Abstract
MicroRNAs are known to regulate developmental processes but their mechanism of regulation remains largely uncharacterized. We show the transcription factor Twist-1 drives the expression of a 7.9-kb noncoding RNA transcript (from the Dynamin-3 gene intron) that encodes a miR-199a and miR-214 cluster. We also show that knocking down Twist-1 with shRNAs decreased miR-199a/214 levels and that Twist-1 bound an E-Box promoter motif to developmentally regulate the expression of these miRNAs. The expression of HIF-1 (known to mediate Twist-1 transcription), miR-199a and miR-214 was maximal at E12.5 and the miRNAs were expressed specifically in mouse cerebellum, midbrain, nasal process and fore- and hindlimb buds. This study shows the expression of the miR199a/214 cluster is controlled by Twist-1 via an E-Box promoter element and supports a role for these miRNAs as novel intermediates in the pathways controlling the development of specific neural cell populations.
INTRODUCTION
MicroRNAs (miRNA) bind the 3′-untranslated region (UTR) of mRNAs and elicit translational repression or mRNA cleavage and this action represents an entirely new level of post-transcriptional regulation. More than 500 human miRNAs together with some of their mRNA targets have now been identified empirically and it is clear that they act to govern fundamental cellular pathways controlling development, cancer and apoptosis (1,2). Although miRNAs are now known to be key regulators of gene expression, the pathways controlling the expression of individual miRNA genes are only beginning to be elucidated. For example, recently, the transcription of miR-146a during development was shown to be suppressed by promyelocytic leukemia zinc-finger (PLZF) protein (3,4); the transcription factor GATA-1 was shown to drive the expression of a single transcript encoding miR-144 and miR-451 during zebra fish development (5). While investigating the control of miR-214 expression we found that the miR-199a and miR-214 genes are located ∼6 kb apart within the intron of the dynamin-3 gene and that Twist-1 had previously been reported to drive the expression of a single noncoding transcript from this region (6). Twist is a nuclear basic helix–loop–helix (bHLH) transcription factor known to be crucial for mesoderm formation in Drosophila (7). In mammals, Twist-1 is expressed in cranial neural crest derivatives and in mesenchymal structures (8–11). Mutations in the Twist-1 gene cause the Saethre-Chotzen syndrome, an autosomal dominant craniosynostosis characterized by abnormal fusion of the cranial sutures (12,13). Studies have also indicated that Twist family members bind preferentially to E-box DNA promoter sequences as homodimers (14) or heterodimers (15). We present data to show that during embryonic development Twist-1 binds an E-box region within the DNM-3 gene intron to drive the expression of a single transcript that is processed to miR-199a and miR-214.
MATERIALS AND METHODS
Generation of recombinant adenovirus constructs
Mouse Twist-1 cDNA was cloned into the PxCx-CMV shuttle vector and recombinant E1-deleted adenoviral constructs produced as described previously (16). The predicted DNM3os promoter was recovered from mouse genomic DNA by PCR. The primer sequences were: promoter (–640:0) forward (F), AAA GGG GGG AGC CCC AAC TTA TCT G; reverse (R), TTC CTG CAC CAG GGG CTT GT; E-box deleted (–640:-357) forward, AGG GGG GAG CCC CAA CTT ATC TGA; reverse, TGG GGC CCC AGT ATG GAA AA. The PCR products were cloned into PCR2.1 plasmids using a TOPO cloning kit (Invitrogen) and the plasmid sequenced. The promoter sequences were finally subcloned into PGL2 Luciferase reporter plasmids (Promega).
