binding protein (PTB). To confirm the identity of the protein and its binding to mper2 3′-UTR, immunoprecipitation was performed on the in vitro binding/UV cross-linked extract using a polyclonal anti-PTB antibody. As shown in Figure 3B, PTB was detected by the antibody, indicating that PTB interacts directly with mper2 3′-UTR (lane 3). PTB binding was also observed when the same experiment was repeated with nuclear extract of the SCN of the hypothalamus (supplementary Figures S2A and S2B); however, it was not clear whether the bound PTB was the ubiquitous PTB isoform or neuronal PTB (nPTB) which is specifically expressed in the brain. We also confirmed binding of PTB to biotin-labelled mper2 3′-UTR by immunoblotting with anti-PTB antibody and biotin-streptavidin affinity purification (Figure 3C). Binding of PTB to full length 3′-UTR or the 3′-end region was dramatically inhibited by competition with unlabeled UTR (Figure 3C lane3, supplementary Figure S2E) and deoxyoligonucleotide #1, which is specific for PTB binding (Figure 3B lane 2, 3C lane 4, underlined in Figure 3E, supplementary Figure S2D, supplementary Table2) (41), but was unaffected by competition with oligonucleotide #2 or #3 (data not shown). Specific binding of PTB to the mper2 3′-UTR end region was also observed in a binding experiment with recombinant purified PTB4 protein (Figure 3D), in which the binding capacity was shown to be concentration-dependent (supplementary Figure S2C). These results suggest that PTB directly binds to mper2 3′-UTR and that binding occurs preferentially on the 3′-end region.
Figure 3.
PTB binds to the pyrimidine tract in the mper2 3′-UTR end sequence. (A) Each in vitro transcribed 3′-UTR was subjected to in vitro binding and UV cross-linking assay with CHO-K1 cytoplasmic extract. The arrow indicates the 57 kDa protein that shows differential binding. (B) In vitro binding/UV cross-linking/immunoprecipitation was performed with the 449nts 3′-end region as described in Materials and Methods. In lane 2 the reaction was co-incubated with 1 μM competitor oligonucleotide #1, which specifically binds to PTB. Lanes 3 and 4 show immunoprecipitation with polyclonal anti-PTB antibody or pre-immune serum (PIS), respectively. (C) The in vitro transcribed 3′-U449 construct was labeled with biotin-UTP and incubated with CHO-K1 cell cytoplasmic extract. Streptavidin-affinity purified samples were separated by SDS-PAGE and subjected to western blotting with anti-PTB antibody. Abundant PTB was detected in the reaction with biotin labeled mRNA (lane 2). PTB binding decreased in the presence of a 3-fold excess of unlabeled mRNA (lane 3) or when 1 μM deoxyoligonucloetide competitor #1 was added (lane 4; specific binding of biotin-labeled oligonucleotide #1 to PTB is shown in lane 6) but was not affected by oligonucleotide #2 (lane 5). (D) The experiments in panels A and B were repeated with purified recombinant PTB4 protein. (E) Period 2 3′-UTR end region of 449 nts. Competitor oligonucleotide sequences are underlined and the authentic poly A signal is indicated in bold.
