Abstract
Genome sequencing projects have been initiated for a wide range of eukaryotes. A few projects have reached completion, but most exist as draft assemblies. As one of the main reasons to sequence a genome is to obtain its catalog of genes, an important question is how complete or completable the catalog is in unfinished genomes. To answer this question, we have identified a set of core eukaryotic genes (CEGs), that are extremely highly conserved and which we believe are present in low copy numbers in higher eukaryotes. From an analysis of a phylogenetically diverse set of eukaryotic genome assemblies, we found that the proportion of CEGs mapped in draft genomes provides a useful metric for describing the gene space, and complements the commonly used N50 length and x-fold coverage values.
INTRODUCTION
It is just over a decade since the first genome sequence of a free-living organism (the bacterium Haemophilus influenzae) was published (1). Since then, the field of genome sequencing has expanded dramatically as reflected in the Genomes OnLine Database (2) which lists almost 100 ‘complete published’ eukaryotic genome sequences in addition to over 1000 ‘ongoing’ genome projects. Early genome sequencing projects used the ‘hierarchical’ or ‘clone-by-clone’ sequencing approach and the first three eukaryotic genomes that were sequenced in this way were Saccharomyces cerevisiae, Caenorhabditis elegans and Arabidopsis thaliana (3–5). Hierarchical sequencing is labor-intensive but produces very high-quality sequence.
Most modern genome projects employ the whole genome shotgun (WGS) sequencing strategy. The Drosophila genome project (6) represented the first attempt at using the WGS method to sequence a moderately large genome sequence (∼130 Mb). WGS genomes are usually described by two statistics: sequence coverage, and N50 length. Sequence coverage is calculated as the ratio of the total amount of sequence produced, divided by the estimated genome size. Such estimates have a degree of uncertainty and can change as genome projects near completion. For example, the initial publication of the Takifugu rubripes genome used an estimated genome size of 380 Mb to calculate coverage as 5.6 × (7). Subsequent assemblies have revised the genome estimate upwards to ∼400 Mb which reduces the coverage of the published assembly slightly to 5.3×. A contrasting example comes from the initial genome assembly of the nematode Trichinella spiralis (ftp://genome.wustl.edu/pub/organism). The release notes for this assembly reveal that the estimated genome size was 270 Mb, but based on the results from sequencing, the genome is now believed to be only ∼65 Mb.
N50 length is calculated by first ordering all contig (or scaffold) sizes and then adding the lengths (starting from the longest contig) until the summed length exceeds 50% of the total length of all contigs. This measure is preferred over measures of ‘average contig size’ due to the high frequency of very short contigs in most genome assemblies, and has been used to compare the quality of different genome assemblies. For example, improvements to the ARACHNE assembler algorithm (8) dramatically improved the assembly of the dog genome sequence when compared to the previous version; specifically the N50 contig size increased 3-fold from 61 kb to 180 kb (9).
One of the most important reasons for sequencing a genome is to determine its catalog of genes. Although sequence coverage and N50 length are useful for describing the base pairs of a genome project, they do not describe the state of the gene space. That is, they do not address whether or not one can identify genes in the sequence. In this article, we report on a novel method for assessing the gene space that utilizes the CEGMA mapping protocol (10) to map a set highly conserved eukaryotic genes that are present in higher eukaryotes. We refine the original set of 458 genes to a subset of 248 that are generally present in low copy number and show that the proportion of these genes that can be mapped in a genome assembly provides a rough approximation for the proportion of all known genes that may be present.
MATERIALS AND METHODS
CEGMA modifications
This article extends our previously described method for obtaining and mapping a set of core genes (10) and includes important changes to address two key issues. First, we removed genes from the set of core genes that are highly paralogous as this reduces our false positive rate when trying to identify the true ortholog of a core gene. Second, in addition to finding full-length orthologs of core genes, we also wanted to provide the ability to find fragments of core genes.