RNA isolation and RT-PCR
Whole embryos were collected from time mated C57Bl6 mice at the embryonic stages (E) 10.5, 11.5, 12.5, 14.5 and 16.5 (plug day was E0.5, B&K, UK). Total RNA and miRNA was extracted from embryos and N2A cells using RNeasy mini (Qiagen) or miRNA extraction kit (Ambion), respectively. RT-PCR for the analysis of gene expression was performed on 500 ng of RNA and cDNA synthesis was carried out using SuperscriptIII (Invitrogen). RT-PCR analysis of the DNM3, Dynamin-3 opposite strand (DNM3os) and GAPDH gene expression was performed using previously described primer sequences (6). Optimized HIF-1 and Twist-1 primers were designed using the vector NTI program (Invitrogen). The primer sequences for RT-PCR were: DNM3, (F) CCG AGA CTT AAT TCC AAA ACA ATA ATG; DNM3 (R), AAA GGA GTC ACT GCT GTT GGG AAA A; DNM3os, (F) GGT CTC ACC CTG CTT GTT AAT CAA GGG GGA; DNM3os, (R) TCC TGT TGT TAC TGG CCC TCA TGC AGC C; GAPDH (F), ACC ACA GTC CAT GCC ATC TC; GAPDH (R), TCC ACC ACC CTG TTG CTG TA; HIF-1 (F), TGA GCG TTT CCT AAT CTC AT; HIF-1 (R), AGC GAC AAA GTG CAT AAA AC; Twist-1 (F), CGA CGA CAG CCT GAG CAA CA; Twist-1 (R), TGC AGC TCC TCG TAC GAC TG. PCR was carried out at 94°C for 45 s, 60°C for 45 s and 72°C for 1 min for 25 cycles, using high-fidelity Taq polymerase (Roche). Parallel amplifications were performed on the templates from which the reverse transcriptase was omitted to ensure that amplification was dependent on the presence of single strand of cDNA.
Western blot
All primary antibodies were diluted in PBS with 5% (v/v) Blocking solution (Roche). The mouse anti-Twist1 (AbCam) Ab was used at a 1:1000 dilution and the mouse anti-α-Tubulin (Sigma) at 1:1000. In all cases, corresponding horseradish peroxidase-linked antibodies were used (Amersham).
RNA isolation and miRNA detection
Small RNAs were isolated using the mirVana miRNA Isolation Kit (Ambion). miRNA expression levels were assessed using Taqman® MiRNA Assays (Applied Biosystems) according to the manufacturer's protocol. Results were normalized against levels of RNU6B (as assessed by the corresponding Taqman® Assay; Applied Biosystems); 10 ng of total RNA were used as the template for the RT reaction in each case.
Luciferase reporter assay
Cells (at 50% confluence) in 24-well plates were transfected using lipofectamine (Invitrogen) mixed with a Firefly luciferase reporter gene construct (200 ng). Cell extracts were prepared 48 h after transfection and the luciferase activity was measured using the Luciferase Reporter Assay System (Promega).
Whole-mount in situ hybridization
DIG (digioxigenin) labeled DNM3os antisense RNA probes were generated from RT-PCR derived templates by in vitro transcription. In order to produce a DNA template for the generation of antisense RNA probes by in vitro transcription, T7 RNA polymerase promoter sequences were added to the 5′end of the PCR product. The primer sequences used are shown below (promoter sequences are in bold): DNM3os (F), GGT CTC ACC CTG CTT GTT AAT CAA GGG GGA, DNM3os (R): GCA GTA ATA CGA CTC ACT ATA GGG TCC TGT TGT TAC TGG CCC T CA TGC AGC C. One hundred nanograms of template DNA was used for in vitro transcription using a DIG-labeling kit (Roche). Pre DIG labeled LNA miR-199a-5p, miR-199a-3p, mir-214 and scrambled control probes were purchased from Exiqon. In situ hybridization with DIG probes and detection with alkline-phosphatase-conjugated antibodies were carried out as described previously (17).
RESULTS
Twist-1 drives the expression of miR-199a/214
The miR-199a/214 gene cluster is located in the opposite (3′–5′) direction to the DNM-3 gene suggesting it is transcribed from its own promoter (Figure 1). Furthermore, it was predicted previously that the expression of a (non-coding) 7.9-kb Dynamin-3 opposite strand (DNM3os) during development may be under the control of Twist-1(6). Hence, we investigated whether Twist-1 drives the expression of DNM3os, miR-199a and miR-214 (Figure 1). To investigate this possibility we used an adenovirus to over-express Twist-1 in N2a neuroblastoma cells and found that DNM3os expression was increased 5- to 6-fold above control levels (Figure 2A). Further RT-PCR experiments then showed that Twist-1 expression also mediated a 5-fold increase in both miR-214 and miR-199a expression (Figure 2B).
