Abstract
The RES complex was previously identified in yeast as a splicing factor affecting nuclear pre-mRNA retention. This complex was shown to contain three subunits, namely Snu17, Bud13 and Pml1, but its mode of action remains ill-defined. To obtain insights into its function, we have performed a structural investigation of this factor. Production of a short N-terminal truncation of residues that are apparently disordered allowed us to determine the X-ray crystallographic structure of Pml1. This demonstrated that it consists mainly of a FHA domain, a fold which has been shown to mediate interactions with phosphothreonine-containing peptides. Using a new sensitive assay based on alternative splice-site choice, we show, however, that mutation of the putative phosphothreonine-binding pocket of Pml1 does not affect pre-mRNA splicing. We have also investigated how Pml1 integrates into the RES complex. Production of recombinant complexes, combined with serial truncation and mutagenesis of their subunits, indicated that Pml1 binds to Snu17, which itself contacts Bud13. This analysis allowed us to demarcate the binding sites involved in the formation of this assembly. We propose a model of the organization of the RES complex based on these results, and discuss the functional consequences of this architecture.
INTRODUCTION
Pre-mRNA splicing is an essential step of gene expression in eukaryotes. Indeed, the accurate removal of introns is critical to reconstitute meaningful coding sequences from the dispersed information present in pre-mRNAs. Biochemical and genetic analyses have demonstrated that pre-mRNA splicing is catalyzed in a large assembly called the spliceosome (1). This large complex assembles around introns through the ordered addition of splicing factors in an energy-dependent process.
In recent years, many splicing factors that contribute to spliceosome formation have been identified. Those can be divided in two types: small nuclear ribonucleoproteins (or snRNPs) and protein splicing factors. The former consist of a short RNA, which appears to be stably associated with a specific set of proteins, whereas factors of the second type are composed exclusively of one or more protein subunits (1). Among the latter, the RES complex was identified in Saccharomyces cerevisiae as an assembly required for efficient splicing and nuclear pre-mRNA retention (2). TAP purification demonstrated that the RES complex is hetero-trimeric and composed of the subunits Snu17, Bud13 and Pml1 (2). Intriguingly, yeast Snu17 had been proposed to be a subunit of the Sf3b splicing factor (3,4), but biochemical purification of the latter demonstrated that it does not contain Snu17 (2).
Genetic analyses demonstrated that all RES subunits were dispensable for normal growth, but were required for growth at high temperature (2,3). Characterization of the molecular processes affected in strains carrying deletions of Snu17, Bud13 or Pml1 demonstrated that they were all required for efficient pre-mRNA splicing. Moreover, this requirement was exacerbated in the case of introns with weak 5′ splice sites. In addition, in such strains, pre-mRNAs were shown to leak from the nucleus to the cytoplasm. Strikingly, Pml1 deletion mutants appeared to display weaker phenotypes than Snu17 or Bud13 deletion mutants, such that in appropriate conditions pre-mRNA leakage occurred in the absence of obvious splicing defect in Pml1-deleted strains. The implication of the RES complex in splicing was consistent with the previous co-purification of Snu17 and Bud13 with the splicing factor Cdc5/Cef1, (5) and the independent observation of the presence of Pml1 and Snu17 in U1·U2·U4/U6·U5 penta-snRNP (6). Implication of the RES complex in RNA processing was further supported by the presence of an RNA recognition motif (RRM) in its Snu17 subunit.
As expected for splicing factors, RES subunit homologues are clearly identifiable in other eukaryotic species, including humans (2). Moreover, purification of spliceosomes assembled in vitro from human cell extracts demonstrated that human RES (hRES) subunit homologues also incorporate into spliceosomes (7,8). These results showed that hRES joins the spliceosome at a late stage of assembly, the complex A to B transition. This corresponds to the joining of the tri-snRNP composed of U4/U6·U5 snRNPs to pre-spliceosome containing the U2 snRNP. Thus, the addition of hRES occurs immediately prior the first catalytic splicing step that leads to intron excision.
Further understanding of the function of the RES complex will require a better knowledge of the structure of its subunits and of its organization. We report here the structure of the Pml1 subunit determined by crystallization and X-ray diffraction. This demonstrated the presence of a FHA domain, which is a module reported to bind phosphothreonine (9–11). The structure was used to design mutations designed to perturb the phosphothreonine-binding cleft. However, using a new sensitive splicing assay based on competition between alternative splice sites, we show that alteration of the phosphothreonine-binding pocket does not affect impair splicing. A description of the RES subunit architecture was also deduced from the analysis of the recombinant complex and derivatives harboring mutated, truncated and/or deleted subunits. This allowed the identification of a potential binding site of Pml1 on Snu17, as well as a C-terminal sequence of Bud13, which is necessary and sufficient to bind to Snu17. On the basis of these results, we propose a model of the subunit architecture of RES and discuss functional implications in splicing and RNA metabolism.
MATERIALS AND METHODS
Cloning and protein expression
To express Pml1 for crystallization trials, C-terminally 6His tagged proteins were used. Two PCR reactions were performed with oligonucleotides: YLR016c-DN24-Nde and YLR016c-as-his-Not or YLR016c-s-Nde and YLR016c-as-his-Not (Supplementary Table 2) and the products were cloned into the vector pET9 (Novagen, Madison, USA) using standard procedures, generating plasmids pET9::Pml1:6His and pET9::DN24Pml1:6His (Supplementary Table 1). Similarly, the primer pairs Bud13-201-NcoI and Bud13-NotI-6H, and Bud13-NcoI and Bud13-201-Not-6H (Supplementary Table 2) were used to generate PCR products encoding the six histidine-tagged Bud13 truncations Bud13-DN201:6His and Bud13DC201:6His, respectively. These PCR fragments were cloned into pET28 (Novagen) to generate the plasmids pET28:Bud13DN201:6H and pET28:Bud13DC201:6H (Supplementary Table 1). For expression of each of these plasmids, Escherichia coli strain Rosetta(DE3)pLysS (Novagen) transformed by these expression plasmids was grown in 2× Yeast Tryptone (2YT) broth until OD600 reached ∼1. Cells were then induced with IPTG (500 μM final concentration) at 30°C for 3 h. To detect the sites of anomalous scattering, selenium was incorporated into DN24Pml1 by expression in media containing seleno-methionine (12).
For operon constructions, coding regions were amplified by PCR and inserted by standard cloning downstream of the T7 promoter of the pBS2454 expression vector. Resulting plasmids, and the oligonucleotides used for their construction, are presented in Supplementary Tables 1 and 2, respectively. In every case, the upstream ORF was fused to a 6His tag to allow purification on Ni2+-NTA. Point mutations were introduced with the QuickChange strategy (Stratagene, La Jolla, USA). Mutagenesis with the Entranceposon system was performed as recommended by the supplier (Finnzyme, Espoo, Finland) on plasmid pBS3160. The mutated Xba I-Sac I region covering the coding sequence of Snu17 and the N-terminal part of Bud13 was recovered and inserted in plasmid pBS3117 to reconstruct a Snu17-Bud13-Pml1 operon. Proteins were expressed in BL21(DE3)pLysS using autoinduction media (13).
To construct the yeast plasmid, the Pml1 ORF and flanking sequence was amplified by PCR and inserted in a yeast replicating vector with a LEU2 marker giving plasmid pBS3012. Point mutations were introduced using the Quick-change system (Stratagene).
Protein purification
Cells expressing operon constructs were broken with a Cell Disruptor (Constant systems) from 200 ml cultures or lyzed with lysozyme in case of small-scale culture (2 ml). Purifications were performed on Ni2+-NTA in batches in 20 mM Tris–Cl pH 8.0, 10 mM imidazole, 500 mM NaCl, 2 mM β-mercaptoethanol, 0.2% NP40. After several washes, proteins were eluted with the same buffer adjusted to 200 mM imidazole. Samples corresponding to the eluted fractions, and in some cases to soluble proteins isolated from the cell extract, were mixed with SDS-containing loading buffer before fractionation on 12% denaturing gels. Proteins were detected by Coomassie staining.
To prepare samples for crystallization, cells were harvested, resuspended in buffer A (20 mM Tris-Cl pH 7.5, 200 mM NaCl), and the cell contents released using a probe-tip sonicator (Branson, Danbury, USA). After clarification of the extract by centrifugation, the extract was purified on a Ni2+-NTA column (Qiagen, Hilden, Germany), and eluted using buffer A which included 200 mM imidazole. The peak fractions were purified on a Superdex 75 column (GE Healthcare, Buckinghamshire, UK), and concentrated using Vivaspin 5000 nominal molecular weight limit cut-off centrifugal concentrators (Vivascience, Goettingen, Germany).