We have previously extracted data from the eukaryotic orthologous groups (KOGs) database (11) for six model organisms: Homo sapiens, Drosophila melanogaster, A. thaliana, C. elegans, S. cerevisiae and Schizosaccharomyces pombe. Although the database comprises groups of proteins (referred to as KOGs) that have varying degrees of conservation amongst the different species, we only considered those KOGs that contained proteins from all six species. From the resultant set of 1788 KOGs, global multiple sequence alignments of all proteins in each KOG were generated. When a KOG contained multiple proteins from the same species, only the one most similar to the global alignment was retained and the alignment was then rebuilt with the remaining sequences. The alignments of each KOG were then assessed and those that contained large insertions and/or divergent proteins were discarded. This last filtering step removed the majority of the KOGs and defined a set of 458 core eukaryotic genes (CEGs) for which we have identifiable orthologs in all of the six species. For this article we have further refined this initial set to reduce the number of genes that may have paralogs. To address the issue of paralogy, we excluded any KOG that contained multiple proteins from three or more species. This produces a set of genes that are single-copy in most species. This additional step removes nearly half of the original set, leaving a final set of 248 CEGs. More restrictive filtering (e.g. multicopy in two or more species) produces a data set that is too small to be of practical use. We do not find any significant similarity among any the 248 proteins (data not shown).
For any new genome sequence our procedure uses a combination of bioinformatics tools to first identify candidate regions that may contain orthologs of the CEGs, and then to make gene predictions of the likely orthologous gene structure. For each protein in the set of 248 CEGs, TBLASTN (blast.wustl.edu) is used to identify matching regions in the new genome sequence and the top five proteins from six species are chosen. The HMMER software package (12) is then used to create a profile hidden Markov model for each CEG (using each multiple sequence alignment of six proteins). Each profile is then processed by GeneWise (13) to produce a gene prediction for each of the regions identified by TBLASTN. These gene predictions are then enhanced by the geneid program (14), which utilizes the exons predicted by GeneWise and integrates them into a suitable complete gene structure (from translational start site to stop codon). Finally, the putative proteins encoded by each geneid prediction are compared to the HMMER profile for each CEG. Only geneid predictions that match above a threshold value are retained (see below). This last filtering step means that potential orthologs will not always be found for all of the CEGs. It also means that as many as five homologs may be identified for each CEG (one for each TBLASTN region); in these scenarios the highest scoring match to the HMMER profile is designated as the most likely ortholog and other matches above the threshold are considered potential paralogs. We record the proportion of mapped CEGs that have at least one potential paralog and define this as the ‘Paralogy index’.
To be sure that we are finding only true homologs of each core gene, we only consider those gene predictions that produce a high enough score when aligned to the HMMER profile of each CEG. In this study, we calculate the threshold in a different way than before (10). We now calculate the threshold by aligning all ‘non-core’ genes to the profile and noting the maximum score. Non-core genes are taken from the latest annotations of all protein-coding genes from the original six species with the set of 248 CEGs removed. Thus, to be considered an ortholog of a core gene, a gene prediction must align to the profile and produce a score that exceeds any that can be produced from the alignment of any non-core gene.
Matches that exceed the threshold usually correspond to full-length proteins, though it is sometimes possible for shorter fragments of a protein to still score higher than the threshold. For example, this can occur when just the functional domains of a core gene are present in a genome assembly. Because the profiles were built using the default hmmbuild parameters, alignments can be global with respect to the HMM and local with respect to the sequence. This means that the fraction of predicted protein that aligns to the HMMER profile varies from 20% to 100%. To avoid predicting short genes, we required that the proportion of the predicted protein that aligns to the profile is at least 70%. Changing or removing this length requirement can allow the mapping protocol to predict either more fragmentary proteins, or fewer but more complete proteins. All results in this article use the 70% length cut-off.
The set of 248 CEGs were divided into four subsets based on their degree of protein sequence conservation. Using BLASTP (blast.wustl.edu), we produced pairwise alignments for all combinations of the six proteins within each CEG. Then, we assigned each CEG to one of four conservation groups based on the average degree of conservation observed in the pairwise alignments from each CEG. Group 1 contains the least conserved of the CEGs and Group 4 the most conserved (Supplementary Table S4). As the different conservation groups have different average lengths (more conserved proteins are shorter) we use the partial hits to count for the presence of the conservation groups to avoid bias related with the mapping protocol.