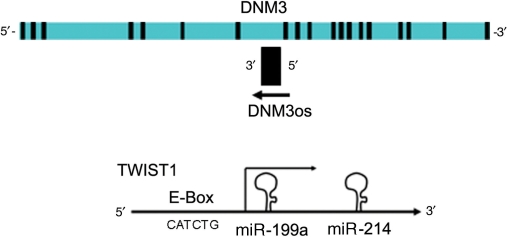
Figure 1.
The miR-199a/214 cluster lies within the DNM3 gene. A schematic diagram of the Dynamin3 gene showing the complementary strand within its intron encodes a single transcript (DNM3os). The inset figure shows the DNM3os transcript encodes miR-199a and miR-214 and that an E-Box promoter drives transcription in a reverse orientation.
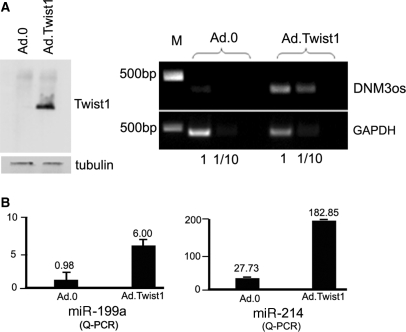
Figure 2.
Twist-1 mediates the expression a single antisense transcript encoding miR-199a and miR-214. (A) Western blot and agarose gel showing that Twist-1 and the DNM3os transcript are expressed respectively in N2a cells when they are transduced with Ad.Twist-1. (B) Q-PCR analysis also shows that the expression of miR-199a and miR-214 are significantly increased (P < 0.001 by t-test in both cases) following transduction with Ad.Twist-1.
NIH-3T3 embryonic fibroblasts express Twist-1 constitutively and we therefore used this cell line to investigate whether Twist-1 regulates miR-199a and miR-214 under normal physiological circumstances. Western blotting and quantitative PCR confirmed that Twist-1 (Figure 3A) and the noncoding RNA DNM3os (Figure 3B) were expressed in NIH-3T3 cells, respectively. To investigate whether under normal physiological conditions Twist-1 was responsible for driving miR-199a and miR-214 expression we used an anti-Twist1 shRNA construct (kindly provided by Dr. Michael C. Naski). The Taqman results showed that anti-Twist-1 shRNA decreased the expression of miR-214 and miR-199a expression while the scrambled shRNA control did not (Figure 3C).
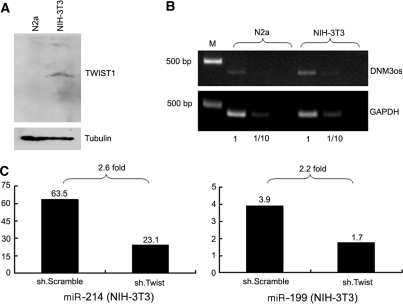
Figure 3.
ShRNA mediated knockdown of Twist-1 results in decreased miR-199a and miR-214 expression. Western blotting (A) and QPCR analysis (B) show that Twist-1 and DNMO3 is constitutively expressed in NIH-3T3 cells. (C) Inhibiting Twist-1 expression by transfecting cells with an anti-Twist-1 shRNA resulted in a >2-fold decrease in the levels of miR-199a and miR-214 (n = 3 in all cases).