Protein interaction studies using gel-filtration were performed by mixing equimolar quantities of either Bud13DN201:6His or Bud13DC201:6His, with 6His:Snu17. After incubation for 30 min on ice, the samples were injected onto a Superdex 75 10/30 gel filtration column which had been pre-equilibrated with buffer A. The elution profile was followed at 280 nm and peak fractions were subjected to SDS–PAGE.
Proteolysis and mass-spectrometry
Samples were subjected to tryptic and chymotryptic digestion as described previously (14). Samples were withdrawn from the reactions at specific time-points and quenched using PMSF. These samples were mixed with sinapinic acid matrix, evaporated onto a mass-spectrometry sample grid and analyzed with an Applied Biosystems (Framingham, MA) Voyager Elite XL MALDI-TOF spectrometer. Mass/charge ratios identified using this technique were analyzed using tools from ‘Protein Prospector’ (15).
Crystallization
Crystallization trials of native and selenium-labeled DN24Pml1 were performed using the hanging drop vapor-diffusion method at 291°K using a 100 nl drop dispensing robot (Cartesian), with protein concentrations varying from 20 to 30 mg ml−1. A sparse-matrix screen allowed initial crystallization conditions to be obtained (Classics screen, Nextal), condition 89 (0,1M Tris-Cl pH 8.5, pH 8.5, 0.2 M LiSO4 and 30% PEG 4000). Grid and additive screening allowed refinement of the conditions to 0,1M Tris-Cl pH 8.5, pH 8.5, either 0.2 M Na-Acetate or 0.2 M LiSO4 and from 20% to 30% PEG 4000. Crystals appear within 3–10 days.
Data collection
Crystals were briefly treated with a cryo-protectant identical to the mother liquor, except that 20% water was replaced by glycerol. This allowed them to be flash-frozen at 100°K prior to exposure to X-rays. Data were collected at 0.979 Å using the oscillation method on the ESRF beamline ID14-EH4 equipped with a Q315r detector (ADSC). One native dataset of 50°, and one selenium labeled dataset of 60° were collected at with 0.33° and 0.5° oscillations, respectively, and then processed using XDS and XSCALE (16), as well as using the CCP4 suite of programs (17). The crystals belong to the trigonal space group P3221, with unit cell parameters a = b = 85.7 Å, c = 97.8 Å and a = b = 85.72 Å, c = 97.84 Å for the selenium-labeled and native crystals, respectively. Statistics from the data collection are summarized in Table 1.
Table 1.
Data collection statistics
| SAD dataseta | Native dataset | |
|---|---|---|
| Wavelength (Å) | 0.979 | 0.979 |
| Unit-cell parameters (Å) | a = b = 85.75, c = 96.99 | a = b = 85.72, c = 97.84 |
| Space group | P3221 | |
| Matthews coefficient (Å3 Da–1) | 3.06 | |
| Corresponding solvent (%) | 59.6 | |
| Resolution limits (Å)b | 40.0–2.5 (2.6–2.5) | 20.0–2.5 (2.6–2.5) |
| Reflections measuredb | 51 959 (5565) | 42 767 (4821) |
| Unique reflections measuredb | 26 864 (2921) | 14 550 (1617) |
| Completeness (%)b | 97.3 (95.0) | 98.2 (99.2) |
| 〈I〉/〈σ(I)〉b | 13.02 (3.34) | 19.83 (5.82) |
| Rsym (%)b,c | 4.3 (50.6) | 3.8 (22.5) |
a Friedel mates were treated as separate reflections.
b Values in parentheses refer to the highest resolution shell (2.6–2.5 Å).
c Rsym = ΣhΣi |〈I〉h – Ih,i|/ΣhΣi Ih,i, where 〈I〉h is the mean intensity for reflection Ih and Ih,i is the intensity of an individual measurement of reflection Ih.
Structure solution
Crystals obtained with protein which was labeled with selenium (12) was used for a single wavelength anomalous dispersion (SAD) experiment. After processing, SOLVE (18) was used to attempt to locate 10 heavy atom sites, using reflections between 30 and 2.5 Å resolution. After solvent flattening and phase improvement of the resulting electron-density map, an initial partial model was built automatically with RESOLVE (19).
Model building and refinement
Initial refinement of the atomic model of Pml1 was performed using simulated annealing using a phased maximum-likelihood target function as implemented in CNS (20). Later rounds of refinement were performed with REFMAC with maximum likelihood analysis, while including the phase information from the SAD experiment (21,22) of the CCP4 suite of programs (17), with intermittent rounds of model building with COOT (23) and O (24). During initial rounds of refinement, phase restraints and non-crystallographic symmetry restraints were included. Additionally, one TLS group was used per protein monomer of the asymmetric unit. Once an Rfree of ∼0.27 was attained, the model was refined against the ‘native’ data set. Validation of the structure was performed using MOLPROBITY (25), and statistics of the refined model of Pml1, after correction of the B-factors to take into account the TLS refinement (26) are summarized in Table 2. The atomic coordinates of the refined model have been deposited into the PDB, and the entry has been assigned the accession code 2JKD.
Table 2.
Refinement statistics
| Number of non-hydrogen atoms (Protein/water/other) | 2364/46/28 |
|---|---|
| Resolution range (Å) | 40.9–2.5 |
| R/Rfree (%) | 21.6/25.9 |
| R.M.S.D. bonds (Å)/angles (°) | 0.01/1.44 |
| 〈B〉(Å2) Protein/water/other | 57.61/54.3/89.4 |
Yeast strains and splicing assays
Isogenic yeast strains BMA64 (wild type), BSY1395 (Δsnu17) and BSY1397 (Δpml1) (2) were used for splicing assays, which were performed at 30°C. Yeast transformations, reporter inductions and β-galactosidase assays were performed as described previously (27).
RESULTS
Crystallization of the Pml1 subunit
To gain insights into the structure of the RES complex we expressed the various subunits individually or in combination and tested for the crystallization of purified proteins. First attempts were unsuccessful. Thus, limited proteolysis was performed on individual subunits with trypsin or chymotrypsin to identify disordered regions. Treatment of tagged Pml1 (calculated molecular weight 23 653.6 Da) yielded stable fragments between 24 and 18 kDa. MALDI-TOF mass spectrometry analysis indicated that the fragments of 21 604, 19 020 and 18 410 Da obtained with trypsin, covered residues 23–210, 46–210 and 52–210. respectively. Crystallization screening with a construct corresponding to Pml1 truncated from its 24 amino-terminal residues, designed to mimic the former of these fragments, successfully yielded diffracting crystals.
Crystal structure of Pml1
After data collection from Pml1 crystals into which seleno-methionine had been incorporated, the data were reduced while treating each reflection of Friedel pairs as having independent intensities. The locations of atoms exhibiting anomalous scattering were determined using SHELX (28), which located 10 credible sites in a SAD experiment. Subsequent heavy atom searches using SOLVE (18) were performed to find these 10 sites and calculate initial phases using the same data. Phase improvement was then performed by density modification using RESOLVE (18), which was also able to construct an initial model by iterations of model building, further density modification and refinement of the model (19).
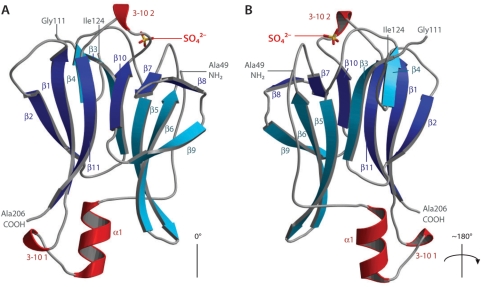
The asymmetric unit consists of two protomers of Pml1, the refined models of which consist of residues 49–112 and 122–206 of chain A, as well as residues 51–111 and 122–206 of chain B (Figure 1). As anticipated from sequence comparisons, the polypeptide chain has the fold of a Forkhead-Associated (FHA) domain, an 11-stranded β-sandwich, with two inserted 3–10 helices, and an N-terminal α-helix. The entire structure has approximate dimensions of 50 × 35 × 40 Å and is composed of two sandwiched mainly anti-parallel β-sheets. The first consists of β-strands (in order along the sheet) 2, 1, 11, 10, 7 and 8, while the second consists of β-strands 4, 3, 5, 6 and 9. The first 10 N-terminal residues of the truncated Pml1 which are ordered form an extended sequence which folds back along β-sheet 2, which is a feature that is particular to this FHA domain structure. The C-terminus also folds back in this direction. Several residues of Pml1 are conserved in close homologues, including amino-acids probably generally involved in ligand binding by FHA domains (see below and Supplementary Figure 1).
Figure 1.
