Abstract
The ubiquitous RNA-binding protein AUF1 promotes the degradation of some target mRNAs, but increases the stability and translation of other targets. Here, we isolated AUF1-associated mRNAs by immunoprecipitation of (AUF1–RNA) ribonucleoprotein (RNP) complexes from HeLa cells, identified them using microarrays, and used them to elucidate a signature motif shared among AUF1 target transcripts. The predicted AUF1 motif (29–39 nucleotides) contained 79% As and Us, consistent with the AU-rich sequences of reported AUF1 targets. Importantly, 10 out of 15 previously reported AUF1 target mRNAs contained the AUF1 motif. The predicted interactions between AUF1 and target mRNAs were recapitulated in vitro using biotinylated RNAs. Interestingly, further validation of predicted AUF1 target transcripts revealed that AUF1 associates with both the pre-mRNA and the mature mRNA forms. The consequences of AUF1 binding to 10 predicted target mRNAs were tested by silencing AUF1, which elevated the steady-state levels of only four mRNAs, and by overexpressing AUF1, which also lowered the levels of only four mRNAs. In total, we have identified a signature motif in AUF1 target mRNAs, have found that AUF1 also associates with the corresponding pre-mRNAs, and have discovered that altering AUF1 levels alone only modifies the levels of subsets of target mRNAs.
INTRODUCTION
In mammalian cells, the expression of stress-response, proliferative, immune and developmental proteins is critically regulated through processes such as pre-mRNA splicing and mRNA transport, stability and translation (1–3). Specific families of mRNA-binding proteins (RBPs) directly influence these processes by binding to the 3′ and 5′ untranslated regions (UTRs) of the transcript. These regulatory UTR sequences are heterogeneous; they often encompass stretches rich in U or AU nucleotides (and hence are termed AU-rich elements or AREs), but other times they are rich in different residues, such as GU or C (4,5). Several RBPs which associate with these regulatory sequences function as mRNA turnover and translation regulatory proteins; thus, they have been collectively termed TTR-RBPs (6). Several TTR-RBPs function primarily as translational inhibitors, such as the T-cell-restricted intracellular antigen-1 (TIA-1) and the TIA-1-related protein TIAR (4,7–10), but can also influence mRNA turnover (11,12). The Hu/elav proteins (HuR, HuB, HuC and HuD), broadly enhance mRNA stability (13,14), but can also enhance or inhibit the translation of several target mRNAs (15–20). Other TTR-RBPs decrease the stability of target mRNAs, including tristetraprolin (TTP), K homology splicing-regulatory protein (KSRP), the CUG triplet RNA-binding protein 1 (CUG-BP) and the butyrate response factor-1 (BRF1) (5,21–23).
The TTR-RBP AUF1 (AU-binding factor 1), also called hnRNPD (heterogeneous nuclear ribonucleoprotein D), has also been implicated in several distinct post-transcriptional regulatory processes. AUF1 was originally found to promote mRNA decay, as determined from studies using cultured cells expressing different levels of AUF1 as well as using AUF1-deficient mice (24–30). However, in some instances AUF1 was also shown to enhance mRNA stability (26,28,31) and to promote translation (11). AUF1 comprises four proteins that arise from alternative splicing (p37, p40, p42, p45) and shuttle between the nucleus and the cytoplasm (27). AUF1 isoforms associate with hsp70/hsc70, with the translation initiation factor eIF4G, and with the poly(A)-binding protein PABP (32). The degradation of AUF1-bound mRNAs requires the dissociation of AUF1 from eIF4G followed by proteasome-mediated destruction (32), and has also been linked to the recruitment of AUF1 to the exosome (33).
All of the AUF1 isoforms contain two RNA recognition motifs (RRMs) through which they bind to a select group of mRNAs (32,34,35). Reported AUF1 target transcripts include many mRNAs that encode stress-response and proliferative proteins such as p21, Cyclin D1, MYC, FOS, GM-CSF, TNF-α, IL-3, parathyroid hormone (PTH), and the growth arrest and DNA damage-inducible (GADD)45α (24–26,28–31,36). AUF1 has been shown to influence the steady-state mRNA levels both in untreated conditions and in response to treatment with damaging and growth-regulatory stimuli. For example, in cells treated with the growth inhibitor prostaglandin A2, irradiated with the genotoxin UVC (short wavelength ultraviolet light) or left without treatment, AUF1 associated with the cyclin D1 mRNA and reduced its stability (36,37). In untreated cells, AUF1 was also found to associate with the p21 mRNA and reduced its half-life (25). Similarly, following treatment with bacterial lipopolisaccharide (LPS), AUF1 was implicated in the degradation of target mRNAs encoding TNF-α, IL-1β and cyclooxygenase-(COX)-2 (38,39). Through its actions on these and other mRNAs, AUF1 was shown to play a central role in the cellular response to mitogenic and immune factors (39–41), differentiation cues (42,43) and carcinogenesis (44).
Given the participation of AUF1 in critical cellular functions and its ubiquitous expression pattern (45), we sought to identify the collective of AUF1 target mRNAs. We employed an approach based on the immunoprecipitation of ribonucleoprotein (RNP) complexes from unstimulated HeLa cells using an anti-AUF1 antibody. The AUF1-associated mRNAs were then identified by microarray hybridization and the existence of a shared sequence among the putative targets was elucidated by computational analysis, as previously described for TIA-1, TIAR and HuR (4,10,46). The signature motif sequence was 29–39-bases long and was highly AU-rich, consistent with the sequences of previously reported AUF1 targets. The usefulness of the signature motif in the prediction of additional AUF1 targets was tested by RNA-binding assays which examined the interaction of AUF1 with individual transcripts. These analyses revealed that AUF1 associates with both the mature mRNA (cytoplasmic) and the unprocessed pre-mRNA (nuclear). Despite extensive evidence that AUF1 promotes target mRNA decay, silencing AUF1 only elevated the levels of some target mRNAs, while AUF1 overexpression only reduced the abundance of a subset of target mRNAs. These findings support the notion that AUF1 only accelerates mRNA decay for a fraction of targets and may regulate the metabolism of other target mRNAs through different post-transcriptional mechanisms.
MATERIALS AND METHODS
Cell culture, treatment and transfection
Human cervical carcinoma cells (HeLa) were cultured in Dulbecco's modified essential medium (Gibco BRL, Gaithersburg, MD) supplemented with 10% fetal bovine serum (HyClone, Logan, Utah) and antibiotics and were maintained at 37°C in a humidified atmosphere containing 5% CO2 in air. For AUF1 knockdown, cells were transfected with pSILENCER plasmids that either expressed an AUF1-directed shRNA or lacked an insert (25). For AUF1 overexpression, cells were transfected with a mixture of four plasmids (pcDNA-derived plasmids expressing p37, p40, p42 or p45) or with pcDNA lacking an insert (36). All transfections were performed by using Lipofectamine 2000; 8 μg plasmid DNA was used per transfection.
cDNA array analysis
RNA in the material obtained after IP reactions using either an anti-AUF1 antibody or IgG was reverse transcribed in the presence of [α-33P]dCTP (MP Biomedicals), and the radiolabeled product was used to hybridize cDNA MGC arrays (http://www.grc.nia.nih.gov/branches/rrb/dna/index/dnapubs.htm#2), containing 9600 genes, as previously reported (4,10,46). The data were analyzed using the Array Pro software (Media Cybernetics, Inc.), then normalized by Z-score transformation and used to calculate differences in signal intensities. Significant values were tested using a two-tailed Z-test and a P of <0.01. The data were calculated from three independent experiments. The complete cDNA array data are available from the authors.