PTB destabilizes mper2 mRNA 3′-UTR
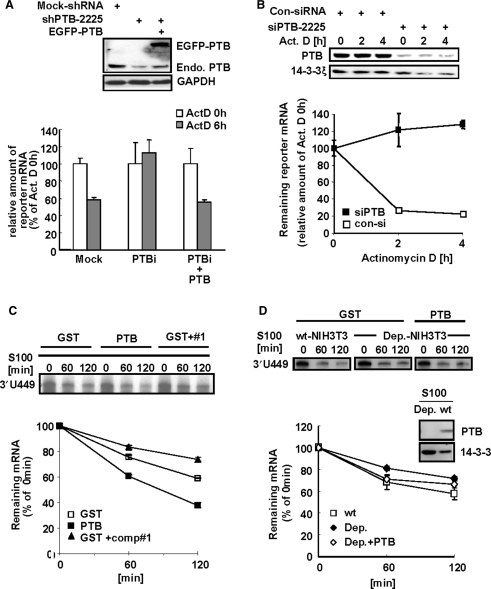
Although PTB acts as a splicing regulator, several reports have shown that PTB can also bind to 3′-UTR regions and stabilize target mRNAs (23,42,43). The effect of PTB on mper2 mRNA stability was examined by transient expression analysis using a 3′-UTR containing reporter and actinomycin D pulse-chase experiments in HeLa cells. Depletion of endogenous PTB was successfully achieved by targeted RNAi and could be ectopically rescued by co-expression of GFP-PTB (Figure 4A upper panel). When cytoplasmic PTB was knocked down by RNA interference, the reporter mRNA was very stable after treatment with actinomycin D compared with controls or cells that were rescued by GFP-PTB (Figure 4A). In addition, the degradation kinetics of reporter mRNA after the actinomycin D pulse were also influenced by PTB depletion in NIH 3T3 cells (Figure 4B upper panel). Transiently expressed reporter mRNAs were stabilized by cotransfection with siRNA targeting PTB (Figure 4B, closed squares), compared with transfection with control siRNA (Figure 4B, open squares). The destabilizing effect of PTB was also observed in an in vitro mRNA decay assay (Figures 4C and 4D). A radio-labeled in vitro transcribed 3′-UTR probe was incubated with an S100 cytoplasmic extract of HeLa or NIH 3T3 cells. When purified PTB4 protein was added to the reaction mixture, degradation occurred more rapidly (Figure 4C, closed squares) than with the GST control (Figure 4C, open squares). In contrast, when competitor oligonucleotide #1 was added (41), PTB binding to mRNA was completely inhibited (supplementary Figure S2D) and degradation kinetics slowed down (Figure 4C, closed triangles). In addition, mRNA decay was slower in the presence of PTB-depleted NIH 3T3 S100 extract (Figure 4D, middle panel, closed diamonds) and recovered when recombinant PTB was added (Figure 4D, open diamonds). Together, these results indicate that PTB has mRNA destabilizing activity when it binds to mper2 3′-UTR.
Figure 4.
PTB destabilizes mper2 3′-UTR containing mRNA. (A) HeLa cells were transiently transfected with firefly luciferase reporter construct containing mper2 3′-UTR, β-gal as a normalizing control, and the pShuttle-shPTB plasmid for endogenous PTB knock down, with or without GFP-tagged PTB, and incubated for 24 h before treatment with 5 μM actinomycin D. Total RNA (1 μg) was reverse-transcribed using oligo-dT primer then quantified by real-time PCR. Each value was normalized to β-gal activity. The open bar indicates the mRNA level at time 0 h of actinomycin D treatment and the grey bar indicates the mRNA level 6 h after actinomycin D treatment. The error bars represent the mean ± SEM of three independent experiments. Knocked down and rescued PTB protein levels were validated by western blotting (upper panel). (B) NIH 3T3 cells were transiently expressed with firefly luciferase reporter containing mper2 3′-UTR, β-gal as a normalizing control, and siPTB for endogenous PTB knock down, then incubated for 24 h followed by 5 μM actinomycin D pulse-chase. Total RNA (1 μg) was reverse transcribed using oligo-dT primer then quantified by real-time PCR. Each value was normalized to β-gal activity. Open and closed squares indicate transfection with control siRNA and siPTB-2225, respectively. The error bars represent the mean ± SEM of three independent experiments. Reduction of PTB protein level by siRNA was validated by western blotting (upper panel). (C) In vitro transcribed radiolabeled 3′-UTR probe was incubated with HeLa S100 cytoplasmic extract with or without recombinant PTB (100 nM) or with competitor oligonucleotide #1 (1 μM) for the indicated time. Phenol-chloroform extracted probes were separated in 7 M urea/6%acrylamide gel and subjected to autoradiography (upper panel). Image intensity of each probe was quantified by a phosphoimager. The error bars represent the mean ± SEM of duplicate measurements. (D) In vitro transcribed 3′-UTR probe was incubated for the indicated times with either wild type (wt) NIH-3T3 S100 cytoplasmic extract or PTB-depleted cell extract (Dep.) with or without rescue with recombinant PTB (100 nM). Phenol-chloroform extracted probes were separated in 7 M urea/6%acrylamide gel and subjected to autoradiography (upper panel). Signal intensity was quantified by a phosphoimager (lower panel). The error bars represent the mean ± SEM of duplicate measurements. PTB depletion of the S100 extract was confirmed by immunoblotting (middle panel).