Reassembling Caenorhabditis briggsae
The published genome of C. briggsae is a 12× WGS assembly (15) that was produced using the Phusion assembler (16). We used the original sequencing reads from this assembly and randomly sampled them to produce new assemblies at defined levels of sequence coverage (2×, 4×, 6×, 8×, and 10×). The 2× assembly derives from 400 000 sequence reads and each subsequent assembly adds another 400 000 reads. The current version of Phusion was used to produce both contigs and scaffolds for each assembly. In addition to mapping CEGs to each of these assemblies, we also determined (using BLAST) how many of the annotated set of 19 256 C. briggsae proteins from the published genome were present.
Generating simulated draft human genomes
We generated six simulated draft human genome assemblies by using the distribution of known contig sizes from the WGS assemblies of guinea pig (1.9× sequence coverage), cow (3×, 6× and 7.1×), chimpanzee (4.2× and 6.6×) and rhesus macaque (5.3×). Estimates of genome size for these species—as measured by the C-value—are all in a narrow range between 3.43 and 3.59 pg of DNA (17). For each simulated draft we iterated through the list of contig sizes and extracted an equal length of sequence from the published human genome sequence. In doing so we effectively sampled random sites from across the genome and ensured that all extracted sequences were not overlapping. We then used our mapping protocol to map orthologs of CEGs against these assemblies.
Analysis of H. sapiens, C. briggsae and Toxoplasma gondii annotations in assemblies
To determine whether a gene from a set of gene annotations was present in any given genome assembly, we required that 65% of the length of a CDS was present in either a contig or scaffold. For C. briggsae and T. gondii we determined the overlap using BLAST, for H. sapiens we used the coordinates of genes in the final (full) assembly and cross-referenced them against our simulated draft genomes to see whether the same sequence region was present. The choice of a 65% cut-off is a trade-off that attempts to mostly only detect full-length (or nearly full-length) annotations, while allowing for the fact that parts of an annotation may be missing in a low-coverage assembly. This is more likely in vertebrate genomes where terminal exons of some longer gene annotations may be missing from shorter contigs. In these situations, using a cut-off value that is too high would mean that we would not count a gene annotation as present, even if we were in fact detecting all of the available sequence from that annotation.
Comparing predicted proteins from other annotation pipelines
As we have described in the CEGMA modifications section, after obtaining the complete predicted gene structures we compare them against the HMMER profile for each CEG. If we already have a set of gene annotations from any other source (e.g. Ensembl), we can also analyze this set of proteins to see which ones match (or do not match) the HMMER profiles derived from the 248 CEGs. We took the available annotations for all the analyzed genomes and compared them against the HMMER profiles using the same protocol as described in the CEGMA modifications section.
CEGs chicken analysis
We used the TBLASTN algorithm to compare a published set of chicken ESTs (18) with the human proteins of the 36 missing CEGs. We find 29 ESTs with at least one HSP with an expected value below 10e–6. From the selected ESTs we tried to map them against the chicken genome sequence using BLASTN. To consider a significant hit it must have at least one HSP with 95% identity over 50 bp.
Genome data
Genome sequences and genome assembly data were downloaded for the following eukaryotes: Anopheles gambiae, Apis melifera, A. thaliana, Bos taurus, Canis familiaris, Cavia porcellus, C. brenneri, C. briggsae, C. elegans, C. remanei, Chlamydomonas reinhartdii, Ciona intestinalis, D. melanogaster, Felis catus, Gallus gallus, Giardia lamblia, H. sapiens, Loxodonta africana, Macaca mulatta, Magnoporthe grisea, Neurospora crassa, Ornithorynchus anatinus, Pan troglodytes, Plasmodium falciparum, Populus trichocarpa, S. cerevisiae, S. pombe, T. rubripes, T. gondii, T. spiralis and Xenopus tropicalis (full details of source data and download sites are listed in Supplementary Table S6).