Twist-1 acts on an E-Box to drive miR-199a/214 expression
Having confirmed Twist-1 was driving miR-214 and miR-199a expression we searched for promoter motifs and noted that a region containing an E-Box (a promoter element known to be bound by Twist-1) was present. This region (–640 to 0 bp of the DNM3os) was isolated from genomic DNA by PCR and cloned next to a luciferase reporter gene (Figure 4A). Further, we also deleted the basic-helix–loop–helix (bHLH)-binding E-box element (CATCTG) known to be essential for Twist-1 transcriptional activity (and also cloned this next to a luciferase reporter gene). The results of reporter assays showed that luciferase activity was increased by Twist-1 in cells transfected with the E-Box-luciferase construct (–640:0); however, no increase in luciferase activity was observed in cells transfected with the (–640:–357) promoter region lacking the E-box (Figure 4B).
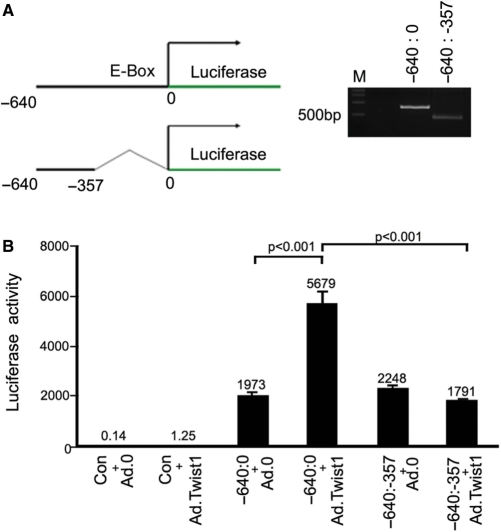
Figure 4.
Promoter assays showing Twist-1 mediates expression via the E-box region. (A) Schematic diagram together with an agarose gel showing the entire (–640 to 0 bp) promoter and E-Box deleted promoter region of DNM3os. (B) Luciferase Promoter assays show that Twist-1 drives expression via the E-Box region (–640:0) within the DNM3os transcript. Promoter activity is at background levels when the E-Box is deleted (–640:357) and when cells are transduced with control vectors.
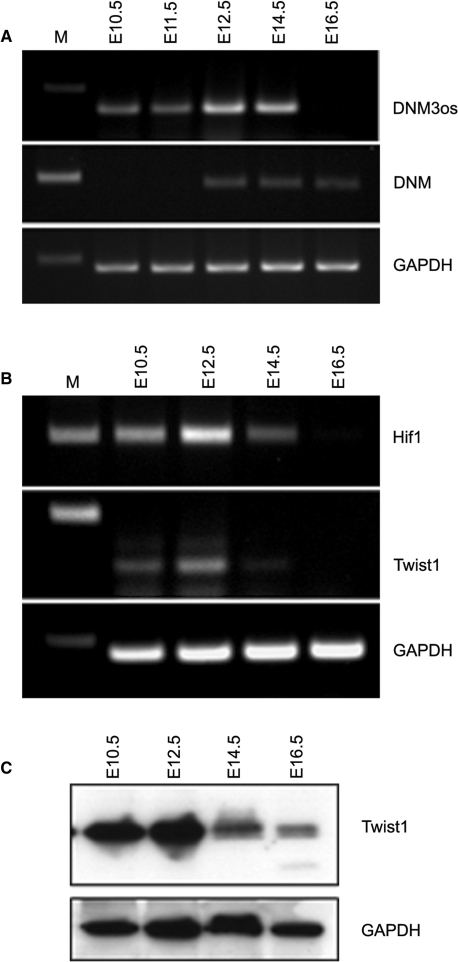
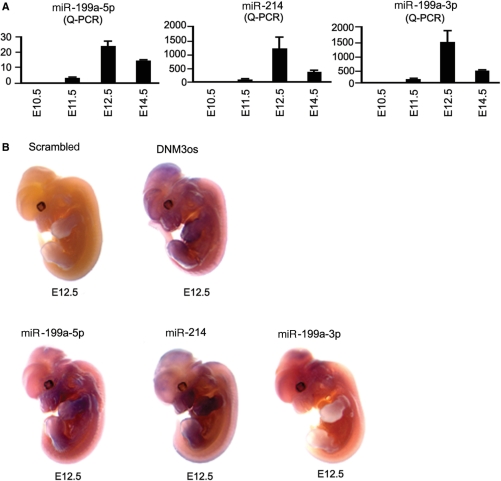
We next sought to confirm that Twist-1 expression during development coincided with that of DNM3os, miR-214 and miR-199a and we analyzed transcript expression at E10.5, E12.5, E14.5 and E16.5. Figure 5A shows that DNM3os mRNA levels were first expressed at E10.5, were maximal at E12.5 and began to decrease at E14.5. The sense DNM3 mRNA transcript was also detected from E12.5 onward (Figure 5A). Western and QT-RTPCR showed that the expression of the Twist-1 transcription factor and mRNA, respectively, was also maximal at E12.5 and decreased at E14.5 (Figure 5B). HIF-1 has been shown to directly regulate Twist-1 expression (18,19) and we also showed that HIF-1 mRNA expression corresponded closely with that of Twist-1 being expressed maximally at E12.5 (Figure 5B). Furthermore, Taqman analyses showed that the expression of miR-199a-5p, miR-199a-3p and miR-214 levels were highest at E12.5 and this corresponded with peak DNM3os and Twist-1 expression (Figure 