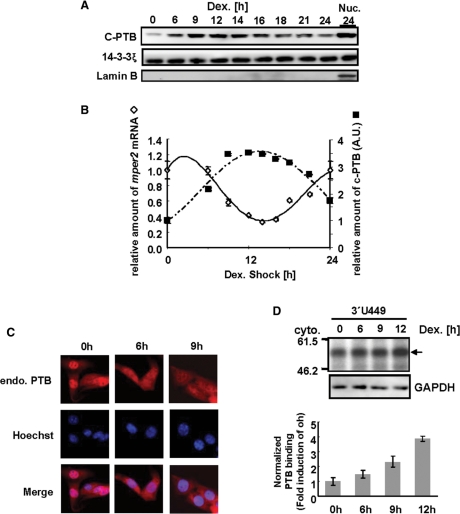
Cytoplasmic PTB shows reciprocal expression pattern with mRNA oscillation
PTB plays a role in mRNA splicing to regulate excision of certain exons (15,44) and PTB levels in the total cell lysate of various cell lines is very high (data not shown). PTB is predominantly localized in the nucleus where mRNA splicing occurs; however, because cellular mRNA decay mainly occurs in the cytoplasm we examined the cytoplasmic PTB level. Interestingly, the amount of cytoplasmic PTB increased according to the specific period of circadian time following 2h exposure of NIH 3T3 cells to dexamethasone (Figure 5A and supplementary Figure S3), but the total level of PTB did not change (data not shown). Furthermore, the pattern of cytoplasmic PTB levels was almost reciprocal to that of mper2 mRNA levels (Figure 5B). Immunostaining analysis confirmed the increase in levels of cytoplasmic PTB (Figure 5C). To examine the amount of cytoplasmic PTB that bound to mRNA, aliquots of the cytoplasmic protein fraction from each time point were coincubated in vitro with radiolabeled mper2 3′-UTR. Compared with other binding proteins, the binding of cytoplasmic PTB to mRNA increased during the phase of mRNA decline (6–12 h) (Figure 5D). Since decay of mature mRNA occurs mainly in the cytoplasm, our results suggest that the increase in cytoplasmic PTB and its binding to target mRNA are responsible for the rapid decay of mper2 mRNA during the declining phase.
Figure 5.
Cytoplasmic PTB shows reciprocal expression pattern with mRNA oscillation. (A) NIH 3T3 cells were treated with 100 nM dexamethasone, harvested at the indicated times, and cytoplasmic extract prepared for western blot analysis. Cytoplasmic PTB was detected with polyclonal anti-PTB antibody, and 14-3-3ξ was included as an internal control. Nuclear lamin B was used as a nuclear marker. The results shown are representative of three independent experiments. (B) Relative levels of cytoplasmic PTB, normalized to 14-3-3ξ, were calculated and plotted on panel B (closed squares) for the 2nd day after treatment. Relative levels of endogenous mper2 mRNA (open diamonds) were also quantified using the mouse TATA box binding protein (mtbp) as an internal control for each time point. The results shown are representative of three independent experiments. Error bars represent the mean ± SEM. (C) Cytoplasmic PTB in NIH 3T3 cells was observed by staining with polyclonal anti-PTB antibody at the indicated times after dexamethasone treatment. (D) NIH 3T3 cell cytoplamic extracts prepared at the indicated times after dexamethasone treatment were subjected to in vitro binding assay with radiolabeled 3′-UTR mRNA. Results shown are representative of three independent experiments. The arrow indicates cytoplasmic PTB binding (upper panel). PTB binding intensities were quantified by normalization to GAPDH levels in the same extracts (lower panel). The error bars represent the mean ± SEM.