Computational analysis
Human UniGene records were first identified from the most strongly enriched AUF1 targets derived from the array analysis; the top 244 transcripts served as the experimental dataset (Supplementary Table S1) for the identification of the AUF1 motif. The 128 transcripts with complete, high-quality 3′UTR sequences were first scanned with RepeatMasker (www.repeatmasker.org) to remove repetitive sequences (Supplementary Data). The remaining sequences were divided into 100-base-long subsequences with 50-base overlap between consecutive sequences and were organized into 50 data sets. Common RNA motifs were elucidated from each of the 50 random data sets. The top 10 candidate motifs from each random data set were selected and used to build the stochastic context-free grammar (SCFG) model, which summarizes the folding, pairing and additional secondary structure features. The SCFG model of each candidate motif was then used to search against the experimental 3′UTR dataset as well as the entire human UniGene 3′UTR data set to obtain the number of hits for each motif. The motif with the highest enrichment in the experimental data set compared with the entire UniGene data set was considered to be the top AUF1 candidate motif. The enrichment was examined by Fisher's exact test. The identified RNA motif for AUF1 forms a stem-loop and appeared in 75% of the transcripts that were found by IP analysis.
The identification of the RNA motif in unaligned sequences was conducted using FOLDALIGN software (47), and the identified motif was modeled by the SCFG algorithm and searched against the transcript data set using the COVE and COVELS software packages (48). The motif logo was constructed using WebLogo (http://weblogo.berkeley.edu/). RNAplot was used to depict the secondary structure of the representative RNA motifs. The computation was performed using the NIH Biowulf computer farm. Both UniGene and Refseq data sets were downloaded from NCBI.
Cell fractionation, RNA purification and western blot analysis
Cells were incubated on ice for 5 min in cytoplasmic lysis buffer containing 20 mM Tris–HCl (pH 7.5), 100 mM KCl, 5 mM MgCl2, 0.3% IGEPAL CA-630, 1000 U/ml of RNaseOUT, and protease inhibitors, then centrifuged (10 000×g) for 10 min. After removing the supernatant (cytoplasmic fraction), the pellet was resuspended in 100 μl IP buffer (100 mM KCl, 5 mM MgCl2, 10 mM Hepes, pH 7.0, 0.5% Nonidet P-40, 1000 U/ml RNaseOUT, plus protease inhibitors), kept on ice 10 min, subjected to three cycles of freezing and thawing, and centrifuged (10000 ×g) for 10 min.
For western blot analysis, whole-cell lysates (10 μg) were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) and transferred onto polyvinylidene difluoride (PVDF) membranes. Hybridizations were carried out using a rabbit polyclonal anti-AUF1 antibody (Upstate Biotech.) or mouse monoclonal anti-GAPDH antibody (Santa Cruz Biotech.). Following incubation with the appropriate secondary antibodies, the signals were detected with the ECL™ reagent (Amersham Biosciences).
Binding assays: IP and biotin pulldown
IP of endogenous AUF1–mRNA complexes was carried out using previously described methods (4,46,49). Briefly, 20 million HeLa cells per sample were lysed and used for IP (1 h at 4°C in the presence of 30 µg either a rabbit polyclonal anti-AUF1 antibody or control rabbit IgG (Upstate). RNA was isolated using acid phenol–chloroform (Ambion).
For biotin pulldown assays, PCR fragments containing the T7 RNA polymerase promoter sequence (T7): CCAAGCTTCTAATACGACTCACTATAGGGAGA were used as a templates for in vitro transcription of the GAPDH 3′UTR, p53 coding region (CR) and, the AQP11, CANX, CGI-149, MATR3, RTKN2, SERP1, VIL2, SNX13, TMEM2 and RAB23 3′UTR using biotinylated CTP. Two μg of biotinylated transcript was incubated with 80 µg of lysate (cytoplasmic or nuclear) for 30 min at room temperature, following which the complexes were isolated with streptavidin-coated magnetic Dynabeads (Dynal) and the pulldown material was analyzed by standard western blotting.
Oligonucleotides used to prepare templates for in vitro biotinylated transcription and pulldown assay
To prepare the coding region of p53, primers (T7)ATGGAGGAGCCGCAGTCAGATCCTAGC and AGAATGTCAGTCTGAGTCAGGC (forward and reverse, respectively) were used.
To prepare the 3′UTR templates, the following primer pairs (forward and reverse, respectively) were used:
(T7) CCTCAACGACCACTTTGTCA and GGTTGAGCACAGGGTACTTTAT for GAPDH,
(T7) GCATAACAACCATACAATTAATAAAAA and AATGAGGCTTTTCTAGCAGCA for AQP11,
(T7) CAAGAAACAGAAAGCCACGA and TCGGTACATTTGAAAGCCTCT for CANX,
(T7) CCAGCCACTGTCTCACAGAT and CCAGCACTGAAGCTCTTGGT for CGI-149,
(T7) ATTGGCAGAAGAACGCAGAC and CATGGCCTAGGGTTTTCTTTT for MATR3,
(T7) CTCCAAGGCAGAAATCCATC and TGCTGTTGCTGACACCTTATG for RTKN2,
(T7) TATCAGGATGGGCATGTGAA and ATGGCACATTGGACTCAACA for SERP1,
(T7) AGGCCGGGACAAGTACAAG and CACCTGCACATGGCATCTTA for VIL2,
(T7) GCAGAAAAGGTGACACTCCA and TGGGTTTAAAGGTATACACTGAGG for SNX13,
(T7) CTTAAGTGCTGGGGGAAAAA and GCAGGAGGGTGAACAGAAAA for TMEM2, and
(T7) ACCCAACAAACAAAGGACCA and CGCTGTCAGATGAAAACTGC for RAB23.
RT followed by conventional PCR and real-time, quantitative (q)RT–PCR
Total RNA or RNA isolated from the IPs was reverse transcribed (RT) using oligo-dT (for mRNA) or random hexamers (for pre-mRNA), and subjected to either PCR or quantitative PCR (qPCR) to assess the abundance of the products using the gene-specific primer pairs listed below. Conventional PCR amplification was visualized by electrophoresis through ethidium bromide-stained 1.5% agarose gels; qPCR analysis was performed using the QuantiTect SYBR Green PCR kit (Qiagen) and an ABI Prism 7000 detection system with the ABI Prism 7000 SDS software.