Structure of the Pml1 FHA domain. (A) Pml1 consists of an 11-stranded β-sandwich with two 3–10 helices and one α-helix. In the first β-sheet, the strands are colored in dark blue, and those of the second sheet are colored cyan, while helical elements of secondary structure are colored in red. The putative phosphopeptide-binding loops are oriented at the top of the diagram. The sulphate ion found in the ‘native’ structure is shown as sticks. (B) The structure has been rotated 180° around the axis shown. The extended N-terminal extension to the FHA fold is found on this face of the molecule and approaches the sulphate ion. This figure was produced using MOLSCRIPT (47) and BOBSCRIPT (48), as was Figure 3.
FHA domains have been shown to mediate interactions with phosphorylated peptides, particularly those phosphorylated on threonine residues (9,10,29), although weaker interactions between phospho-tyrosine peptides and FHA domains have also been reported, with dissociation constants in the micromolar range (30). The resolution of the crystal structure of Rad53-FHA1 in complex with an in vitro selected phosphorylated peptide ligand, demonstrated the molecular basis of their recognition (10). The recognition of target peptides was shown to be mediated by a cleft formed by loop sequences that intersperse the β-strands. The loops of Pml1 form an analogous cleft, which may bind a target peptide in a similar manner. The least well-defined electron-density in Pml1 is located in the loop-region, and the structured residues in this zone have elevated B-factors compared to the rest of the molecule. While Pml1 has a mean B-factor of 58 Å2, residues 85–92 and 191–196 have average temperature factors of above 75 Å2. This could reflect flexibility of these loops required to accommodate conformational changes during ligand binding.
A DALI search (31) revealed that the most similar proteins are: antigen Ki-67, protein kinase Rad53, Afadin, Kinase-associated Protein Phosphatase (KAPP) and Bifunctional Protein Kinase/Phosphatase (Table 3; see Supplementary Figure 2 for a superposition of these models). Although these structural neighbors are all FHA domains, significant differences exist, particularly in the loop regions and termini. There are certain residues that are structurally conserved amongst these proteins, and many of these are involved in maintaining the integrity of the fold and are probably generic to the FHA domain. Other notable residues, which are structurally conserved amongst these proteins, are located in the phosphopeptide-binding site.
Table 3.
Structurally similar protein structures
| PDB | Z-score | RMSD | Aligned | % ID | Protein |
|---|---|---|---|---|---|
| 1r21 | 12.7 | 2.0 | 100 | 29 | Ki-67 |
| 1g6g | 11.9 | 2.3 | 103 | 21 | Rad53 FHA1 |
| 1wln | 11.1 | 2.3 | 105 | 20 | Afadin 6 |
| 1mzk | 11.1 | 2.3 | 103 | 21 | Kinase-associated Protein Phosphatase |
| 2brf | 9.4 | 2.9 | 98 | 15 | Poly-nucleotide Phosphatase |
| 1ujx | 8.8 | 2.9 | 101 | 19 | Polynucleotide Kinase 3′ Phosphatase |
| 1qu5 | 8.5 | 3.8 | 112 | 14 | Rad53 FHA2 |
| 1ygs | 8.1 | 2.6 | 104 | 7 | Smad4 |
| 1qwt | 6.7 | 3.0 | 109 | 6 | Interferon regulatory factor 3 |
| 1lgp | 5.1 | 2.9 | 69 | 23 | CHFR mitotic checkpoint protein |
Comparison of the structure of Pml1 with the structure of the N-terminal FHA domain (FHA1) of Rad53 bound to a phosphopeptide demonstrates the potential utility of the conserved residues in binding putative ligands by Pml1 (29). In that structure, the O1 of the pThr is contacted by the γ-oxygen of Thr106, a residue which is identical in the structures of the Ki67 FHA domain and FHA2 of Rad53 (32,33), and is conserved as Ser162 in the structure of Pml1 (Figures 2 and S1). Ser137 of Pml1 is also conserved as Ser85 in FHA1 of Rad53, which makes contacts with the O2 of the phosphate via the γ-oxygen of the serine side chain. On the opposite side of the pThr in the Rad53-peptide complex, the guanidino-group of Arg70 forms a hydrogen bond with a γ-oxygen of the phosphate moiety. This residue is also highly conserved (present as Arg108 in Pml1 has given), and is probably involved in conferring specificity towards phosphorylated target peptides. Accordingly, mutation of this residue to alanine in Rad53 reduced the affinity of such pThr-containing target peptides approximately 10-fold (9). Additionally, Asn86 of Rad53-FHA1 forms hydrogen bonds with the pThr from its main chain nitrogen, as well as via a δ-nitrogen. The structurally equivalent residue of Pml1 is Lys138, so this side chain would also be able to contact the phosphopeptide in a similar way, if the putative target peptide was located sufficiently nearby (Figure 3B).
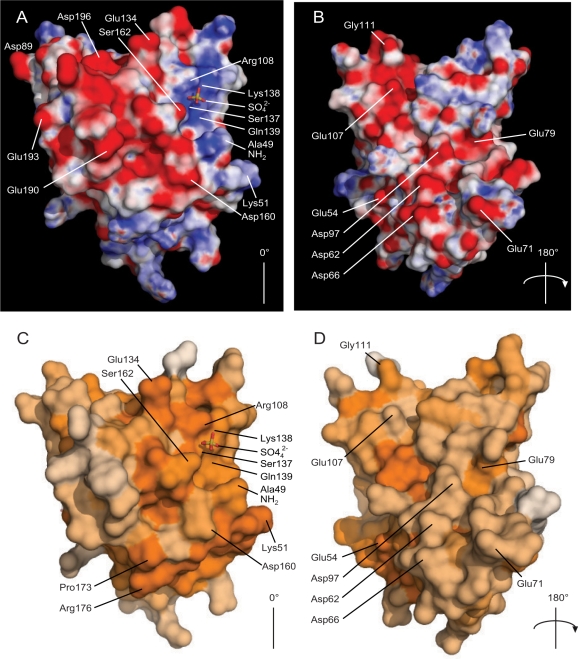
Figure 2.
Electrostatic and conservation surfaces of Pml1. Pml1 is shown in two views, (A) and (C), depicting the sulphate-binding cleft, and after having been rotated 180° around the axes shown (B) and (D) to show the opposite face. A, B): Electrostatic properties of Pml1. Electrostatic potentials were calculated using APBS (49), with a protein dielectric constant of 2.0 and a solvent dielectric of 80 and mapped onto the surface of Pml1. Electrostatic potentials are shown as a gradient from −15 kT/e (negatively charged areas, red) <0 kT/e neutral, white <15 kT/e (positively charged regions, blue). In (A), the basic cleft in which the sulphate ion is situated (shown as sticks, as in Figure 1), is located at the top right of the molecule, with the N-terminal Ala49 just below it. An acidic region is also apparent on the left section of the molecule, which runs over the top, to the opposite face (B). (C, D) Conservation of surface residues of Pml1. Conservation of residues, using the same alignment as Figure S1 for the calculation (34), in steps from white (not conserved), to dark orange (sequence identity). A notable region of conserved residues is apparent around the sulphate-binding site. This figure was produced using PyMol (50).
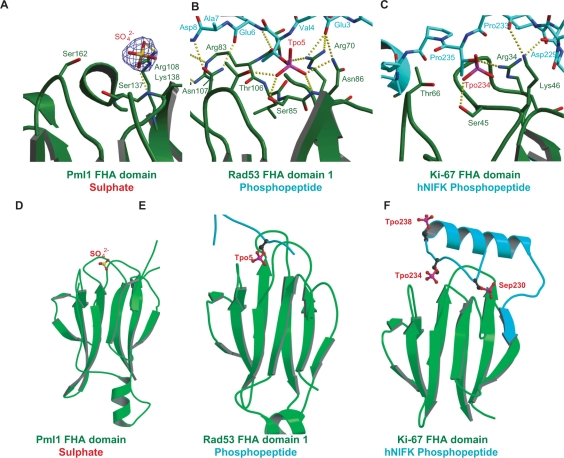
Figure 3.
A potential phosphopeptide-binding site of Pml1. The sulphate-bound form of Pml1 (A, D) is compared with two ligand-bound FHA domains: Rad53:phosphopeptide (10); (B, E) and Ki-67:hNIFK phosphopeptide (29); (C, F). In each panel, the FHA domain-containing proteins are colored green, and the phospho-peptides are colored turquoise. Phosphorylated threonine (Tpo) and serine (Sep) residues are shown as balls and sticks, as is the sulphate of Pml1. Atoms are colored as follows: carbon; black, phosphorous; pink, oxygen; red and sulfur; yellow. The sulphate is situated approximately in a similar position to Tpo5 and of the Rad53:phosphopeptide structure and Tpo234 of the Ki-67:hNIFK phosphopeptide structures. Panels A, B and C show overviews of each of the structures. Panels D, E and F depict close-up views of the sulphate of Pml1, the pThr of the Rad53:phosphopeptide structure and Tpo234 of the Ki67 phosphopeptide, respectively. In panel A, the electron density from a 2Fo-Fc map is shown around the sulphate ion in the phosphopeptide-binding cleft of Pml1 is contoured at 1.5σ.