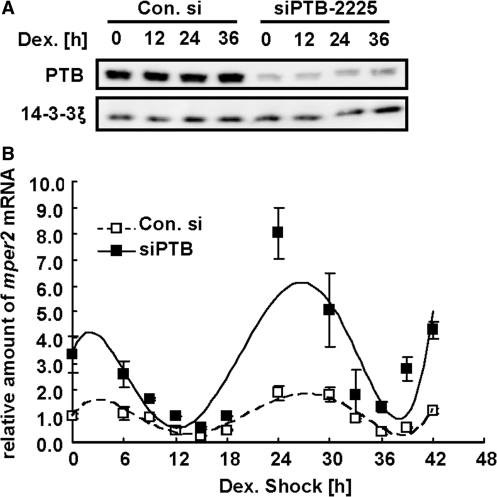
PTB down regulation affects mper2 mRNA circadian peak amplitude
The influence of PTB on mper2 mRNA stability and oscillation was further examined by measuring the mper2 mRNA oscillation profile in NIH 3T3 cells when PTB was downregulated. Transfection wih siRNA efficiently knocked down endogenous PTB proteins throughout the experimental circadian period (Figure 6A). Interestingly, PTB-depleted cells showed a significant increase in peak amplitude of mper2 mRNA expression (Figure 6B), but did not exhibit changes in mper2 mRNA oscillation pattern other than a slight advance of peak time compared with cells treated with control siRNA. The mRNA level of bmal1, the transcription activator of the mper2 gene, was not significantly different between PTB-depleted cells and control cells (data not shown), indicating that PTB acts specifically at the post-transcription level on the 3′-UTR of mper2 mRNA. Taken together, these results show that the cytoplasmic level of PTB plays a crucial role in regulating mper2 mRNA stability and damping down the oscillation rhythm, critical functions for the maintenance of a robust mRNA oscillation pattern.
Figure 6.
PTB down regulation affects mper2 mRNA circadian oscillation amplitude. (A) Western blot showing reduction of endogenous PTB in total cell extract from NIH 3T3 cells by PTB siRNA. 14-3-3ξ was used as an internal control. The results shown are representative of three independent experiments. (B) Effect of PTB depletion on endogenous mper2 mRNA oscillation. NIH 3T3 cells were transfected with control siRNA (open squares) or siPTB (closed squares) and grown to 80% confluency prior to treatment with 100 nM dexamethasone to synchronize the circadian oscillation (time 0 h). Total RNA (1 μg) from each time point was subjected to quantitative real time RT–PCR with primers specific to mper2 or mtbp (TATA box binding protein) as a normalizing control. Results shown are representative of three independent experiments. The error bars represent the mean ± SEM.
DISCUSSION
Post-transcriptional regulation in circadian per2 mRNA oscillation
Autonomous circadian biorhythms have been studied for many years; however, we still do not fully understand the regulation of such rhythms and it is generally assumed that other molecular regulatory mechanisms must be involved in addition to transcriptional negative feedback. Although post-transcriptional regulation of circadian rhythm in Drosophila was reported a decade ago (30), its role in the mammalian circadian system has only recently been revealed. Several reports have demonstrated that post-transcriptional regulation is involved in active mRNA decay in pineal gland melatonin biosynthesis and in the oscillation of the core circadian gene mper3 (31,32). Translational regulation and miRNA involvement were also reported in circadian rhythmicity (37,45,46). The present study investigates the functional role of mRNA decay in circadian mper2 gene regulation. mper2 mRNA decay kinetics were verified using a modified synchronization method, double-dexamethasone shock, in which an additional 100 nM dexamethasone shock was applied 24 h prior to the standard treatment at 0 h. This technique gave reproducible mRNA (Figure 1A) and cytoplasmic PTB (Figure 5A) profiles and the mRNA profile induced by our modified treatment was not significantly different from that reported previously (40). Although transcriptional activation and repression of circadian genes is the main regulatory mechanism (47), our results indicate that mRNA stability also plays an important role, and implicate the 3′-UTR as a major regulatory element in this process. We have shown that a reporter gene with full length 3′-UTR but lacking a circadian promoter did not oscillate, indicating that 3′UTR-mediated regulation of mRNA stability is not sufficient to generate circadian oscillation. Although translational repression in mper1 mRNA oscillation has been described (45), this is the first evidence that active mRNA decay plays an important role in maintaining the mper2 oscillation.