Oligonucleotides for RT–PCR and RT–qPCR-mediated detection of mRNAs in IP materials and in total RNA
Oligonucleotide pairs (forward and reverse, respectively) to detect mature mRNAs were as follows:
GGCTGCACTGGATTCATCA and TGAGGCTTTTCTAGCAGCATT for AQP11 mRNA, ATCGTTCATTGCCTTTCCAG and ATGGAGAGTGGGCATTTGAC for CANX mRNA, AAGTCAGGAGCTGACCAGGA and CCAGAGGATCTGGGTTTGAA for CGI-149 mRNA, GTTTGAACCCGTTTTGGTTG and TCACGGGATTCAAGTGACAA for MATR3 mRNA, AGAAGATGCTGCAGGGAAAA and ACTCTGAGGGCACAACTGCT for RTKN2 mRNA, AAAGCTACCGTGGTGGAGTG and ATGGCACATTGGACTCAACA for SERP1 mRNA, AACCATGGCACTTGATGTGA and TGCACGAGAAGGAATGAGTG for VIL2 mRNA, AGAGAAAACTGCCCCCATCT and CACAGCCCTTAAAGCACACA for SNX13 mRNA, AAAAGGAGCGCTTTCTAGGG and GGCATTTTGCTGCTTCTTTC for TMEM2 mRNA, and CCTGTGAAGTGTGGTGATGC and TGTGGGACTGACAGCTCTTG for RAB23 mRNA.
Oligonucleotides used for RT–qPCR-mediated detection of pre-mRNAs in IP materials and in total RNA
Oligonucleotide pairs designed to amplify fragments spanning intron–exon junctions were as follows:
GGCTGCACTCATCACCTTTT and TGTCGGCCCTACTGGTTAAG for AQP11 pre-mRNA, TCTCCCAAGGTTTGAAATGG and CGAATTTTGCTTTCCCTTTG for CANX pre-mRNA, CTGATCGTCCATGCTTTCAA and AGGGTGTCTGATGGGAACAG for CGI-149 pre-mRNA, GGTTATCCCCATCTGTGCTC and TGCTTGATCAACTCAATGTCATC for MATR3 pre-mRNA, CAAGTTTCATAGGTGAATCAATGC and TAGTGCAGCGCATGATCTTT for RTKN2 pre-mRNA, TTTGAATAATCTGGAAAATTGCTG and CAGGCCTGTCTTCACTTCTT for SERP1 pre-mRNA, CCTCATGTTCTCGTTGTGGA and TTGCCCTTCCTGTTTTCCTA for VIL2 pre-mRNA, CAGCCCCAATAATGTGCTTC and TTGTCTCCCAGTAGCTTTTGC for SNX13 pre-mRNA, TTTCCACTGAATCCCAAAAA and TTGTTATCTGGGGCAAAAGG for TMEM2 pre-mRNA, and CGGAGTGACTTCCACCAGAT and CCACTGAACCGTATGCCTAAA for RAB23 pre-mRNA.
RESULTS
Sequence and structure of a predicted AUF1 signature motif
A collection of mRNAs associated with AUF1 was previously identified in HeLa cells (25). Briefly, pools of AUF1-associated mRNAs were isolated by immunoprecipitation (IP, Figure 1A) under conditions that preserved the integrity of RNP complexes. The RNA in the IP material (associated with AUF1 in the AUF1 IP samples or bound non-specifically to the reagents in the IgG IP samples) was then extracted and subjected to RT. The resulting products were hybridized to human cDNA arrays (MGC arrays, Figure 1A, http://www.grc.nia.nih.gov/branches/rrb/dna/dna.htm#). A total of 244 array spots (∼2.5% of the total spots on the array, Supplementary Table S1) had Z ratios >1.45 when the signals in AUF1 IP arrays were compared with the signals in the IgG IP arrays and were thus deemed to represent specific AUF1-associated transcripts.
Figure 1.
Sequence and structure of the predicted AUF1 signature motif, as identified among AUF1-associated transcripts. (A) Left, Schematic of the experimental approach. HeLa cell lysates were subjected to immunoprecipitation (IP) with either IgG or anti-AUF1 antibodies. The collections of RNAs isolated from each IP reaction were identified using microarrays [Materials and Methods section and (25)] and were used to identify a signature sequence shared by the transcripts. Right, representative AUF1 signals in IP samples, as detected by western blot analysis; Input, aliquot of lysate before IP; H.C., heavy immunoglobulin chain. (B) Probability matrix (graphic logo) of the AUF1 motif, elucidated from the array-derived experimental dataset, indicating the relative frequency with which each nucleotide is likely to be found at each position within the motif. (C) Specific sequence and secondary structure of eight representative examples of the AUF1 motif in specific mRNAs; the corresponding gene names are shown.
These 244 transcripts (the ‘experimental dataset’) were used for further analysis. First, they were subjected to computational analysis to identify conserved primary RNA sequences and secondary structures (Materials and methods section). Among the 100 possible candidate motifs initially obtained from the experimental dataset, one motif comprising 29–39 nucleotides had the highest frequency of hits per kb in the experimental dataset over the entire Unigene database. A graphic representation of the relative frequency of nucleotides at each position (the motif logo) revealed that the most frequent nucleotides were A and U, jointly comprising a striking ∼79% of the sequence (Figure 1B). While the numbers of As and Us were similar, U residues appeared to cluster in the 5′ and 3′ regions of the motif, while A residues were predominant in the center of the motif. This signature motif differs markedly from U-rich motifs identified for HuR, TIA-1 and a C-rich motif identified for TIAR (4,10,46), but does agree with the AU-richness of many AUF1 target mRNAs [e.g. (50)]. Depicted in Figure 1C are eight examples of the secondary structure of the AUF1 signature motif on specific mRNAs.
The AUF1 signature sequence predicts AUF1 target mature mRNAs and pre-mRNAs
Using the AUF1 motif in Figure 1B, we queried the Unigene database for additional putative AUF1 target mRNAs. A total of 7572 transcripts were identified, comprising 6.2% of the complete Unigene database. Table 1 lists a subset of transcripts and the positions of the individual AUF1 motif hits within the 3′UTR (the entire list can be found as Supplementary Table S2). The number of motif hits in the 3′UTRs of each transcripts is also indicated. Importantly, among a list of 15 previously described AUF1 target mRNAs, 10 were found to have at least one hit of the AUF1 motif (Table 2), supporting the predictive value of the motif. Five other prominent AUF1 targets (p21, MYC, IL-1β, IL-6 and PTH mRNAs) did not have specific hits for this RNA motif, indicating that the signature motif does not identify the entirety of AUF1 target mRNAs; other motifs also identified many but not all target mRNAs for given RBPs (4,10,46).
Table 1.