RESULTS
Identifying proteins for examining gene space
Our strategy for examining gene space is to determine how well one can map complete proteins in unfinished genomes. Ideal proteins would be easily identifiable, present in all eukaryotes, and single copy. We had previously developed the CEGMA mapping protocol (10) using data from the KOGs database (11). Utilizing complete protein catalogs from six model organisms (H. sapiens, D. melanogaster, C. elegans, A. thaliana, S. cerevisiae and S. pombe), we produced a set of 458 CEGs whose entire coding sequence can be reliably mapped in higher eukaryotes (by which we mean plants, animals and fungi). To find genes that tend to be single copy, we selected those genes that are present as a single copy in the KOGs database in at least four of the six species. Each KOG cluster contains one or more genes that are reciprocal best matches among these genomes. Some clusters contain genes that tend to duplicate while other clusters tend to exist in single copies. For example, KOG0157 (Cytochrome P450 CYP4/CYP19/CYP26 subfamilies), contains 87 genes in A. thaliana, 14 in D. melanogaster, 33 in H. sapiens, 24 in C. elegans, 2 in S. cerevisiae and 1 in S. pombe. However, KOG0261 (RNA polymerase III, large subunit) has only one orthologous protein in each species. After enriching for single-copy genes, the dataset was reduced to 248 proteins. Approximately 90% of these genes are single-copy in D. melanogaster, C. elegans, S. cerevisiae and S. pombe. In H. sapiens and A. thaliana, multi-gene families were significantly reduced and 50% are single-copy genes in the set of 248 CEGs (Table 1).
Table 1.
Reducing the number of orthologs in the original set of 458 CEGs
| 458 CEGs |
248 CEGs |
|||||
|---|---|---|---|---|---|---|
| Average number of orthologs per CEG | Percentage CEGs with more than one ortholog | Percentage CEGs with more than two orthologs | Average number of orthologs per CEG | Percentage CEGs with more than one ortholog | Percentage CEGs with more than two orthologs | |
| Arabidopsis thaliana | 2.49 ± 1.89 | 65.7 | 34.7 | 2.04 ± 1.47 | 52.4 | 21.3 |
| Caenorhabditis elegans | 1.34 ± 0.80 | 22.4 | 6.7 | 1.17 ± 0.55 | 11.1 | 2.3 |
| Drosophila melanogaster | 1.32 ± 0.69 | 22.9 | 6.5 | 1.16 ± 0.45 | 12.7 | 2.8 |
| Homo sapiens | 2.84 ± 2.67 | 62.4 | 37.3 | 2.13 ± 1.73 | 49.6 | 23.2 |
| Saccharomyces cerevisiae | 1.31 ± 0.65 | 23.8 | 4.8 | 1.10 ± 0.35 | 8.8 | 0.8 |
| Schizosaccharomyces pombe | 1.20 ± 0.49 | 17.2 | 3.2 | 1.11 ± 0.39 | 8.8 | 2.0 |
For the sets of 458 and 258 CEGs, the average number of orthologs per CEG, and percentages of CEGs with more than one and two orthologs are listed. SDs are shown for the average number of orthologs per CEG.
The CEGMA mapping protocol was run using the reduced set of 248 CEGs against the six model organism genomes from which the set of core genes were originally identified, and as expected, CEGMA correctly identified all of the CEGs in these species (data not shown). The sole exception to this was the failure to predict one core gene in C. elegans. On inspection, we found that the gene in question (WormBase gene ID: WBGene00007698) is interrupted by the presence of another gene in one of its introns. CEGMA correctly predicts the first half of the gene upstream of the gene-containing-intron but not the rest of the gene. This suggests that the CEGMA may have problems with other genes whose structure is interrupted by large insertions of other genes. Non-canonical splice sites, selenocysteine codons, regulated frame-shifting, and other rare features may also confound CEGMA, but since most core genes do not exhibit these features, their effect should be minimal.
Mapping core genes in genomes of varying coverage
To determine the properties of the gene space in genome assemblies with varying levels of sequence coverage, we mapped the 248 core genes against multiple assemblies of C. briggsae, T. gondii and H. sapiens. For C. briggsae we randomly sampled reads at 2×, 4×, 6×, 8× and 10× coverage from the original 12× project (15) and re-assembled them with an updated version of the Phusion assembler. For T. gondii, we used the six assemblies (0.7×, 1×, 2×, 4×, 6× and 10×) that were available online (the latter two assemblies are scaffold-based, the rest are contig-based). Finally, for H. sapiens, we created simulated draft genomes based on the known distribution of contig and scaffold sizes for several non-human WGS assemblies (see Methods section). These draft genomes are not true assemblies because they consist of sampled finished sequence rather than contigs built up from shotgun sequencing reads. As a result, the simulated human genomes do not mimic all the properties present in WGS assemblies. We find that the number of genes found by CEGMA in the simulated drafts is consistent with the original non-human genomes (Supplementary Table S1). Therefore, while the simulated draft human genomes may not faithfully mirror WGS contigs, the content of the gene-space is similar.