Circadian mRNA decay is regulated by the 3′-UTR and its binding partners
In general, mRNA decay is mediated by interactions between a specific cis-acting element in the mRNA and its binding partners. Sophisticated regulation of gene expression involves various factors acting at the post-transcriptional level, e.g. in splicing regulation, mRNA export, mRNA degradation and surveilence mechanisms such as nonsense-mediated mRNA decay (NMD) and nonstop-mediated mRNA decay (NSD) (48), in addition to translational regulation. Those regulatory mechanisms involve many kinds of RNA binding protein including hnRNPs and splicing-related proteins, and various non-coding RNAs such as miRNA. We have shown that many RNA binding proteins interact directly or indirectly with mper2 mRNA (Figure 3A), suggesting that a single mRNA can have many different binding partners. The combination of binding proteins on a mRNA molecule appears to determine its fate. With respect to the fine regulation of circadian mRNA oscillation, it is important to determine which proteins bind to the mRNA, the function of these proteins, how they interact with each other, and how they trigger changes in target mRNA metabolism. This study investigated the significance of mRNA decay and dampening-down effect in the regulation of circadian oscillation with particular emphasis on the role of cytoplasmic PTB.
The role of increased level of cytoplasmic PTB
We showed that PTB can translocate according to the circadian phase to perform cytoplasmic functions such as mRNA degradation. To further elucidate the effects of PTB on mRNA degradation, it is necessary to examine what causes the increase in cytoplasmic PTB levels and the function of PTB in the cytoplasm. Several reports have suggested that PTB increases target mRNA stability (23,42); however, we found that PTB had an opposite effect on mRNA stability. This difference in action of PTB may be the result of complex formation with available RNA binding proteins and generation of stoichiometric change within the mRNA-protein complex. UV cross-linking analysis of mouse SCN nuclear protein revealed changes in the profile of mRNA binding proteins during the circadian period (data not shown), indicating that complex formation between mRNA and its binding partners can change during circadian phases. Considering that nucleocytoplasmic shuttling and a broad range of RNA-binding specificity are general characteristics of members of the hnRNP family, the complex relationships between mRNA and RNA binding proteins such as PTB may influence mRNA stability. We cannot exclude the possibility that other functions of PTB affect circadian rhythm; since PTB has a broad range of binding capacity to mRNA, the increase of peak amplitude caused by depletion of PTB may not be solely caused by inhibition of mRNA decay.
Circadian oscillation is flexible but enduring
Our experiments demonstrated robust and reproducible mper2 mRNA oscillation following synchronization with the modifed dexamethasone treatment. Many reports have demonstrated that certain physiological treatments or gene disruptions affect different aspects of circadian oscillation such as peak amplitude change in mRNA or protein oscillation, phase shift in locomotor activity, or even elimination of oscillation (49–51). We found that the oscillation of mper2 mRNA continued with increased peak amplitude when the mper2 mRNA binding protein PTB was depleted. Similar results were reported at both transcriptional and post-transcriptional levels for mRNA regulation of another circadian gene, aanat (arylalkylamine N-acetyltranferase; serotonin N-acetyltransferase). When nocturnal aanat transcription was inhibited by ICER (inducible cAMP early repressor), robust oscillation continued with increased peak amplitude and no affect on circadian phase (52). In addition, reduction of RNA-binding proteins hnRNP R, Q and L, which have a destabilizing effect on aanat mRNA, also resulted in increased mRNA amplitude (32). As various regulation mechanisms are involved in the modulation of overt circadian gene oscillation, regulatory mechanisms besides PTB-mediated mRNA degradation are probably also involved in circadian mper2 mRNA oscillation. We previously reported that the 3′-UTR binding protein hnRNP Q can bind to an IRES (internal ribosomal entry site) element in the 5′-UTR and modulate nocturnal AANAT translation (35), showing that a single binding protein can function in various post-transcriptional mechanisms. Our present study demonstrates that PTB has a destabilizing effect on mper2 mRNA, but we assume that additional, as yet uncharacterized, RNA binding partners also act in a concerted manner to influence the fate of mper2 mRNA.
Circadian oscillation may provide buffering capacity against drastic changes in the molecular environment. As circadian oscillation is the result of complicated regulation and interconnected regulatory circuits, the oscillatory pattern may not be readily disrupted. Circadian rhythm is linked to cellular energy metabolism and the cell cycle, therefore an organism will face serious physiological problems if the regulation of circadian rhythm is disturbed. As is the case for many other fundamental physiological phenomena, the complete regulatory mechanism controlling circadian rhythms remains to be revealed.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
FUNDING
Korea Science and Engineering Foundation (R01-2006-000-10194-0, R17-2008-027-01000-0); National Core Research Center (R15-2004-033-06001-0); Brain Korea 21 Program of the Korea Ministry of Education.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Woo-Jae Kim and Dr Sung -Key Jang (POSTECH) for kindly providing the recombinant PTB protein.