Overall, Pml1 possesses a phosphopeptide-binding site with many of the hallmarks exemplified by Rad53-FHA1, which serves as a model for FHA domains bound to ligand peptides. Other structures of co-crystals of FHA domains with phosphorylated ligands support this general conclusion, even if minor differences suggest that the mechanism of peptide binding may differ from one FHA domain to the next (see Discussion section).
Sulphate ions are located in a conserved, charged cleft of Pml1
The crystals from which the ‘native’ data sets were collected were grown in mother liquor which contained lithium sulphate (Table 1). During refinement, strong electron density was seen at equivalent positions of the two Pml1 monomers of the asymmetric unit, which were modeled as SO42– ions (Figures 2 and 3). Electron density at these positions was not seen in the case of the model refined against the SAD dataset, however. These ions occupy positions corresponding approximately to the phosphate moiety of pThr of the Rad53-FHA1 structure (Figure 3A, B, D and E). Indeed, the interaction network around the sulphate is typical of the number and type of bonds found contacting the phosphate ion of the pThr in structural precedents, and so these sulphate ions mimic the phosphate of pThr.
Electrostatic potential representations of the surface of Pml1 highlight two main features (Figure 2A and B). First, the charged cleft in which the sulphate ion is situated is relatively basic, and is lined by the residues Arg108, Lys138 and Gln139. The charge of the cavity appears to provide a suitable electrostatic environment for the SO42– ion, or indeed a potential phospho-threonine. The presence of such a charged region around the ligand-binding site has been noted previously in the case of the FHA1 of Rad53 (11). Mapping the sequence conservation amongst close homologues of Pml1 (Figure S1) onto its surface (34) illustrates the locations of the conserved residues (Figure 2C and D). Residues such as Arg108, Lys138 and Gln139, which are located within the cleft, appear to be among the most conserved on the surface of the protein. Additionally, Ser162, which is in close proximity to the SO42– ion is also highly conserved (Figures S1 and 3A), which implies that it is necessary for Pml1 function. The second major feature of the electrostatic potential surface is an almost continuous negatively charged stripe which passes from one face of the molecule, along residues Asp160, Glu190, Glu193, Asp196, Glu134, over to another region on to the opposite face of the molecule (glutamates 54, 71, 79 and 107 as well as aspartates 62, 66 and 97).
The crystal structure of Pml1 has given us clues that it possesses a relatively conserved, charged binding site, but various questions remain unanswered. For example, the identity of the ligand which most likely binds to the conserved cleft is not currently known. Consequently, it is not known either whether its phosphorylation is absolutely required for interaction, despite the circumstantial evidence provided by sulphate ions found in this structure.
An alternative 5′ splice-site competition assay reveals that the FHA phosphothreonine-binding cleft of Pml1 is not required for splicing
The data presented above indicate that, through the Pml1 subunit, the RES complex may interact with partners in a phosphorylation-regulated manner. To test the implication of such a potential interaction in splicing, we constructed Pml1 mutants carrying substitutions of residues R108 to A, S137 to R or S137 to A. These mutations were designed based on the structure of the Pml1 subunit to avoid the perturbation of the overall structure of Pml1, while sterically hindering binding to the FHA pThr-binding pocket. These mutations were constructed in yeast shuttle vectors containing the Pml1 ORF and natural flanking sequences, including its putative promoter and terminator. A plasmid expressing wild-type Pml1 was also constructed as a control. These plasmids were introduced into a Pml1 deletion strain with splicing and pre-mRNA leakage reporters, which had been used previously to characterize the phenotypes of the RES mutants. However, β-galactosidase analysis failed to provide a definitive conclusion as to the role of the Pml1 pThr-binding site on splicing or pre-mRNA leakage. This result is probably a consequence of the moderate phenotypes exhibited by the Pml1 mutant, which may have been further weakened by heterogeneity in plasmid numbers (resulting from interaction/competition between the reporter plasmids and Pml1 expressing plasmids in this specific test), thus affecting the β-galactosidase assay results.
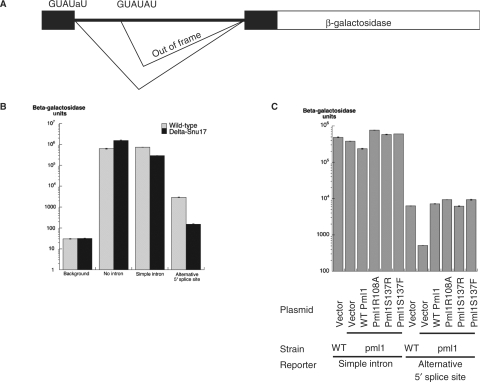
To overcome the limited sensitivity of this first analysis, we designed a potentially more sensitive assay by testing the effect of RES mutations on a reporter carrying competing 5′ splice sites (Figure 4A). Indeed, as RES inactivation had been shown previously to affect preferentially introns carrying weak 5′ splice sites, we reasoned that RES inactivation might shift the usage of alternative splice sites from a weak site to a strong one. To test this hypothesis, a reporter carrying a GUAUaU mutant upstream site and a wild-type GUAUGU downstream site, as well as controls, were introduced into a Snu17 deletion mutant. A Snu17 mutant was selected to validate the assay, as it demonstrates a stronger splicing phenotype than Pml1. Splicing of the reporter was followed by analyzing β-galactosidase production, with increases reflecting usage of the upstream site and decreases usage of the downstream site, because the latter one produces an out-of frame mRNA. Notably, while deletion of Snu17 resulted in only a 2.5-fold reduction in the splicing of a wild-type intron carrying a single 5′ splice site [consistent with previous analyses (2)], we observed a 20-fold reduction of the usage of the upstream splice site in a Snu17 mutant using the alternative splicing reporter (Figure 4B). These results demonstrated that Snu17 is endowed with the ability to alter splice-site choice, and that the duplicated splice-site reporter indeed provides a more sensitive assay for RES splicing activity.
Figure 4.
A sensitive assay based on alternative 5′ splice-site choice shows that the Pml1 phophopeptide-binding pocket does not affect splicing. (A) Structure of the reporter used for the splicing assays. The reporter is based on the insertion of the RP51A intron and flanking sequence in the lacZ coding sequence encoding β-galactosidase (51). The 5′ splice site was duplicated with the upstream copy being mutated at position 5 from G to A, while the downstream copy is wild type. Usage of the upstream 5′ splice site produces in-frame messages encoding β-galactosidase while usage of the downstream 5′ splice site produces out of frame mRNAs. (B) Histogram reporting the results of β-galactosidase assays from the reporters encoding no β-galactosidase (‘background’), the LacZ reporter gene with no intron (‘no intron’), the LacZ reporter gene interrupted by a simple intron (‘simple intron’) or the LacZ reporter gene interrupted by an intron with alternative 5′ splice sites depicted in (A) (‘alternative 5′ splice sites’) expressed either in a wild-type yeast strain (hatched bars) or in a strain in which Snu17 had been deleted (black bars). (C) Plasmids without an insert (‘Vector’), encoding Pml1 (‘WT Pml1’) or containing Pml1 mutated at the putative pThr-binding site (‘Pml1R108A’, ‘Pml1S137A’ and ‘Pml1S137F’) were assayed to detect effects on splicing. These were tested in wild-type yeast strains (‘WT’) or in a strain in which Pml1 had been deleted (‘Δpml1’), using either the reporter encoding LacZ reporter gene interrupted by a simple intron (‘simple intron’) or LacZ reporter gene with alternative 5′ splice sites depicted in (A) (‘alternative 5′ splice sites’). A histogram reporting the results of these β-galactosidase assays is depicted.
The same reporter was thus used to analyze splicing in the Pml1 mutant strains. β-galactosidase assays demonstrated that Pml1 inactivation also induces a 12-fold drop in the usage of the upstream site of this reporter, while it had no significant effect on the splicing of a wild-type intron under the same conditions, in agreement with previous analyses. The Pml1R108A, Pml1S137F and Pml1S137R mutants did not result in any alteration of the splicing pattern using this sensitive assay, suggesting that the interaction of the FHA domain of Pml1 with a phosphorylated ligand is unlikely to be involved in pre-mRNA splicing, suggesting its implication in another function. This is consistent with previous analyses that revealed that the RES complex was a multifunctional entity affecting several steps of the gene expression pathway (2).