Putative AUF1 target mRNAs based on the presence of the AU-rich signature motif in the 3′UTR
| Position in 3′UTR | # hits | Name | Symbol | UniGene |
|---|---|---|---|---|
| 885–917, 1621–1694, 1768–1801, 2565–2594, 3285–3317, 3678–3748, 4480–4512 | 10 | Insulin-like growth factor 1 | IGF1 | Hs#S3218917 |
| 415–453, 1469–1501, 2035–2067, 2083–2115, 2224–2256, 2738–2770, 2825–2857 | 7 | Cathepsin S | CTSS | Hs#S1728864 |
| 866–898, 1632–1664, 2787–2819, 3215 3247, 3248–3280, 4460–4492 | 6 | Rhotekin 2 | RTKN2 | Hs#S4553519 |
| 301–333, 354–395, 1340–1370, 1385–1417, 1510–1542, 4407–4461 | 6 | Protocadherin 19 | PCDH19 | Hs#S4812509 |
| 61–93, 259–289, 433–465, 776–808, 1911–1946 | 5 | Stress–associated ER protein 1 | SERP1 | Hs#S2139162 |
| 20–52, 63–94, 116–147, 548–580, 1025–1072 | 5 | Matrin 3 | MATR3 | Hs#S3219061 |
| 779–810, 891–923, 981–1013, 1106–1138, 1611–1643 | 5 | Calcyphosphine 2 | CAPS2 | Hs#S3619535 |
| 474–513, 989–1021, 1472–1504, 1594–1666 | 4 | NAG14 protein | NAG14 | Hs#S3817889 |
| 108–144, 856–888, 1608–1652, 1791–1823 | 4 | Transmembrane protein 2 | TMEM2 | Hs#S2138764 |
| 264–296, 1073–1104, 1111–1143, 1331–1363 | 4 | Calnexin | CANX | Hs#S3218949 |
| 87–122, 424–456, 485–584 | 3 | Chemokine (C–C motif) ligand 8 | CCL8 | Hs#S1730823 |
| 120–150, 550–582, 982–1081 | 3 | Sorting nexin 13 | SNX13 | Hs#S4001833 |
| 1261–1312, 2067–2099, 2183–2215 | 3 | CGI–149 protein | CGI–149 | Hs#S2140320 |
| 724–756, 1112–1143, 1664–1724 | 3 | RAB23, RAS oncogene family | RAB23 | Hs#S2140367 |
| 753–784, 1156–1255 | 2 | Villin 2 | VIL2 | Hs#S1728180 |
| 173–272 | 1 | Cytochrome c–1 | CYC1 | Hs#S1726589 |
| 255–354 | 1 | Aquaporin 11 | AQP11 | Hs#S7089975 |
| 462–561 | 1 | Huntingtin interacting protein C | HYPC | Hs#S15644799 |
| 71–170 | 1 | Cleavage/polyadenylation factor 3 | CPSF3 | Hs#S2140592 |
| 1353–1452 | 1 | Extracellular signal–regulated kinase 8 | ERK8 | Hs#S4435687 |
Partial list of genes bearing the AU–rich signature motif (Figure 1) in the 3′UTR of the corresponding transcripts. The complete list of predicted target transcripts is available (Supplementary Table S2). The relative position(s) and number of the AU–rich motif hits in the 3′UTR of each transcript are indicated.
Table 2.
Reported AUF1 target mRNAs bearing the AU–rich signature motif
| Position in 3′UTR | # hits | Name | Symbol | UniGene | Reference |
|---|---|---|---|---|---|
| 410–442, 1378–1411, 1581–1615 | 3 | Cyclin D1 | CCND1 | Hs#S3987156 | (25,37) |
| 45–99 | 1 | Cyclin B1 | CCNB1 | Hs#S3618391 | (unpublished) |
| 291–326, 457–488 | 2 | Gadd45A | GADD45A | Hs#S1731372 | (36) |
| 492–529, 592–624 | 2 | p16 | CDKN2A | Hs#S4001849 | (53) |
| 210–248 | 1 | IL8 | IL8 | Hs#S3218902 | (54) |
| 648–680 | 1 | TNFa | TNF | Hs#S3218906 | (55) |
| 1783–1831, 2389–2429 | 2 | Estrogen receptor a | ESR1 | Hs#S1726719 | (56) |
| 423–454 | 1 | TS | TS | Hs#S1728128 | (57) |
| 939–970, 1433–1473, 1807–1840, 1885–1917, 2189–2220 | 5 | Cyclooxygenase–2 | PTGS2 | Hs#S1730801 | (38) |
| 227–265, 3976–4013, 4373–4407, 4957–4989 | 4 | Bcl–2 | BCL2 | Hs#S1730182 | (58) |
List of known AUF1 target mRNAs, out of a total of 15 best–characterized AUF1 target mRNAs previously reported. The positions of the motif hits within the 3′UTRs are indicated. Overlapping motifs are listed as one.
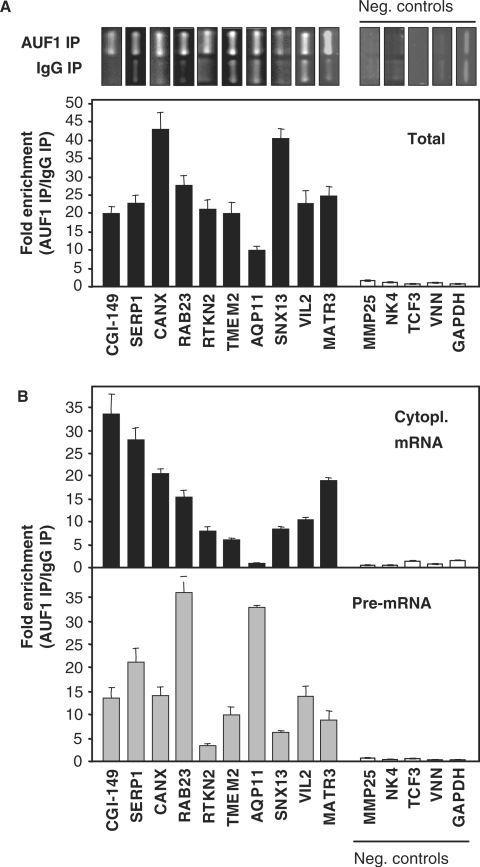
In order to test if the AUF1 signature motif could be used to predict AUF1 target RNAs, we monitored the association of AUF1 with several transcripts that were computationally found to have at least one occurrence of the AUF1 motif. In total, 10 transcripts were chosen based on the presence of different numbers of motif hits in their 3′UTRs: one hit (AQP11), two hits (VIL2), three hits (RAB23, SNX13, CGI-149), four hits (TMEM2, CANX), five hits (MATR3, SERP1) and six hits (RTKN2). Initially, we assayed the interaction between AUF1 and predicted target mature mRNAs by RNP IP analysis using whole-cell preparations (‘Total’). Transcript identification was performed by RT of RNA isolated from IP, followed by either conventional PCR (Figure 2A, top) or quantitative, real-time PCR analysis (Figure 2A, bottom). All of the predicted targets (black bars) were found to be significantly (5–40-fold) enriched in AUF1 IP relative to IgG IP, although there was no clear correlation between magnitude of enrichment and the number of motif hits. Negative control transcripts (clear bars) were not enriched.
Figure 2.