REFERENCES
- 1.Zylka MJ, Shearman LP, Weaver DR, Reppert SM. Three period homologs in mammals: Differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron. 1998;20:1103–1110. doi: 10.1016/s0896-6273(00)80492-4. [DOI] [PubMed] [Google Scholar]
- 2.Albrecht U, Sun ZS, Eichele G, Lee CC. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell. 1997;91:1055–1064. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 3.Izumo M, Sato TR, Straume M, Johnson CH. Quantitative analyses of circadian gene expression in mammalian cell cultures. Plos Computat. Biol. 2006;2:1248–1261. doi: 10.1371/journal.pcbi.0020136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings MH, et al. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 5.Shearman LP, Zylka MJ, Weaver DR, Kolakowski L.F., Jr, Reppert SM. Two period homologs: circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron. 1997;19:1261–1269. doi: 10.1016/s0896-6273(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 6.Albrecht U, Zheng BH, Larkin D, Sun ZS, Lee CC. mPer1 and mPer2 are essential for normal resetting of the circadian clock. J. Biol. Rhythms. 2001;16:100–104. doi: 10.1177/074873001129001791. [DOI] [PubMed] [Google Scholar]
- 7.Akashi M, Ichise T, Mamine T, Takumi T. Molecular mechanism of cell-autonomous circadian gene expression of Period2, a crucial regulator of the mammalian circadian clock. Mol. Biol. Cell. 2006;17:555–565. doi: 10.1091/mbc.E05-05-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Terazono H, Mutoh T, Yamaguchi S, Kobayashi M, Akiyama M, Udo R, Ohdo S, Okamura H, Shibata S. Adrenergic regulation of clock gene expression in mouse liver. Proc.Natl Acad. Sci. USA. 2003;100:6795–6800. doi: 10.1073/pnas.0936797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 10.Chen-Goodspeed M, Lee CC. Tumor suppression and circadian function. J. Biol. Rhythms. 2007;22:291–298. doi: 10.1177/0748730407303387. [DOI] [PubMed] [Google Scholar]
- 11.Lamont EW, Diaz LR, Barry-Shaw J, Stewart J, Amir S. Daily restricted feeding rescues a rhythm of period2 expression in the arrhythmic suprachiasmatic nucleus. Neuroscience. 2005;132:245–248. doi: 10.1016/j.neuroscience.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 12.Wilusz CJ, Wilusz J. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet. 2004;20:491–497. doi: 10.1016/j.tig.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 13.Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, Di Padova F, Lin SC, Gram H, Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 14.Valcarcel J, Gebauer F. Post-transcriptional regulation: the dawn of PTB. Curr. Biol. 1997;7:R705–R708. doi: 10.1016/s0960-9822(06)00361-7. [DOI] [PubMed] [Google Scholar]
- 15.Spellman R, Rideau A, Matlin A, Gooding C, Robinson F, McGlincy N, Grellscheid SN, Southby J, Wollerton M, Smith CW. Regulation of alternative splicing by PTB and associated factors. Biochem. Soc. Trans. 2005;33:457–460. doi: 10.1042/BST0330457. [DOI] [PubMed] [Google Scholar]
- 16.Wagner EJ, Garcia-Blanco MA. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 2001;21:3281–3288. doi: 10.1128/MCB.21.10.3281-3288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, Shiue L, Ares M., Jr, Black DL. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castelo-Branco P, Furger A, Wollerton M, Smith C, Moreira A, Proudfoot N. Polypyrimidine tract binding protein modulates efficiency of polyadenylation. Mol. Cell. Biol. 2004;24:4174–4183. doi: 10.1128/MCB.24.10.4174-4183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soderberg M, Raffalli-Mathieu F, Lang MA. Inflammation modulates the interaction of heterogeneous nuclear ribonucleoprotein (hnRNP) I/polypyrimidine tract binding protein and hnRNP L with the 3′untranslated region of the murine inducible nitric-oxide synthase mRNA. Mol. Pharmacol. 2002;62:423–431. doi: 10.1124/mol.62.2.423. [DOI] [PubMed] [Google Scholar]