Production of recombinant RES and subunit interaction pattern
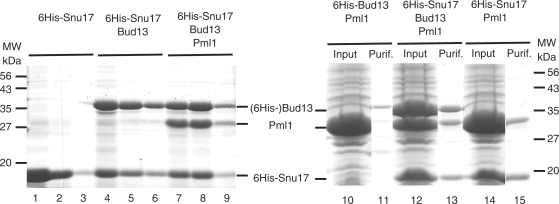
To better understand the structure and function of the RES complex, the next step was to elucidate how Pml1 interacts with the two other subunits of the RES complex, namely Snu17 and Bud13. The RES complex can be purified from TAP-tagged strains of S. cerevisiae (2). This allowed us to estimate the size of the complex purified from yeast by size exclusion chromatography to be roughly 80 kDa, consistent with a 1:1:1 stoichiometry of the three subunits (calculated molecular weight: 72 kDa). However, the yields were too low for further structural analyses. Therefore, we decided to resort to recombinant protein production for the analysis of complex organization. For this purpose, we constructed a set of artificial operons in E. coli expression vectors. These plasmids contained the coding sequence of one RES subunit fused to an N-terminal 6His-tag either alone or in tandem with the coding sequence(s) of one or both of the other RES subunits. Proteins expressed in E. coli were purified on Ni2+-NTA beads and analyzed by gel electrophoresis. Production of the 6His-tagged Snu17 protein was efficient (Figure 5, lanes 1–3), and co-expression of 6His-Snu17 with either Bud13, or with both Bud13 and Pml1 resulted in the recovery of complexes containing the two or the three proteins after affinity purification on Ni2+-NTA beads (Figure 5, lanes 4–9). Control experiments confirmed that untagged proteins were unable to interact with the affinity media alone (data not shown). Thus, this experiment demonstrated that the RES complex can be produced in a recombinant manner in E. coli, where it assembles efficiently. Alternatively, we were able to reconstitute the entire RES complex by incubating together independently expressed subunits (data not shown).
Figure 5.
RES subunit interactions. Coomassie stained gels of input material and purified fraction are presented. On the left hand side, aliquots of three consecutive elution fractions from Ni2+-NTA columns were fractionated on a 12% denaturing gel. Proteins were purified from cells expressing the 6His-Snu17 construct (lanes 1–3), the 6His-Snu17 Bud13 operon (lanes 4–6) or the 6His-Snu17 Bud13 Pml1 operon (lanes 7–9). Positions of migration of molecular weight markers are indicated on the left, while protein bands are identified on the right. On the right hand side, proteins present in the soluble input material (lanes 10, 12 and 14) and the first elution fractions (lanes 11, 13 and 15) have been fractionated on a 12% denaturing gels. Operons expressing 6His-Bud13 Pml1 factors (lanes 10 and 11), the 6His-Snu17 Bud13 Pml1 factors (lanes 12 and 13) or the 6His-Snu17 Pml1 factors (lanes 14 and 15) were used. Protein bands are identified on the left and the positions of migration of molecular weight markers are indicated on the right.
Analysis of the proteins that co-purified after co-expression in E. coli also indicated that Bud13 interacts directly with Snu17 (see earlier section). We produced and analyzed other operons, in order to determine the subunit interaction pattern. Expression and purification of 6His-tagged Bud13 was less efficient than 6His-Snu17, but still allowed the recovery of the tagged protein (data not shown). Vectors encoding untagged Pml1, together with 6His-Bud13, led to the production of a large amount of soluble Pml1 (Figure 5, lane 10) that did not co-purify with his-tagged Bud13 (lane 11), suggesting either that the two proteins do not interact, or that they interact only weakly. In contrast, co-expression of 6His-Bud13 with Snu17 led to the co-purification of the two proteins (data not shown), confirming that they interact directly. Finally, co-expression of 6His-Snu17 with Pml1 led to the efficient co-purification of the two proteins, providing evidence that these two subunits interact directly (Figure 5, lane 15). Overall, these data have demonstrated that Snu17 is the central component of the RES complex, where it interacts independently with Bud13 and Pml1. Conversely, we have been unable to observe a strong direct interaction between Bud13 and Pml1.
Identification of sequences Snu17 of required for interaction with its partners
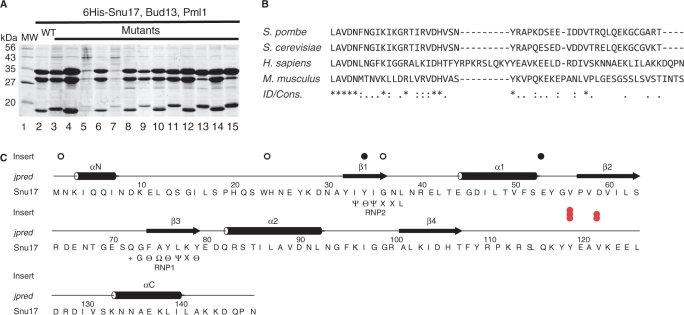
We used a transposon-based mutagenesis strategy (Entranceposon, Finnzyme) to construct a collection of operons encoding 6His-Snu17, Bud13 and Pml1, carrying random insertions of 15 nucleotides in the region covering the Snu17 coding sequence as well as the N-terminal half of Bud13 (see Materials and Methods section). Because of the sequence of the inserted nucleotides, this mutagenesis strategy preserves the reading frames and avoids the creation of truncated proteins due to the undesired introduction of stop codons. Small-scale protein purifications were then performed from cultures of 49 randomly selected mutants and recovered proteins analyzed by gel electrophoresis (Figure 6A and data not shown). Analysis of the Coomassie-stained SDS-PAGE experiments allowed us to classify the mutations into four main groups: (i) efficient co-purification of the three RES subunits as observed with the wild-type operon (e.g. lanes 4, 6, 8, 10, 12 and 14), (ii) efficient co-purification of the three RES subunits but with altered mobility of the Snu17 subunit (e.g. lanes 3, 9, 11), (iii) strongly reduced levels of all three RES proteins (e.g. lanes 5 and 7) or (iv) normal levels of Bud13, usual levels of Snu17 of altered mobility and a reduced yield of Pml1 (e.g. lanes 13 and 15).
Figure 6.
Identification of a conserved sequence element, PIPR, in Snu17 required for efficient interaction with Pml1. (A) Protein profiles of fraction from small-scale purification of factors encoded by a wild-type 6His-Snu17 Bud13 operon (lane 2) and 13 mutant operons carrying a 5 amino-acid insertion in the region encoding 6His-Snu17 and the N-terminus of Bud13 (lanes 3–15) are depicted. (Other mutants are not shown.) Note the reduced Pml1 level observed in lanes 13 and 15. A molecular weight marker was loaded in lane 1 and the corresponding molecular weights are indicated on the left. (B) Conservation of the C-terminal region of Snu17 located after the RRM domain between various species (S. pombe, S. cereviseae, H. sapiens and M. musculus) that corresponds to the Pml1 Interaction Promoting Region prepared using the ClustalW algorithm (52). On the line entitled ‘ID/Cons’. (identity/conservation), asterisks indicate absolutely conserved positions, colons denote strong conservation, and periods indicate weak conservation. (C) Sequence of the 6His-Snu17 protein fusion and location of the sequenced 5 amino-acid insertion resulting in normal protein recovery (open circles, for example the first, second and third open circles correspond respectively to protein profiles depicted in lanes 7, 9 and 1 of panel A), background protein recovery (black dots), or low Pml1 recovery but normal 6His-Snu17 and Bud 13 recovery [red dots, for example the first red dot corresponds to profiles shown in lanes 13 and 15 of (A)]. Elements of secondary structure predicted by Jpred (53) are indicated above the sequence. The topology of these elements in an RRM domain is β1, α1, β2, β3, α2, β4 (indicated above the elements of secondary structure. Snu17 is predicted to have additional α-helices at the N-and C-termini (αN and α-C). Sequences which conform to the RNP1 and RNP2 sequences of canonical RRM domains are represented below the primary structure. The symbols used are as used in (36); + corresponds to basic amino acids, Θ aromatic residues, Ψ aliphatic residues and Ω to small amino acids (see text for a further discussion).
Many mutations were mapped by virtue of a unique restriction site introduced within the inserted nucleotide sequence. The clustering of such mutations indicated that the first category corresponded to insertions in the N-terminus of the Bud13 coding sequence (or intergenic regions), whereas members of the second category were consistent with insertions in Snu17 that did not alter interaction with its partners. The third category consisted of mutations of Snu17 that prevented its expression and therefore co-purification of its partners. Finally, the fourth category corresponded to Snu17 mutations which specifically perturbed the Snu17–Pml1 interaction, while preserving the Snu17–Bud13 interaction. For several representative mutants and for all mutants which specifically blocked the interaction with Pml1, the precise sites of insertion were determined by DNA sequencing. Mutants that did not affect complex formation were located in the N-terminal extension preceding the Snu17 RRM or extended loops connecting the predicted β-sheets and α-helices of the RRM (Figure 6C). Mutants which reduced the recovery of the RES complex either disrupted structural elements of the RRM domain, or altered the 6His tag. Interestingly, all mutants which were detrimental to the interaction with Pml1 were grouped within a stretch of residues located immediately downstream of the RRM domain (Figure 6C, red dots).