Analysis of the association of AUF1 with predicted target mRNAs bearing AU-rich motif. (A) Analysis of the interaction between AUF1 and 10 putative target transcripts chosen randomly from Table 1. Following RNP IP, the association of putative target mRNAs with AUF1 in whole-cell (‘Total’) HeLa cell lysates was assessed by monitoring the enrichment of the mRNAs in AUF1 IP samples relative to IgG IP samples. Conventional PCR (top, gel images) and real-time, quantitative (q)PCR (bottom, graph) were used to quantify this enrichment. In the qPCR analysis, the levels of housekeeping GAPDH mRNA were used to normalize differences in sample input. (B) RNP IP analyses were conducted as described in the legend of Figure 2A, except that cytoplasmic lysates (top graph, ‘Cytopl. mRNA’) and nuclear lysates (bottom graph, ‘Pre-mRNA’) were used. For pre-mRNA detection, specific primer pairs were used to amplify DNA regions that spanned intron–exon junctions. In (A) and (B), negative control transcripts (‘Neg. controls’) not AUF1 targets) are depicted as white bars. In (A) and (B), the graphed data show the means + the standard error of the means (SEM) from three independent measurements.
Further analysis revealed that AUF1 associated with all of the target mRNAs (except with the AQP11 mRNA) when using cytoplasmic lysates (Figure 2B, top), and when using nuclear lysates (data not shown). Therefore, we hypothesized that AUF1 might associate with such target transcripts in pre-mRNA form (the pre-spliced version of the mRNA). To test this possibility, we designed primer pairs that would amplify fragments spanning intron–exon junctions of the corresponding pre-mRNAs, and would thereby reveal if the pre-mRNA version of the mRNA was also a target of AUF1. While the overall abundance of the pre-mRNA transcripts was much lower than that of the mature mRNAs, all of the target pre-mRNA transcripts were found enriched in the AUF1 IP compared with the IgG IP (Figure 2B, bottom), suggesting a model of co-transcriptional or early post-transcriptional loading of AUF1 onto newly synthesized target transcript. The relative enrichments of pre-mRNAs and the corresponding mature, cytosolic mRNA were generally different (compare top and bottom graphs in Figure 2B). In both experimental systems, the negative control transcripts were not found to interact as pre-mRNAs, supporting the specificity of the interactions (Figure 2B).
To obtain independent evidence that AUF1 interacted specifically with the 3′UTRs of these mRNAs, biotinylated segments spanning the 3′UTRs of target mRNAs containing at least one hit of the AUF1 motif (Figure 3, top) were prepared by in vitro transcription (Materials and methods section). Following incubation with nuclear or cytoplasmic HeLa cell lysates, the corresponding RNP complexes were pulled down using streptavidin-coated paramagnetic beads, whereupon the identity and abundance of the various AUF1 isoforms in the resulting RNP complexes were assayed by western blot analysis. In keeping with reports that AUF1 isoforms are not uniformly present in both cytoplasmic and nuclear compartments, p45 was preferentially seen in the nuclear lysates, while the p40/p42 doublet was seen in both sets of pulldown samples; p37 showed low abundance in both compartments (Figure 3, bottom). These results suggest that AUF1 binds to the 3′UTR of target mRNAs, both in the nucleus and in the cytoplasm.
Figure 3.
Schematic of the RNA segments spanning sections of the 3′UTRs of putative AUF1 target mRNAs that were amplified and used as templates for in vitro transcription in the presence of biotinylated CTP (Materials and methods section). The amplified regions contained one or several AUF1 motif hits; the complete number of hits is shown in Table 1; all numbers refer to positions within the 3′UTR. The biotinylated transcripts were incubated with HeLa cytoplasmic lysates or nuclear lysates and the presence of AUF1 in the resulting RNP complexes was tested by western blot analysis. Biotinylated transcripts spanning the p53 coding region (p53CR) and the GAPDH 3′UTR as well as beads without biotinylated RNA were included as controls to detect background signals.
Taken together, these data indicate that the presence of at least one hit of the AUF1 signature motif described in Figure 1 can be used to successfully predict if a given mRNA or pre-mRNA associates with AUF1, as measured by RNP IP analysis of endogenous mRNAs and by analysis of binding to biotinylated transcripts.
Functional consequences of the interaction of AUF1 with target mRNAs
Given the broadly recognized influence of AUF1 in promoting target mRNA decay, we sought to study the influence of AUF1 upon the subset of mRNAs bearing hits of the signature motif identified here. We investigated this question by modulating the levels of AUF1, then measuring the steady-state levels of the target mRNAs.
First, we reduced AUF1 levels by transfecting HeLa cells with plasmid that either expressed a silencing hairpin (sh)RNA directed towards all four AUF1 isoforms [pSIL-AUF1shRNA (25)] or contained no insert (pSIL). Western blot analysis revealed the efficiency of the silencing (Figure 4A). Unexpectedly, however, lowering AUF1 levels did not augment the levels of all target mRNAs, as would be expected if AUF1 universally promoted their decay. Only RTKN2, TMEM2, AQP11 and MATR3 showed slightly (<50%) elevated levels; the remaining six AUF1 target mRNAs remained at levels similar to those seen in cells transfected with the control vector (pSIL) (Figure 4B). The levels of a positive control transcript, Cyclin D1 mRNA (‘Pos.’), increased upon silencing AUF1, while negative control transcripts (‘Neg.’, GAPDH, MMP25, TCF3 mRNAs) remained unchanged (Figure 4B).
Figure 4.
Effect of silencing AUF1 on the steady-state levels of target mRNAs. (A) The levels of AUF1 in HeLa cells were monitored by western blot analysis 48 h after transfection with either a plasmid that expressed a silencing hairpin (sh) targetting AUF1 (pSIL-AUF1shRNA) or a control plasmid pSILENCER (pSIL). The relative intensities of the combined AUF1 signals, calculated after densitometry analysis, are indicated. (B) Forty-eight hours after silencing as described in (A), the steady-state levels of AUF1 target mRNAs (left), normalization control 18S rRNA, positive control CCND1 (cyclin D1, ‘Pos.’) and negative controls (‘Neg.’), were measured by RT–qPCR analysis. The data show the means and SEM from three independent experiments.
Second, we increased AUF1 protein levels by transiently transfecting a mixture of four plasmids, each expressing one AUF1 isoform (Figure 5B), while the control group was transfected with the corresponding empty plasmid. We then tested the consequences of this intervention on the steady-state abundance of AUF1 target mRNAs. Here, we also found that AUF1 overexpression led to a moderate reduction (<40%) in the levels of some predicted targets (CGI-149, CANX, RAB23 and SNX13 mRNAs) and had little influence on the levels of the remaining mRNAs. As in Figure 5B, the positive control transcript (Cyclin D1 mRNA) was reduced after AUF1 overexpression, while negative control transcripts (GAPDH, MMP25 and TCF3 mRNAs) also remained unchanged (Figure 5B), suggesting that the modest reduction in the levels of AUF1 mRNAs was specific.
Figure 5.
Effect of overexpressing AUF1 on the steady-state levels of target mRNAs. (A) The levels of AUF1 in HeLa cells were monitored by western blot analysis 48 h after transfection with a cocktail of plasmids that expressed p37, p40, p42 and p45 (collectively shown as pCDNA-AUF1) or with a control plasmid (pCDNA). The relative intensities of the combined AUF1 signals, calculated after densitometry analysis, are indicated. (B) Forty-eight hours after AUF1 overexpression as described in (A), the steady-state levels of AUF1 target mRNAs (left), normalization control 18S rRNA, positive control CCND1 (cyclin D1, ‘Pos.’) and negative controls (‘Neg.’), were measured by RT–qPCR analysis. The data show the means and SEM from three independent experiments.