6A). Finally, we used in situ hybridization to observe the expression of the single-stranded DNM3os, miR-199a-3p, miR-199a-5p and miR-214. The results showed that DNM3os was strongly expressed in the cerebellum, midbrain, nasal process and fore- and hindlimb buds and that expression of miR-214 mirrored that of DNM3os. However, miR-199a-3p was most strongly expressed in the hindbrain, while miR-199a-5p was strongly expressed in the limb buds (Figure 6B).
Figure 5.
DNM3os, DNM3, HIF-1 and Twist-1 are expressed together during development. The expression of DNM3os, DNM3 (A) and Hif1 and Twist-1 (B) was measured in the whole-cell embryo by RT-PCR at various stages of embryonic development. Western blot showing Twist-1 protein expression at the different stages of development is also shown (C).
Figure 6.
DNM3os, miR-199a and miR-214 are transcribed in specific tissues during embryonic development. (A) miR-199a-5p, miR-199a-3p and miR-214 levels were measured by quantitative PCR in whole embryos at embryonic stage E10.5, E11.5, E12.5, E14.5 and E16.5. (B) Shows the expression of DNM3os, miR-199a-5p, miR-199a-3p and miR-214 expression in E12.5 embryos visualized following whole-mount in situ hybridization.
DISCUSSION
It was observed previously that a noncoding 7.9-kb DNM3os fragment is expressed during development and that its expression may be under the control of Twist-1(6). Here we present promoter deletion studies to show that the bHLH transcription factor Twist-1 drives the expression of the 7.9-kb antisense transcript via an E-Box within the Dynamin-3 gene intron and that the transcript encodes the miR-199a and miR-214 genes. In addition, we show that inhibiting the expression of Twist-1 resulted in decreased miR-199a and miR-214 levels, indicating that under normal physiological conditions Twist-1 can drive the expression of the miR-199a/214 cluster. We further show that mir-199a and mir-214 are developmentally regulated, with expression beginning at E11.5, being maximal at E12.5 and falling at E14.5. The expression of the transcription factor HIF1 (known to drive the expression of Twist-1) Twist-1, and miR-199a and miR-214 were coincident during development. Together, these data strongly suggest that Twist-1 drives DNM3os, miR-199a and miR-214 expression during development. Furthermore, in situ studies on E12.5 embryos showed that the miRNAs were expressed specifically in mouse cerebellum, midbrain, nasal process and fore and hind limb buds.