Comparison of Snu17 sequences from various Saccharomyces species indicates that this region is particularly well conserved (data not shown). A more moderate conservation is also detectable in other fungi (e.g. S. pombe) and mammals (Figure 6B). This observation indicates that these amino acids are probably part of the Pml1-binding site. This sequence was named PIPR for ‘Pml1 Interaction Promoting Region’. We note that residual binding of Pml1 was still observed with Snu17 mutants carrying insertions in the PIPR sequence, suggesting that Pml1 may also contact other structural elements of the Snu17 factor.
To confirm the results obtained with insertion mutants, we constructed operons which express various truncated versions of 6His-Snu17, Bud13 and Pml1. Deletion of the last 10 residues of Snu17 that are not conserved did not affect its interaction with either Bud13 or Pml1. In contrast, removal of the last 35 residues of Snu17, which include the PIPR, lead to only residual binding of Pml1, but normal association with Bud13 (data not shown). This confirms the importance of the conserved region located downstream of the RRM for efficient Pml1 binding. However, the C-terminal tail of Snu17, encompassing the PIPR (residues 111–149) was unable to pull-down significant quantities of Bud13 or Pml1 (data not shown), confirming that other structural elements of Snu17 are required for efficient interaction with Pml1.
Taken as a whole, these data indicate that Snu17 contains in its C-terminal tail a specific motif, PIPR, which is required for efficient Pml1 binding. This element probably constitutes part of the Pml1-binding site, but is not sufficient in itself for binding, suggesting that other Snu17 residues contribute to the Pml1 interaction. However, we cannot exclude the possibility that the PIPR residues affect indirectly Pml1 binding through an effect on Snu17 folding.
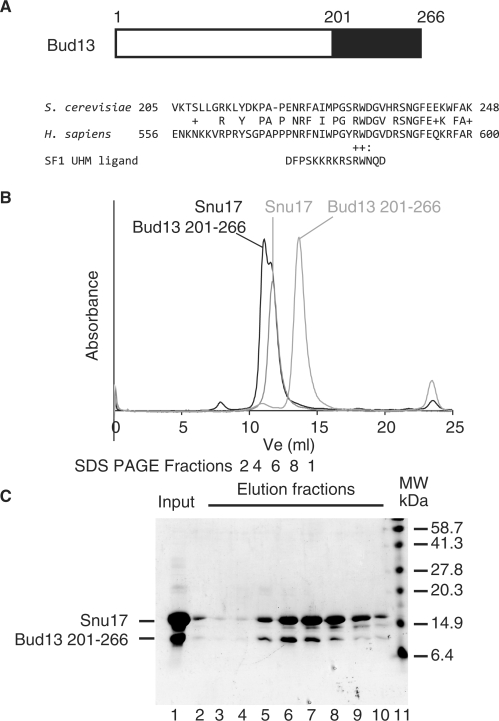
The C-terminal region of Bud13 mediates interaction with Snu17 via a ULM-like motif
Mutagenesis of the N-terminal half of Bud13 failed to identify a region that mediates Snu17 binding (see in the earlier section). Bud13 appears to be poorly conserved between species, apart from its C-terminal region (residues 201–266 of the yeast protein, Figure 7A). Analysis of this sequence revealed the presence of conserved tryptophan residues, which in some factors have been shown to mediate interactions with RRMs, as for example in the case of the interaction occurring between the third RRM of U2AF65 and SF1 (35). Such RRMs are collectively termed UHMs (for U2AF Homology Motifs) (36), while their cognate ligands are called ULMs (for UHM ligand motifs). The residues flanking tryptophan 232 of Bud13 bear a significant similarity to those flanking the tryptophan residue of SF1 that mediate contacts with the U2AF RRM (Figure 7A).
Figure 7.
The conserved C-terminal region of Bud13 is sufficient to mediate interaction with Snu17 and shows similarity with UHM ligand motifs (ULM). (A) Schematic representation of yeast Bud13 showing the location of the most conserved region (amino acids 201–266) as a black rectangle. An alignment of part of the sequence of this region with the sequence of the human homologue (hBud13/MGC13125) is presented below. Underneath this alignment, the sequence of the ULM of the SF1 splicing factor, which as been shown to be sufficient to bind to the third RRM of U2AF65 (35), is presented to demonstrate sequence similarity. (B) A recombinant peptide encompassing residues 201–266 of Bud13 was mixed with recombinant 6His-Snu17 (Input, lane 1) and the two factors were shown to interact by co-fractionation on a Superdex 75 column (GE Healthcare) (see Elution fractions, lanes 2–10). Control fractionation demonstrated that the Bud13 peptide which contains its first 201 residues elutes later under the same conditions. (C) SDS–PAGE of samples corresponding to the complex between Snu17 and the Bud13 fragment 201–266. The sample which had been injected onto the gel-filtration column was migrated on lane 1, while lanes 2–10 correspond to the peak fractions indicated beneath the chromatograms in B). Standard molecular weight markers were migrated alongside for comparison (lane 11). Lane 2 most likely contains carry-over from the input protein.
Despite the fact that Snu17 has not been positively identified as an RRM domain of the UHM subfamily, we tested whether the C-terminal region of Bud13 could promote the interaction with Snu17. For this purpose, we mixed equimolar quantities of the purified Snu17 protein with a recombinant peptide encompassing residues 201–266 of Bud13 and detected the formation of a complex by size exclusion chromatography. Fractionation of the polypeptides present in the various fractions by gel electrophoresis revealed the co-fractionation of the two polypeptides, indicative of complex formation (Figure 7B). As a control, the N-terminal region of Bud13 (residues 1–201) was shown to be unable to interact with Snu17 (data not shown). These experiments indicated that the C-terminal region of Bud13 interacts with Snu17 and this may occur through the interaction of a ULM-like motif with the Snu17 RRM. We observed, however, that the mutation Bud13 arginine 231 to aspartate or tryptophan 232 to alanine did not abolish its interaction with Snu17 (Supplementary Figure 3 and data not shown), in contrast to the results reported for the U2AF65–SF1 interaction (35). This suggests that the mode of interaction of Bud13 and Snu17 may only be distantly related to the one observed of ULM an UHM, consistent with the fact that the Snu17 RRM was not identified as a UHM domain.
DISCUSSION
The X-ray crystallographic structure of the FHA domain of Pml1 indicates that it possesses many of the characteristics required to bind a peptide ligand harboring a pThr residue. This conclusion is further supported by the observation of a sulphate ion in the pThr-binding pocket in the crystal structure. Strikingly, the most N-terminal ordered residue, Ala49, is ∼7.5 Å from the SO42– ion. It will be interesting to determine whether the additional disordered residues at the N-terminus of Pml1 participate in the interaction with ligands. Pml1 possesses a stripe of basic residues which surround the bound sulphate ions, a feature which has been observed previously in the case of the FHA1 of Rad53 (11). However, the Pml1 surface also displays a long acidic stripe, which is composed mainly of glutamate and aspartate residues. The function of this region is still unclear, but may be related to interaction with other partners with significant negatively charged exposed areas.
Although the core features of pThr-peptide recognition appear to be conserved among the known structures of FHA domain-containing proteins, there are also major differences in the modes of peptide binding among them. For instance, the length of the bound peptide differs for different FHA domains. In the case of Rad53 FHA1, which was discussed above, the size of the cavity into which the peptide's +3 residue fits is predicted to influence the specificity of the interaction. In the corresponding structure, the selected aspartate at the +3 position of the ligand forms a hydrogen bond with Rad53 Arg83 (10). The structurally equivalent residue in Pml1 is Thr135, and this residue is displaced away from the peptide-binding cleft by ∼2.2 Å when the two FHA domains are superposed. The peptide-binding cleft of Pml1 is larger than that of Rad53, and Pml1 may therefore recognize a bulkier side chain than aspartate at the pThr +3 position.
The most similar structure to Pml1 is that of the FHA domain of Ki-67 (Table 3) (32), whose structure has also been solved in complex bound to a phosphorylated peptide derived from hNIFK (29) (Figure 3C and F). This interaction differs greatly from its structural precedents in that the target sequence is much longer than thought necessary for other FHA domains and multiple sites must be phosphorylated for optimal binding. Although the primary interactions are centered on the pThr234, two other phosphorylation events, one on Ser230 and another on Thr238, are required for optimal affinity. Also, a β-sheet of Ki-67 is extended by the contribution of an extra β-strand from hNIFK (29). It will be interesting to determine whether such a β-sheet extension also occurs upon binding of Pml1 to a putative ligand. Only slight clashes occur when Pml1 is superimposed onto Ki-67:hNIFK, and these occur between the hNIFK peptide and the loop regions of Pml1, indicating that this mode of peptide binding is compatible with the FHA domain of Pml1.