Collectively, these data indicate that AUF1 reduces the steady-state levels of some, but not all AUF1 target mRNAs. Instead, the abundance of a subset of AUF1-associated transcripts is not directly altered following the modulation of AUF1 levels.
DISCUSSION
The signature motif identified among AUF1-associated mRNAs was prominently AU-rich (38% A, 41% U, 7% G, 14% C), and formed a stem with four internal bulges and one loop at the end (Figure 1). Using this motif, we identified many other putative target mRNAs which had at least one hit of the AUF1 motif (Supplementary Table S2). Importantly, out of the 15 best-characterized AUF1 target mRNAs, 10 had at least one hit of the AUF1 motif in their 3′UTRs (Table 2). As noted for signature sequences identified earlier for other RNA-binding proteins (HuR, TIA-1 and TIAR) (4,10,46), the specific motifs were not capable of identifying every single target mRNA. For these RBPs, some mRNAs that were demonstrated targets were absent from the lists of RNAs bearing signature sequences. There are several reasons why a signature motif does not identify every target, as seen for the AUF1 signature motif. It is possible that slight sequence variations within the probability matrix will be needed in order to detect all of the possible targets. Alternatively, it is possible that AUF1 (and other RBPs) recognize and/or associate with two or more entirely different sequence motifs. Finally, as we observed with TIAR (4), it is possible that under different culture conditions, AUF1 may exhibit different affinity for RNA sequences. It also remains unanswered whether the AUF1 signature motif accommodates one or both RRMs.
Since AUF1 promotes the decay of many target mRNAs [those encoding cyclin D1, p21, GADD45α, MYC, TNFα, thymidylate synthase, IL-1β, etc. (Table 2)], we hypothesized that the AUF1 motif-bearing transcripts identified here might also be degraded by AUF1-mediated action. In this regard, we did not expect to find that the levels of most of the 10 transcripts tested remained unaffected after AUF1 silencing or overexpression (Figures 4 and 5). While AUF1 silencing elevated the levels of some mRNAs such as RTKN2, TMEM2, AQP11 and MATR3, plausibly because their half-lives increased as did the half-life of positive control CCND1 mRNA [(25,37), Figure 4B], it is possible that the degree of AUF1 silencing was insufficient to elevate the levels of the other six transcripts. Similarly, while AUF1 overexpression reduced the levels of mRNAs such as CGI-149, CANX, RAB23 and SNX13, likely by lowering their half-lives as it lowered the CCND1 mRNA half-life [(25,37), Figure 5B], it is also possible that the degree of AUF1 overexpression was not enough to decrease the levels of the other mRNAs studied (Figure 5B). In addition, the most labile AUF1 targets actually may not be represented at all in this analysis. If the binding of AUF1 to a given mRNA triggers its degradation, one can imagine that those targets would be degraded rapidly before or during lysis or during the IP reaction, and thus would not be available for isolation by cDNA analysis (and hence would not be included for the motif identification). A further consideration regarding the stability of AUF1 target mRNAs is that in some instances, AUF1 action as a decay-promoting RNP was more apparent after exposure of cells to agents such as methylmethane sulfonate, UVC, prostaglandin A2 or LPS (25,36,37); despite extensive efforts to test the effect of AUF1 overexpression or silencing in stress-treated cells, the AUF1 target mRNA levels did not show any striking differences following exposure to stress agents (data not shown).
Instead, it is more likely that the binding of AUF1 to some target mRNAs does not promote their decay, but serves to perform other functions. For example, it was particularly interesting to find that AUF1 associated with predicted target mRNAs in their pre-mRNA form, as shown in Figure 2B. In fact, it is likely that AUF1 associates with some target transcript co-transcriptionally or immediately after transcription, before splicing has ended. In this capacity, AUF1 could assist with the nuclear processing of target transcripts (e.g. splicing or maturation of the 5′ and 3′ ends) or with their nuclear transit. These findings also support an earlier report by Chen and colleagues showing that AUF1 is a nucleo-cytoplasmic shuttling protein that first associates with target mRNAs in the nucleus and later influence their fate in the cytoplasm (51). In addition, AUF1 was reported to have other post-transcriptional actions. In one instance, AUF1 functioned as an enhancer of MYC translation; it competed with the translational inhibitor TIAR for binding to the MYC mRNA, thereby relieving TIAR's suppressive influence on MYC translation (11). In other reports, AUF1 was shown to stabilize labile transcripts bearing the JUN, FOS and GM-CSF instability sequences (31) and the PTH mRNA (26). Subsequent work by Raineri and coworkers considered each AUF1 isoform individually. They obtained evidence that p37 and p42 stabilized a labile reporter mRNA, while p40 and p45 promoted its degradation (28). Here, we have studied all four AUF1 isoforms jointly, since they share the two RRM regions and therefore they likely share target mRNAs. Besides this consideration, it would have been possible to overexpress AUF1 isoforms individually, but there are no suitable antibodies to IP each isoform specifically, and there are no siRNAs that will individually silence one isoform and not others. However, as these studies progress, it will be important to analyze systematically the subset of target transcripts that each isoform binds to, and the specific influence of each isoform upon the collection of mRNAs with which it complexes. It will also be interesting to find out if treatment with various stimuli will trigger the dissociation, as previously reported (36), or the association (37) of AUF1 isoforms, individually or as a group, to target transcripts.
Paradoxically, it is notable that the sequence identified for AUF1 target mRNAs is, in fact, AU-rich. Similar systematic analyses were performed earlier on RBPs that were traditionally believed to bind AU-rich element-bearing mRNAs (collectively termed ‘ARE-RBPs’ in some instances). These studies were not found to have AU-rich shared motifs (25,37). Instead, such RBPs were found to have a heterogeneous set of signature motifs on their respective target mRNAs [C-rich, U-rich and GU-rich sequences (4,5,46,52)]. The complexity of the RNA motifs identified for the various RBPs is in keeping with the specificity and versatility needed to regulate the post-transcriptional fate of the target transcripts. It also helps to explain the dynamic post-transcriptional control of mRNAs that are targets of multiple RBPs (e.g. p21, COX-2, TNF-α, Bcl-2 and many others). As a result of our expanding knowledge of RBP target sequences, it is becoming possible to study how these RBPs jointly affect mRNA metabolism. Interesting examples of competitive, cooperative, and independent simultaneous binding of RBPs to shared target transcripts are rapidly emerging in the literature.
In closing, the AU-rich signature sequence identified among AUF1 target mRNAs provides a valuable starting point for the systematic analysis of AUF1 function. As AUF1 does not appear to stabilize universally the subset of mRNAs bearing this motif, future work must investigate if AUF1 instead influences their translation or perhaps modulates their transport. Further studies are also warranted to identify possible additional signature motifs for AUF1 targets. As our understanding of AUF1 function increases, so will our ability to recognize its role in processes such as cell proliferation, differentiation, carcinogenesis and the immune and stress responses (39–44), and our capacity to intervene in these biological events.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This research was supported entirely by the Intramural Research Program of the NIA-IRP, National Institutes of Health. Funding for open access charge: Project number Z01-AG000392.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank H.H. Kim (NIA) for assistance with this work and K. G. Becker (NIA) and the NIA Array Facility for providing cDNA arrays for analysis.