Previous studies have examined the role of miRNAs in ES cells and embryonic development by knocking out the enzymes (Dicer, Drosha and DGCR8) that are involved in miRNA biogenesis. These studies have revealed a crucial role for miRNAs in ES cell differentiation, and the control of development (20). MiRNA-specific studies have shown that miR-214 regulates the specification of zebra fish muscle cell types by targeting a negative regulator of Hedgehog signaling (21) and miR-199a has been shown to regulate COX-2 expression in mouse uterus during implantation (22). We have previously shown that during establishment of the brain coronal suture, Twist-1 knockout results in abnormal cell proliferation and osteoblast differentiation. Results that suggest miR-199a and mir-214 may play a role in mediating coronal sutural formation (13). Together, these results support a role for miR-199a/214 in specifying cell fate during embryonic development. MiRBase shows that there are three genomic loci for the miR-199a gene and only one for miR-214 within the DNM-3 intron. It is therefore possible that Twist-1 expression will be found to be associated with miR-214 expression in adult tissues. Interestingly, the altered expression of miRNAs and Twist-1 in mature tissues is known to lead to changes in the expression of cell cycle proteins and tumorigenesis (23,24). Such studies have shown that miR-214 induces cell survival and cisplatin resistance via the control of PTEN expression and subsequent activation of the Akt pathway. MiR-199a targets the 3′UTR of the Met proto-oncogene and its expression results in the downregulation of Met and its down stream target ERK2 (22). Furthermore, the expression of miR-214 and miR-199a have been shown to mirror each other in human epithelial ovarian cancer tissue (24,25), and human solid cancer tissues (26). Significantly, Twist-1 expression has also been shown to play a role in tumour development, progression and survival (27). Together, these data suggest that miR-199a and miR-214 sculpt developmental processes by mediating effects on cell cycle proteins. Furthermore, it will be important to investigate whether that the ectopic expression of miR-199a/214 during tumirogenesis is due to aberrant Twist-1 expression.
FUNDING
The Wellcome Trust. Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared
REFERENCES
- 1.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 2.Krutzfeldt J, Poy MN, Stoffel M. Strategies to determine the biological function of microRNAs. Nat. Genet. 2006;38(Suppl):S14–S19. doi: 10.1038/ng1799. [DOI] [PubMed] [Google Scholar]
- 3.Labbaye C, Spinello I, Quaranta MT, Pelosi E, Pasquini L, Petrucci E, Biffoni M, Nuzzolo ER, Billi M, Foa R, et al. A three-step pathway comprising PLZF/miR-146a/CXCR4 controls megakaryopoiesis. Nat. Cell Biol. 2008;10:788–801. doi: 10.1038/ncb1741. [DOI] [PubMed] [Google Scholar]
- 4.Kelly KF, Daniel JM. POZ for effect—POZ-ZF transcription factors in cancer and development. Trends Cell Biol. 2006;16:578–587. doi: 10.1016/j.tcb.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Dore LC, Amigo JD, Dos Santos CO, Zhang Z, Gai X, Tobias JW, Yu D, Klein AM, Dorman C, Wu W, et al. A GATA-1-regulated microRNA locus essential for erythropoiesis. Proc. Natl Acad. Sci. USA. 2008;105:3333–3338. doi: 10.1073/pnas.0712312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loebel DA, Tsoi B, Wong N, Tam PP. A conserved noncoding intronic transcript at the mouse Dnm3 locus. Genomics. 2005;85:782–789. doi: 10.1016/j.ygeno.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Thisse B, Stoetzel C, Gorostiza-Thisse C, Perrin-Schmitt F. Sequence of the twist gene and nuclear localization of its protein in endomesodermal cells of early Drosophila embryos. EMBO J. 1988;7:2175–2183. doi: 10.1002/j.1460-2075.1988.tb03056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bate M, Rushton E, Currie DA. Cells with persistent twist expression are the embryonic precursors of adult muscles in Drosophila. Development. 1991;113:79–89. doi: 10.1242/dev.113.1.79. [DOI] [PubMed] [Google Scholar]