We investigated the function of the FHA domain of Pml1 by mutating residues in such a way that binding of a pThr ligand would be inhibited, while the general structure of the domain would be maintained. The phenotypes of the three mutants that we produced were tested in yeast. For this purpose, we developed a new, sensitive assay, based on competing 5′ splice sites. First, this assay was validated using the Snu17 mutant demonstrating that Snu17 was able to alter 5′ splice-site selection in the artificial model system. This observation is consistent with earlier analyses that had shown that Snu17 was required to allow the Mer1-dependent regulation of splicing of the AMA1, MER2 and MER3 transcripts during meiosis (37).
In several cases, human homologues of yeast factors endowed with the ability to control splice-site selection of artificial reporters were shown to be bona fide splicing regulators of alternative splicing in mammalian cells (38–40). It is thus tempting to suggest that human Snu17, and more generally the human RES complex, is a good candidate to be an alternative splicing regulator. Moreover, using this assay, we were able to demonstrate that the absence of Pml1 had a strong effect on splicing, but that mutation of the pThr-binding site of its FHA domain did not affect splice-site choice. These results infer that the pThr-binding pocket of Pml1, which corresponds to one of the most conserved regions of this protein, is probably involved in another function, supporting the multifunctional nature of this assembly (2). However, we cannot definitively exclude the possibility that it was insufficiently perturbed by the mutations tested here to affect its binding. The pThr-binding site of Pml1 could be involved in the control of pre-mRNA leakage, as this is one of the other functions ascribed to the RES complex (2) or could, for example, control the intracellular localization of the RES complex. Unfortunately, given that the corresponding phenotype is weak, we were unable to obtain a definitive validation of the former hypothesis in strains where Pml1 mutants are expressed from plasmid copies.
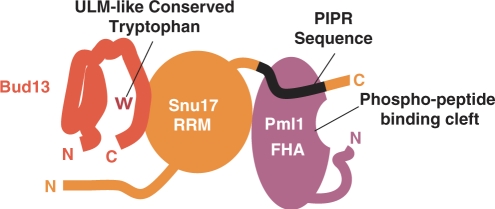
Dissection of the RES complex indicates that it is organized around a central Snu17 subunit, which binds independently to Bud13 and Pml1 (Figure 8). We have not obtained precise information with respect to the site of interaction of Snu17 on Pml1, except that the first 24 residues of this factor that are disordered are not required for binding, since protein co-expression has showed that this sequence is dispensable both for binary complex formation with Snu17 and for formation of the ternary RES complex (data not shown). Snu17 will therefore probably interact with the FHA domain of Pml1, since this domain covers most of the remainder of its sequence. The absence of quantitative phosphorylation of Snu17 produced in yeast (data not shown), and the strong interaction observed between recombinant factors produced in the absence of a native kinase in E. coli, indicate that the Snu17–Pml1 interaction is unlikely to involve the pThr-binding pocket of Pml1. This conclusion is further supported by the observation that elimination of the Pml1 subunit from the RES complex results in a splicing defect that is not detected for mutants of its pThr-binding pocket. Rather, the location of other Pml1 surface residues that are conserved may indicate a plausible location of the Snu17-binding site. Indeed, analysis of the Pml1 X-ray structure reveals that a conserved stripe of residues along Gln139, Leu50, Ser102 and Glu54 is present on one face of the molecule. Further experimentation will be required to test whether this represents residues recognized by Snu17. Insertion mutagenesis identified a sequence element located downstream of the RRM domain of Snu17 as a major determinant for interaction with Pml1 (Figure 8). This element, PIPR, appears to be necessary for a strong interaction, although residual association of Pml1 and Snu17 was still detected in its absence, which indicates that at least one other contact point is likely to exist. BLAST searches (41) failed to identify peptides similar to PIPR in proteins other than Snu17 homologues, suggesting that this motif does not participate in interactions other than between Snu17 or Pml1 and their homologues.
Figure 8.
Schematic model of the RES complex. We propose a model of the RES complex. In the model, Snu17 (orange) is the core subunit with an RRM-domain structure. Bud13 (red) interacts with Snu17 via its UHM Ligand Motif (ULM)-like sequence within its C-terminus, potentially through a conserved tryptophan. The binding of Pml1 (purple) to Snu17 is mediated primarily by a sequence within the Snu17 C-terminal tail, termed the Pml1 Interaction Promoting Region (PIPR).
Concerning the Bud13–Snu17 interaction, sequence comparison and deletion analysis indicated that the conserved C-terminal region of Bud13 is necessary and sufficient for interaction with Snu17. In contrast, we have not been able to identify any Snu17 mutation that specifically affected its interaction with Bud13. All mutations and truncations of the N- and C- terminal extensions which flank the Snu17 RRM that we identified did not perturb the interaction with Bud13. This suggests that the Snu17 RRM domain may be important to mediate this interaction (Figure 8). This possibility is further supported by the sequence similarity between Bud13 and ULM motifs, which suggests that the mode of interaction between Bud13–Snu17 could share some similarity with the one observed between SF1 and the third RRM of U2AF65, or between the fifth ULM of SF3b155 and the UHM of SPF45 (35,42). However, mutations that have been shown to disrupt the SF1–U2AF65 interaction (35) failed to abrogate the Snu17-Bud13 interaction (Supplementary Figure 3). It is likely, therefore, that such mutant only weakened the Snu17–Bud13 binding, and that this association differs in some manner from typical UHM–ULM interaction. RRM domains possess two short characteristic sequences, the RNP1 and RNP2 motifs, and it has been shown that their sequences can be used to determine whether they are of the canonical or UHM-type (36). By these criteria Snu17 should not be classified as a UHM, since the sequence which corresponds to the RNP1 motif (QGFAYLKY, residues 72–78), contains aromatic residues (Phe73, Tyr75 and Tyr78) at all three positions which are characteristic to the consensus RRM domain sequence (+GΘΩΘΨXΘ where + corresponds to basic amino acids, X any residues, Θ aromatic residues, Ψ aliphatic residues and Ω to small amino acids) rather than amino acids proposed to be characteristic of UHM (XGXΨΘΨXΘ) (36) (Figure 6C). As Snu17 consists primarily of an RRM domain, it is possible that it contacts RNA specifically. However, in common with RRM domains of the UHM type, Snu17 has an acidic isoelectric point, which should be suboptimal for RNA binding. However, a reliable categorization of the nature of the RRM domain of Snu17 and its interaction with Bud13 will have to await the solution of the corresponding atomic structure.
Altogether, our data indicate that Snu17 occupies a central position and interacts on the one hand with Pml1 and on the other hand with Bud13 (Figure 8). In the light of the phenotypes elicited by the deletion mutants presented here, it is tempting to speculate that Snu17 would contact target RNAs, Bud13 would interacts with the splicing machinery, while Pml1 would affect primarily nuclear mRNA retention. However, our data indicate that the various functions of the RES complex are not entirely separable, suggesting that the presence of Bud13 or Pml1 would be essential to maintain full complex activity. The structural model proposed here for the RES complex will facilitate future study of its function in various RNA-based processes. In addition, our analyses have demonstrated that the demanding task of deciphering the structural architecture of complexes can be solved, at least in part, by combining crystallographic studies with co-expression of protein in E. coli to reconstitute the complex and various mutagenesis strategies to gain an initial understanding of the interaction network occurring between the subunits. Given the large number of complexes identified in the recent years thanks to the development of by high-throughput affinity purification methods (43–46), our strategy may prove to be a useful tool for the analysis of the organization, structure and function of other cellular assemblies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported in part by La Ligue contre le Cancer ‘Equipe Labellisée 2008’ to B.S., the CNRS, the European Union 6th Framework programs ‘3D-Repertoire’ (LSHG-CT-2005-512028) to H.v.T. and B.S., and ‘EURASNET’ (LSHG-CT-2005-518238) to B.S. Funding for open access charge: European Union.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Christophe Marchand and Pierre le Marechal are thanked for access to the mass spectrometry service at Université Paris Sud. Data collection was performed at the ESRF beamline ID14-4, during which Dr Raimond Ravelli provided excellent support. We also acknowledge Michael Sattler and Anders Friberg as well as members of our laboratories for stimulating discussions.