REFERENCES
- 1.Wilkie GS, Dickson KS, Gray NK. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem. Sci. 2003;28:182–188. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 2.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 3.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 4.Kim HS, Kuwano Y, Zhan M, Pullmann R, Jr, Mazan-Mamczarz K, Li H, Kedersha N, Anderson P, Wilce MC, Gorospe M, et al. Elucidation of a C-rich signature motif in target mRNAs of RNA-binding protein TIAR. Mol. Cell. Biol. 2007;27:6806–6817. doi: 10.1128/MCB.01036-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vlasova IA, Tahoe NM, Fan D, Larsson O, Rattenbacher B, Sternjohn JR, Vasdewani J, Karypis G, Reilly CS, Bitterman PB, et al. Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol. Cell. 2008;29:263–270. doi: 10.1016/j.molcel.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pullmann R, Jr, Kim HH, Abdelmohsen K, Lal A, Martindale JL, Yang X, Gorospe M. Analysis of turnover and translation regulatory RNA-binding protein expression through binding to cognate mRNAs. Mol. Cell. Biol. 2007;27:6265–6278. doi: 10.1128/MCB.00500-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gueydan C, Droogmans L, Chalon P, Huez G, Caput D, Kruys V. Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor α mRNA. J. Biol. Chem. 1999;274:2322–2326. doi: 10.1074/jbc.274.4.2322. [DOI] [PubMed] [Google Scholar]
- 8.Anderson P, Kedersha N. Stressful initiations. J. Cell Sci. 2002;115:3227–3234. doi: 10.1242/jcs.115.16.3227. [DOI] [PubMed] [Google Scholar]
- 9.Anderson P, Kedersha N. Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones. 2002;7:213–221. doi: 10.1379/1466-1268(2002)007<0213:vstroe>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.López de Silanes I, Galbán S, Martindale JL, Yang X, Mazan-Mamczarz K, Indig FE, Falco G, Zhan M, Gorospe M. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol. Cell. Biol. 2005;25:9520–9531. doi: 10.1128/MCB.25.21.9520-9531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liao B, Hu Y, Brewer G. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat. Struct. Mol. Biol. 2007;14:511–518. doi: 10.1038/nsmb1249. [DOI] [PubMed] [Google Scholar]
- 12.Yamasaki S, Stoecklin G, Kedersha N, Simarro M, Anderson P. T-cell intracellular antigen-1 (TIA-1)-induced translational silencing promotes the decay of selected mRNAs. J. Biol. Chem. 2007;282:30070–30077. doi: 10.1074/jbc.M706273200. [DOI] [PubMed] [Google Scholar]
- 13.Antic D, Keene JD. Embryonic lethal abnormal visual RNA-binding proteins involved in growth, differentiation, and posttranscriptional gene expression. Am. J. Hum. Genet. 1997;61:273–278. doi: 10.1086/514866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol. Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antic D, Lu N, Keene JD. ELAV tumor antigen, Hel-N1, increases translation of neurofilament M mRNA and induces formation of neurites in human teratocarcinoma cells. Genes Dev. 1999;13:449–461. doi: 10.1101/gad.13.4.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mazan-Mamczarz K, Galban S, López de Silanes I, Martindale JL, Atasoy U, Keene JD, Gorospe M. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc. Natl Acad. Sci. USA. 2003;100:8354–8359. doi: 10.1073/pnas.1432104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lal A, Kawai T, Yang X, Mazan-Mamczarz K, Gorospe M. Antiapoptotic function of RNA-binding protein HuR effected through prothymosin alpha. EMBO J. 2005;24:1852–1862. doi: 10.1038/sj.emboj.7600661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kullmann M, Gopfert U, Siewe B, Hengst L. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5'UTR. Genes Dev. 2002;16:3087–3099. doi: 10.1101/gad.248902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colegrove-Otero LJ, Devaux A, Standart N. The Xenopus ELAV protein ElrB represses Vg1 mRNA translation during oogenesis. Mol. Cell. Biol. 2005;25:9028–9039. doi: 10.1128/MCB.25.20.9028-9039.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meng Z, King PH, Nabors LB, Jackson NL, Chen CY, Emanuel PD, Blume SW. The ELAV RNA-stability factor HuR binds the 5'-untranslated region of the human IGF-IR transcript and differentially represses cap-dependent and IRES-mediated translation. Nucleic Acids Res. 2005;33:2962–2979. doi: 10.1093/nar/gki603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Min H, Turck CW, Nikolic JM, Black DL. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 1997;11:1023–1036. doi: 10.1101/gad.11.8.1023. [DOI] [PubMed] [Google Scholar]
- 22.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 23.Stoecklin G, Colombi M, Raineri I, Leuenberger S, Mallaun M, Schmidlin M, Gross B, Lu M, Kitamura T, Moroni C. Functional cloning of BRF1, a regulator of ARE-dependent mRNA turnover. EMBO J. 2002;21:4709–4718. doi: 10.1093/emboj/cdf444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brewer G. An A + U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol. Cell. Biol. 1991;11:2460–2466. doi: 10.1128/mcb.11.5.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sela-Brown A, Silver J, Brewer G, Naveh-Many T. Identification of AUF1 as a parathyroid hormone mRNA 3’-untranslated region-binding protein that determines parathyroid hormone mRNA stability. J. Biol. Chem. 2000;275:7424–7429. doi: 10.1074/jbc.275.10.7424. [DOI] [PubMed] [Google Scholar]
- 27.Sarkar B, Lu JY, Schneider RJ. Nuclear import and export functions in the different isoforms of the AUF1/heterogeneous nuclear ribonucleoprotein protein family. J. Biol. Chem. 2003;278:20700–20707. doi: 10.1074/jbc.M301176200. [DOI] [PubMed] [Google Scholar]
- 28.Raineri I, Wegmueller D, Gross B, Certa U, Moroni C. Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res. 2004;32:1279–1288. doi: 10.1093/nar/gkh282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fialcowitz EJ, Brewer BY, Keenan BP, Wilson GM. A hairpin-like structure within an AU-rich mRNA-destabilizing element regulates trans-factor binding selectivity and mRNA decay kinetics. J. Biol. Chem. 2005;280:22406–22417. doi: 10.1074/jbc.M500618200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loflin P, Chen CY, Shyu A.-B. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev. 1999;13:1884–1897. doi: 10.1101/gad.13.14.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu N, Chen C, Shyu A.-B. Versatile role for hnRNPD isoforms in the differential regulation of cytoplasmic mRNA turnover. Mol. Cell. Biol. 2001;21:6960–6971. doi: 10.1128/MCB.21.20.6960-6971.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laroia G, Cuesta R, Brewer G, Schneider RJ. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science. 1999;284:499–502. doi: 10.1126/science.284.5413.499. [DOI] [PubMed] [Google Scholar]