- 9.Hebrok M, Wertz K, Fuchtbauer EM. M-twist is an inhibitor of muscle differentiation. Dev. Biol. 1994;165:537–544. doi: 10.1006/dbio.1994.1273. [DOI] [PubMed] [Google Scholar]
- 10.Fuchtbauer EM. Expression of M-twist during postimplantation development of the mouse. Dev. Dyn. 1995;204:316–322. doi: 10.1002/aja.1002040309. [DOI] [PubMed] [Google Scholar]
- 11.Gitelman I. Twist protein in mouse embryogenesis. Dev. Biol. 1997;189:205–214. doi: 10.1006/dbio.1997.8614. [DOI] [PubMed] [Google Scholar]
- 12.El Ghouzzi V, Le Merrer M, Perrin-Schmitt F, Lajeunie E, Benit P, Renier D, Bourgeois P, Bolcato-Bellemin AL, Munnich A, Bonaventure J. Mutations of the TWIST gene in the Saethre-Chotzen syndrome. Nat. Genet. 1997;15:42–46. doi: 10.1038/ng0197-42. [DOI] [PubMed] [Google Scholar]
- 13.Yoshida T, Phylactou LA, Uney JB, Ishikawa I, Eto K, Iseki S. Twist is required for establishment of the mouse coronal suture. J. Anat. 2005;206:437–444. doi: 10.1111/j.1469-7580.2005.00411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee MS, Lowe GN, Strong DD, Wergedal JE, Glackin CA. TWIST, a basic helix-loop-helix transcription factor, can regulate the human osteogenic lineage. J. Cell Biochem. 1999;75:566–577. doi: 10.1002/(sici)1097-4644(19991215)75:4<566::aid-jcb3>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 15.Spicer DB, Rhee J, Cheung WL, Lassar AB. Inhibition of myogenic bHLH and MEF2 transcription factors by the bHLH protein Twist. Science. 1996;272:1476–1480. doi: 10.1126/science.272.5267.1476. [DOI] [PubMed] [Google Scholar]
- 16.Lee YB, Cosgrave AS, Glover CP, Bienemann A, Heywood D, Hobson RJ, Uney JB. Increased utility in the CNS of a powerful neuron-specific tetracycline-regulatable adenoviral system developed using a post-transcriptional enhancer. J. Gene Med. 2005;7:576–583. doi: 10.1002/jgm.694. [DOI] [PubMed] [Google Scholar]
- 17.Wilkinson DG, Nieto MA. Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol. 1993;225:361–373. doi: 10.1016/0076-6879(93)25025-w. [DOI] [PubMed] [Google Scholar]
- 18.Gort EH, van HG, Verlaan I, Groot AJ, Plasterk RH, Shvarts A, Suijkerbuijk KP, van LT, van der WE, Raman V, et al. The TWIST1 oncogene is a direct target of hypoxia-inducible factor-2alpha. Oncogene. 2008;27:1501–1510. doi: 10.1038/sj.onc.1210795. [DOI] [PubMed] [Google Scholar]
- 19.Yang MH, Wu MZ, Chiou SH, Chen PM, Chang SY, Liu CJ, Teng SC, Wu KJ. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat. Cell Biol. 2008;10:295–305. doi: 10.1038/ncb1691. [DOI] [PubMed] [Google Scholar]
- 20.Blakaj A, Lin H. Piecing together the mosaic of early mammalian development through microRNAs. J. Biol. Chem. 2008;283:9505–9508. doi: 10.1074/jbc.R800002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flynt AS, Li N, Thatcher EJ, Solnica-Krezel L, Patton JG. Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat. Genet. 2007;39:259–263. doi: 10.1038/ng1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chakrabarty A, Tranguch S, Daikoku T, Jensen K, Furneaux H, Dey SK. MicroRNA regulation of cyclooxygenase-2 during embryo implantation. Proc. Natl Acad. Sci. USA. 2007;104:15144–15149. doi: 10.1073/pnas.0705917104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim S, Lee UJ, Kim MN, Lee EJ, Kim JY, Lee MY, Choung S, Kim YJ, Choi YC. MicroRNA miR-199a* regulates the MET proto-oncogene and the downstream extracellular signal-regulated kinase 2 (ERK2) J. Biol. Chem. 2008;283:18158–18166. doi: 10.1074/jbc.M800186200. [DOI] [PubMed] [Google Scholar]
- 24.Yang H, Kong W, He L, Zhao JJ, O’Donnell JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 25.Iorio MV, Visone R, Di LG, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 26.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]