REFERENCES
- 1.Matlin AJ, Moore MJ. Spliceosome assembly and composition. Adv. Exp. Med. Biol. 2007;623:14–35. doi: 10.1007/978-0-387-77374-2_2. [DOI] [PubMed] [Google Scholar]
- 2.Dziembowski A, Ventura AP, Rutz B, Caspary F, Faux C, Halgand F, Laprévote O, Séraphin B. Proteomic analysis identifies a new complex required for nuclear pre-mRNA retention and splicing. EMBO. J. 2004;23:4847–4856. doi: 10.1038/sj.emboj.7600482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gottschalk AB, Neubauer CG, Lührmann R, Fabrizio P. A novel yeast U2 snRNP protein, Snu17p, is required for the first catalytic step of splicing and for progression of spliceosome assembly. Mol. Cell Biol. 2001;21:3037–3046. doi: 10.1128/MCB.21.9.3037-3046.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Q, Rymond BC. Rds3p is required for stable U2 snRNP recruitment to the splicing apparatus. Mol. Cell Biol. 2003;23:7339–7349. doi: 10.1128/MCB.23.20.7339-7349.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohi MD, Gould KL. Characterization of interactions among the Cef1p-Prp19p-associated splicing complex. RNA. 2002;8:798–815. doi: 10.1017/s1355838202025050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stevens SW, Ryan DE, Ge HY, Moore RE, Young MK, Lee TD, Abelson J. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol. Cell. 2002;9:31–44. doi: 10.1016/s1097-2765(02)00436-7. [DOI] [PubMed] [Google Scholar]
- 7.Deckert J, Hartmuth K, Boehringer D, Behzadnia N, Will CL, Kastner B, Stark H, Urlaub H, Lührmann R. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell Biol. 2006;26:5528–5543. doi: 10.1128/MCB.00582-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bessonov S, Anokhina M, Will CL, Urlaub H, Lührmann R. Isolation of an active step I spliceosome and composition of its RNP core. Nature. 2008;452:846–850. doi: 10.1038/nature06842. [DOI] [PubMed] [Google Scholar]
- 9.Durocher D, Henckel J, Fersht AR, Jackson SP. The FHA domain is a modular phosphopeptide recognition motif. Mol. Cell. 1999;4:387–394. doi: 10.1016/s1097-2765(00)80340-8. [DOI] [PubMed] [Google Scholar]
- 10.Durocher D, Taylor IA, Sarbassova D, Haire LF, Westcott SL, Jackson SP, Smerdon SJ, Yaffe MB. The molecular basis of FHA domain: phosphopeptide binding specificity and implications for phospho-dependent signaling mechanisms. Mol. Cell. 2000;6:1169–1182. doi: 10.1016/s1097-2765(00)00114-3. [DOI] [PubMed] [Google Scholar]
- 11.Yuan C, Yongkiettrakul S, Byeon IJ, Zhou S, Tsai MD. Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53. J. Mol. Biol. 2001;314:563–575. doi: 10.1006/jmbi.2001.5140. [DOI] [PubMed] [Google Scholar]
- 12.Doublié S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 1997;276:523–530. [PubMed] [Google Scholar]
- 13.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Exp. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 14.Mazza C, Segref A, Mattaj IW, Cusack S. Large-scale induced fit recognition of an m(7)GpppG cap analogue by the human nuclear cap-binding complex. EMBO J. 2002;21:5548–5557. doi: 10.1093/emboj/cdf538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clauser KR, Baker P, Burlingame AL. Role of accurate mass measurement (+/− 10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal. Chem. 1999;71:2871–2882. doi: 10.1021/ac9810516. [DOI] [PubMed] [Google Scholar]
- 16.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 1993;26:795–800. [Google Scholar]
- 17.Collaborative Computational Project N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 18.Terwilliger TCBJ. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 1999;55(Pt 4):849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Terwilliger TC. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr. D Biol. Crystallogr. 2003;59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Cryst. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 21.Pannu NS, Murshudov GN, Dodson EJ, Read RJ. Incorporation of prior phase information strengthens maximum-likelihood structure refinement. Acta Cryst. D. 1998;54:1285–1294. doi: 10.1107/s0907444998004119. [DOI] [PubMed] [Google Scholar]
- 22.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 23.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 24.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 25.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howlin B, Butler SA, Moss DS, Harris GW, Driessen H.PC. TLSANL: TLS parameter-analysis program for segmented anisotropic refinement of macromolecular structures. J. Appl. Crystallogr. 1993;26:622–624. [Google Scholar]
- 27.Séraphin B, Kandels-Lewis S. 3′ splice site recognition in S. cerevisiae does not require base pairing with U1 snRNA. Cell. 1993;73:803–812. doi: 10.1016/0092-8674(93)90258-r. [DOI] [PubMed] [Google Scholar]
- 28.Sheldrick GM. SHELX: Applications to Macromolecules. Dordrecht: Kluwer Academic; 1998. [Google Scholar]
- 29.Byeon IJ, Li H, Song H, Gronenborn AM, Tsai MD. Sequential phosphorylation and multisite interactions characterize specific target recognition by the FHA domain of Ki67. Nat. Struct. Mol. Biol. 2005;12:987–993. doi: 10.1038/nsmb1008. [DOI] [PubMed] [Google Scholar]
- 30.Wang P, Byeon IJ, Liao H, Beebe KD, Yongkiettrakul S, Pei D, Tsai MD. II. Structure and specificity of the interaction between the FHA2 domain of Rad53 and phosphotyrosyl peptides. J. Mol. Biol. 2000;302:927–940. doi: 10.1006/jmbi.2000.4095. [DOI] [PubMed] [Google Scholar]
- 31.Holm LSC. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 32.Li H, Byeon I.JL, Ju Y, Tsai MD. Structure of human Ki67 FHA domain and its binding to a phosphoprotein fragment from hNIFK reveal unique recognition sites and new views to the structural basis of FHA domain functions. J. Mol. Biol. 2004;335:371–381. doi: 10.1016/j.jmb.2003.10.032. [DOI] [PubMed] [Google Scholar]
- 33.Liao H, Byeon IJ, Tsai MD. Structure and function of a new phosphopeptide-binding domain containing the FHA2 of Rad53. J. Mol. Biol. 1999;294:1041–1049. doi: 10.1006/jmbi.1999.3313. [DOI] [PubMed] [Google Scholar]
- 34.Deprez C, Lloubes R, Gavioli M, Marion D, Guerlesquin F, Blanchard L. Solution structure of the E.coli TolA C-terminal domain reveals conformational changes upon binding to the phage g3p N-terminal domain. J. Mol. Biol. 2005;346:1047–1057. doi: 10.1016/j.jmb.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 35.Selenko P, Gregorovic G, Sprangers R, Stier G, Rhani Z, Krämer A, Sattler M. Structural basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP. Mol. Cell. 2003;11:965–976. doi: 10.1016/s1097-2765(03)00115-1. [DOI] [PubMed] [Google Scholar]
- 36.Kielkopf CL, Lucke S, Green MR. U2AF homology motifs: protein recognition in the RRM world. Genes Dev. 2004;18:1513–1526. doi: 10.1101/gad.1206204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spingola M, Armisen J, Ares MJr. Mer1p is a modular splicing factor whose function depends on the conserved U2 snRNP protein Snu17p. Nucleic Acids Res. 2004;32:1242–1250. doi: 10.1093/nar/gkh281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puig O, Gottschalk A, Fabrizio P, Séraphin B. Interaction of the U1 snRNP with nonconserved intronic sequences affects 5' splice site selection. Genes Dev. 1999;13:569–580. doi: 10.1101/gad.13.5.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Forch P, Puig O, Kedersha N, Martinez C, Granneman S, Séraphin B, Anderson P, Valcarcel J. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol. Cell. 2000;6:1089–1098. doi: 10.1016/s1097-2765(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 40.Puig O, Bragado-Nilsson E, Koski T, Séraphin B. The U1 snRNP-associated factor Luc7p affects 5' splice site selection in yeast and human. Nucleic Acids Res. 2007;35:5874–5885. doi: 10.1093/nar/gkm505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Corsini L, Bonnal S, Basquin J, Hothorn M, Scheffzek K, Valcarcel J, Sattler M. U2AF-homology motif interactions are required for alternative splicing regulation by SPF45. Nat. Struct. Mol. Biol. 2007;14:620–629. doi: 10.1038/nsmb1260. [DOI] [PubMed] [Google Scholar]
- 43.Gavin A-C, Bösche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon A.-M, Cruciat C.-M, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 44.Gavin A-C, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dümpelfeld B, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 45.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Séraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 46.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 47.Kraulis PJ. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 48.Esnouf RM. An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph Model. 1997:15. doi: 10.1016/S1093-3263(97)00021-1. 132–134, 112–133. [DOI] [PubMed] [Google Scholar]
- 49.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Delano WL. The PyMOL Molecular Graphics System. San Carlos, CA: Delano Scientific; 2002. [Google Scholar]
- 51.Teem JL, Rosbash M. Expression of a beta-galactosidase gene containing the ribosomal protein 51 intron is sensitive to the rna2 mutation of yeast. Proc. Natl Acad. Sci. USA. 1983;80:4403–4407. doi: 10.1073/pnas.80.14.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.