- 33.Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 34.Zhang W, Wagner BJ, Ehrenman K, Schaefer AW, DeMaria CT, Crater D, DeHaven K, Long L, Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol. Cell. Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shyu AB, Wilkinson MF. The double lives of shuttling mRNA binding proteins. Cell. 2000;102:135–138. doi: 10.1016/s0092-8674(00)00018-0. [DOI] [PubMed] [Google Scholar]
- 36.Lal A, Abdelmohsen K, Pullmann R, Kawai T, Yang X, Galban S, Brewer G, Gorospe M. Posttranscriptional derepression of GADD45α by genotoxic stress. Mol. Cell. 2006;22:117–128. doi: 10.1016/j.molcel.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 37.Lin S, Wang W, Wilson GM, Yang X, Brewer G, Holbrook NJ, Gorospe M. Downregulation of cyclin D1 expression by prostaglandin A2 is mediated by enhanced cyclin D1 mRNA turnover. Mol Cell. Biol. 2000;20:7903–7913. doi: 10.1128/mcb.20.21.7903-7913.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cok SJ, Acton SJ, Sexton AE, Morrison AR. Identification of RNA-binding proteins in RAW 264.7 cells that recognize a lipopolysaccharide-responsive element in the 3-untranslated region of the murine cyclooxygenase-2 mRNA. J. Biol. Chem. 2004;279:8196–8205. doi: 10.1074/jbc.M308475200. [DOI] [PubMed] [Google Scholar]
- 39.Lu JY, Sadri N, Schneider RJ. Endotoxic shock in AUF1 knockout mice mediated by failure to degrade proinflammatory cytokine mRNAs. Genes Dev. 2006;20:3174–3184. doi: 10.1101/gad.1467606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen ZJ, Esnault S, Malter JS. The peptidyl-prolyl isomerase Pin1 regulates the stability of granulocyte-macrophage colony-stimulating factor mRNA in activated eosinophils. Nat. Immunol. 2005;6:1280–1287. doi: 10.1038/ni1266. [DOI] [PubMed] [Google Scholar]
- 41.Fawal M, Armstrong F, Ollier S, Dupont H, Touriol C, Monsarrat B, Delsol G, Payrastre B, Morello DA. A “liaison dangereuse” between AUF1/hnRNPD and the oncogenic tyrosine kinase NPM-ALK. Blood. 2006;108:2780–2788. doi: 10.1182/blood-2006-04-014902. [DOI] [PubMed] [Google Scholar]
- 42.Dobi A, Szemes M, Lee C, Palkovits M, Lim F, Gyorgy A, Mahan MA, Agoston DV. AUF1 is expressed in the developing brain, binds to AT-rich double-stranded DNA, and regulates enkephalin gene expression. J. Biol. Chem. 2006;281:28889–28900. doi: 10.1074/jbc.M511858200. [DOI] [PubMed] [Google Scholar]
- 43.Nagaoka K, Tanaka T, Imakawa K, Sakai S. Involvement of RNA binding proteins AUF1 in mammary gland differentiation. Exp. Cell Res. 2007;313:2937–2945. doi: 10.1016/j.yexcr.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 44.Gouble A, Grazide S, Meggetto F, Mercier P, Delsol G, Morello D. A new player in oncogenesis: AUF1/hnRNPD overexpression leads to tumorigenesis in transgenic mice. Cancer Res. 2002;62:1489–95. [PubMed] [Google Scholar]
- 45.Lu JY, Schneider RJ. Tissue distribution of AU-rich mRNA-binding proteins involved in regulation of mRNA decay. J. Biol. Chem. 2004;279:12974–12979. doi: 10.1074/jbc.M310433200. [DOI] [PubMed] [Google Scholar]
- 46.López de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl Acad. Sci USA. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gorodkin J, Heyer LJ, Stormo GD. Finding the most significant common sequence and structure motifs in a set of RNA sequences. Nucleic Acids Res. 1997;25:3724–3732. doi: 10.1093/nar/25.18.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eddy SR, Durbin R. RNA sequence analysis using covariance models. Nucleic Acids Res. 1994;22:2079–2088. doi: 10.1093/nar/22.11.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tenenbaum SA, Lager PJ, Carson CC, Keene JD. Ribonomics: identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods. 2002;26:191–198. doi: 10.1016/S1046-2023(02)00022-1. [DOI] [PubMed] [Google Scholar]
- 50.DeMaria CT, Brewer G. AUF1 binding affinity to A +U-rich elements correlates with rapid mRNA degradation. J Biol Chem. 1996;271:12179–12184. doi: 10.1074/jbc.271.21.12179. [DOI] [PubMed] [Google Scholar]
- 51.Chen CY, Xu N, Zhu W, Shyu AB. Functional dissection of hnRNP D suggests that nuclear import is required before hnRNP D can modulate mRNA turnover in the cytoplasm. RNA. 2004;10:669–680. doi: 10.1261/rna.5269304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dember LM, Kim ND, Liu KQ, Anderson P. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J. Biol. Chem. 1996;271:2783–2788. doi: 10.1074/jbc.271.5.2783. [DOI] [PubMed] [Google Scholar]
- 53.Wang W, Martindale JL, Yang X, Chrest FJ, Gorospe M. Increased stability of the p16 mRNA with replicative senescence. EMBO Rep. 2005;6:158–164. doi: 10.1038/sj.embor.7400346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Palanisamy V, Park NJ, Wang J, Wong DT. AUF1 and HuR proteins stabilize interleukin-8 mRNA in human saliva. J. Dent. Res. 2008;87:772–776. doi: 10.1177/154405910808700803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilson GM, Sutphen K, Chuang KY, Brewer G. Folding of A +U-rich RNA elements modulates AUF1 binding. Potential roles in regulation of mRNA turnover. J. Biol. Chem. 2001;276:8695–8704. doi: 10.1074/jbc.M009848200. [DOI] [PubMed] [Google Scholar]
- 56.Ing NH, Massuto DA, Jaeger LA. Estradiol up-regulates AUF1p45 binding to stabilizing regions within the 3′-untranslated region of estrogen receptor alpha mRNA. J. Biol. Chem. 2008;283:1764–1772. doi: 10.1074/jbc.M704745200. [DOI] [PubMed] [Google Scholar]
- 57.Pullmann R, Jr, Abdelmohsen K, Lal A, Martindale JL, Ladner RD, Gorospe M. Differential stability of thymidylate synthase 3′-untranslated region polymorphic variants regulated by AUF1. J. Biol. Chem. 2006;281:23456–23463. doi: 10.1074/jbc.M600282200. [DOI] [PubMed] [Google Scholar]
- 58.Lapucci A, Donnini M, Papucci L, Witort E, Tempestini A, Bevilacqua A, Nicolin A, Brewer G, Schiavone N, Capaccioli S. AUF1 Is a bcl-2 A + U-rich element-binding protein involved in bcl-2 mRNA destabilization during apoptosis. J. Biol. Chem. 2002;277:16139–16146. doi: 10.1074/jbc.M201377200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.