- 20.Jang SK, Wimmer E. Cap-independent translation of encephalomyocarditis virus RNA: structural elements of the internal ribosomal entry site and involvement of a cellular 57-kD RNA-binding protein. Genes Dev. 1990;4:1560–1572. doi: 10.1101/gad.4.9.1560. [DOI] [PubMed] [Google Scholar]
- 21.Luz N, Beck E. A cellular 57 kDa protein binds to two regions of the internal translation initiation site of foot-and-mouth disease virus. FEBS Lett. 1990;269:311–314. doi: 10.1016/0014-5793(90)81182-n. [DOI] [PubMed] [Google Scholar]
- 22.Knoch KP, Bergert H, Borgonovo B, Saeger HD, Altkruger A, Verkade P, Solimena M. Polypyrimidine tract-binding protein promotes insulin secretory granule biogenesis. Nat. Cell Biol. 2004;6:207–214. doi: 10.1038/ncb1099. [DOI] [PubMed] [Google Scholar]
- 23.Tillmar L, Welsh N. Glucose-induced binding of the polypyrimidine tract-binding protein (PTB) to the 3'-untranslated region of the insulin mRNA (ins-PRS) is inhibited by rapamycin. Mol. Cell. Biochem. 2004;260:85–90. doi: 10.1023/b:mcbi.0000026059.56089.e4. [DOI] [PubMed] [Google Scholar]
- 24.Xie J, Lee JA, Kress TL, Mowry KL, Black DL. Protein kinase A phosphorylation modulates transport of the polypyrimidine tract-binding protein. Proc. Natl Acad. Sci. USA. 2003;100:8776–8781. doi: 10.1073/pnas.1432696100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okamura H, Miyake S, Sumi Y, Yamaguchi S, Yasui A, Muijtjens M, Hoeijmakers J.HJ, van der Horst G.TJ. Photic induction of mPer1 and mPer2 in Cry-deficient mice lacking a biological clock. Science. 1999;286:2531–2534. doi: 10.1126/science.286.5449.2531. [DOI] [PubMed] [Google Scholar]
- 26.Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. PERIOD2 :: LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl Acad. Sci. USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoo SH, Ko CH, Lowrey PL, Buhr ED, Song EJ, Chang S, Yoo OJ, Yamazaki S, Lee C, Takahashi JS. A noncanonical E-box enhancer drives mouse Period2 circadian oscillations in vivo. Proc. Natl Acad. Sci. USA. 2005;102:2608–2613. doi: 10.1073/pnas.0409763102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bae K, Weaver DR. Light-induced phase shifts in mice lacking mPER1 or mPER2. J. Biol. Rhythms. 2003;18:123–133. doi: 10.1177/0748730403252248. [DOI] [PubMed] [Google Scholar]
- 29.Frisch B, Hardin PE, Hamblen-Coyle MJ, Rosbash M, Hall JC. A promoterless period gene mediates behavioral rhythmicity and cyclical per expression in a restricted subset of the Drosophila nervous system. Neuron. 1994;12:555–570. doi: 10.1016/0896-6273(94)90212-7. [DOI] [PubMed] [Google Scholar]
- 30.So WV, Rosbash M. Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. EMBO J. 1997;16:7146–7155. doi: 10.1093/emboj/16.23.7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwak E, Kim TD, Kim KT. Essential role of 3'-untranslated region-mediated mRNA decay in circadian oscillations of mouse Period3 mRNA. J. Biol. Chem. 2006;281:19100–19106. doi: 10.1074/jbc.M511927200. [DOI] [PubMed] [Google Scholar]
- 32.Kim TD, Kim JS, Kim JH, Myung J, Chae HD, Woo KC, Jang SK, Koh DS, Kim KT. Rhythmic serotonin N-acetyltransferase mRNA degradation is essential for the maintenance of its circadian oscillation. Mol. Cell. Biol. 2005;25:3232–3246. doi: 10.1128/MCB.25.8.3232-3246.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet. 2005;37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 34.Chae HD, Park TJ, Lee YK, Lee TG, Kim KT. Rapid and simple measurement of serotonin N-acetyltransferase activity by liquid biphasic diffusion assay. Neurochem. Int. 1999;35:447–451. doi: 10.1016/s0197-0186(99)00086-8. [DOI] [PubMed] [Google Scholar]
- 35.Kim TD, Woo KC, Cho S, Ha DC, Jang SK, Kim KT. Rhythmic control of AANAT translation by hnRNP Q in circadian melatonin production. Genes Dev. 2007;21:797–810. doi: 10.1101/gad.1519507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gherzi R, Lee KY, Briata P, Wegmuller D, Moroni C, Karin M, Chen CY. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell. 2004;14:571–583. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 37.Hahm B, Cho OH, Kim JE, Kim YK, Kim JH, Oh YL, Jang SK. Polypyrimidine tract-binding protein interacts with HnRNP L. FEBS Lett. 1998;425:401–406. doi: 10.1016/s0014-5793(98)00269-5. [DOI] [PubMed] [Google Scholar]
- 38.Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu. Rev. Physiol. 2001;63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- 39.Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schutz G, Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- 40.Ohno T, Onishi Y, Ishida N. A novel E4BP4 element drives circadian expression of mPeriod2. Nucleic Acids Res. 2007;35:648–655. doi: 10.1093/nar/gkl868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shih SC, Claffey KP. Regulation of human vascular endothelial growth factor mRNA stability in hypoxia by heterogeneous nuclear ribonucleoprotein L. J. Biol. Chem. 1999;274:1359–1365. doi: 10.1074/jbc.274.3.1359. [DOI] [PubMed] [Google Scholar]
- 42.Pautz A, Linker K, Hubrich T, Korhonen R, Altenhofer S, Kleinert H. The polypyrimidine tract-binding protein (PTB) is involved in the post-transcriptional regulation of human inducible nitric oxide synthase expression. J. Biol. Chem. 2006;281:32294–32302. doi: 10.1074/jbc.M603915200. [DOI] [PubMed] [Google Scholar]
- 43.Tillmar L, Welsh N. Hypoxia may increase rat insulin mRNA levels by promoting binding of the polypyrimidine tract-binding protein (PTB) to the pyrimidine-rich insulin mRNA 3′-untranslated region. Mol. Med. 2002;8:263–272. [PMC free article] [PubMed] [Google Scholar]
- 44.Coutinho-Mansfield GC, Xue Y, Zhang Y, Fu XD. PTB/nPTB switch: a post-transcriptional mechanism for programming neuronal differentiation. Genes Dev. 2007;21:1573–1577. doi: 10.1101/gad.1575607. [DOI] [PubMed] [Google Scholar]
- 45.Kojima S, Matsumoto K, Hirose M, Shimada M, Nagano M, Shigeyoshi Y, Hoshino S, Ui-Tei K, Saigo K, Green CB, et al. LARK activates posttranscriptional expression of an essential mammalian clock protein, PERIOD1. Proc. Natl Acad. Sci. USA. 2007;104:1859–1864. doi: 10.1073/pnas.0607567104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng HY, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S, et al. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ukai-Tadenuma M, Kasukawa T, Ueda HR. Proof-by-synthesis of the transcriptional logic of mammalian circadian clocks. Nat. Cell Biol. 2008;10:1154–1163. doi: 10.1038/ncb1775. [DOI] [PubMed] [Google Scholar]
- 48.Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- 49.Sakamoto K, Ishida N. Light-induced phase-shifts in the circadian expression rhythm of mammalian Period genes in the mouse heart. Eur.J. Neurosci. 2000;12:4003–4006. doi: 10.1046/j.1460-9568.2000.00302.x. [DOI] [PubMed] [Google Scholar]
- 50.Nikaido T, Akiyama M, Moriya T, Shibata S. Sensitized increase of period gene expression in the mouse caudate/putamen caused by repeated injection of methamphetamine. Mol. Pharmacol. 2001;59:894–900. doi: 10.1124/mol.59.4.894. [DOI] [PubMed] [Google Scholar]
- 51.Oster H, Yasui A, van der Horst G.TJ, Albrecht U. Disruption of mCry2 restores circadian rhythmicity in mPer2 mutant mice. Genes Dev. 2002;16:2633–2638. doi: 10.1101/gad.233702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Foulkes NS, Borjigin J, Snyder SH, Sassone-Corsi P. Transcriptional control of circadian hormone synthesis via the CREM feedback loop. Proc. Natl Acad. Sci. USA. 1996;93:14140–14145. doi: 10.1073/pnas.93.24.14140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.