In general, as the sequence coverage increases, the N50 length of contigs and scaffolds also increases, as does the number of mapped CEGs (Table 2). An exception to this is the 12 × C. briggsae assembly, whose scaffold N50 length is shorter than the 8× and 10× reassemblies, and approximately equal to the 6× reassembly. We believe that this is because of improvements made to the Phusion assembler since the published 12× assembly was generated. The other exceptions are the 4.2× and 6.6× simulated human genomes, whose scaffold lengths are exceptionally long. This is because these simulated drafts were based on chimpanzee genome assemblies, which used a reference genome (human) to aid their construction (19).
Table 2.
Assembly statistics and results of mapping 248 CEGs in C. briggsae, H. sapiens and T. gondii
| Contigs |
Scaffolds |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Species | Assembly details | n | N50 length (Kb) | Total number of genes (%) | Number of mapped CEGs (%) | n | N50 length (Kb) | Total number of genes (%) | No. of mapped CEGs (%) |
| Caenorhabditis briggsae | 2× | 34 456 | 2.3 | 9912 (46.1) | 110 (44.3) | 10 297 | 14.2 | 11 006 (57.0) | 132 (53.2) |
| 108 Mb | 4× | 20 421 | 7.4 | 15 372 (79.7) | 200 (80.6) | 2268 | 16.4 | 17 738 (91.9) | 226 (91.1) |
| 19 296 genes | 6× | 11 399 | 16.4 | 17 470 (90.5) | 227 (91.5) | 1028 | 465 | 18 809 (97.4) | 238 (95.9) |
| 8× | 7 363 | 28.9 | 18 311 (94.8) | 231 (93.1) | 971 | 983 | 19 071 (98.8) | 241 (97.2) | |
| 10× | 5614 | 37.4 | 18 578 (96.3) | 243 (97.9) | 675 | 1032 | 19 106 (99.0) | 245 (98.8) | |
| CB25 12× | 5341 | 40.7 | 18 530 (96.0) | 239 (96.4) | 899 | 474 | 19 141 (99.1) | 244 (98.3) | |
| Toxoplasma gondii | 0.7× | 39 143 | 0.8 | 889 (11.4) | 10 (4.0) | – | – | – | – |
| 63 Mb | 1× | 45 663 | 1.1 | 1 499 (19,2) | 19 (7.6) | – | – | – | – |
| 7 793 genes | 2× | 36 333 | 2.8 | 3 813 (48.9) | 82 (33.0) | – | – | – | – |
| 4× | 10 594 | 13.9 | 6358 (81.5) | 163 (65.3) | – | – | – | – | |
| 6× | 4 198 | 95.7 | 7 557 (96.9) | 199 (80.2) | 586 | 1000 | 7745 (99.3) | 212 (85.6) | |
| 10× | 3 922 | 397 | 7 678 (98.3) | 207 (83.5) | 669 | 2474 | 7793 (100) | 213 (85.9) | |
| Homo sapiens | draft 1.9× | 590 603 | 3.1 | 4 963 (20.9) | 52 (21.0) | 130 283 | 51.9 | 7930 (33.4) | 105 (42.3) |
| 3 253 Mb | draft 3× | 795 203 | 4.1 | 7 414 (31.2) | 88 (35.5) | 449 727 | 13.5 | 10 189 (43.0) | 125 (50.4) |
| 23 713 genes | draft 4.2× | 435 593 | 13.1 | 12 006 (50.6) | 142 (57.2) | 81 459 | 2425 | 19 333 (81.5) | 225 (90.7) |
| draft 5.3× | 368 201 | 14.7 | 13 009 (54.8) | 148 (59.7) | 28 863 | 692 | 20 557 (86.7) | 228 (91.9) | |
| draft 6× | 296 517 | 19.1 | 12 739 (53.7) | 149 (60.1) | 62 471 | 436 | 18 769 (79.2) | 212 (85.4) | |
| draft 6.6× | 292 555 | 28.8 | 15 592 (65.7) | 179 (72.2) | 77 769 | 8217 | 20 978 (88.5) | 238 (95.9) | |
| draft 7.1× | 131 620 | 44.3 | 16 205 (68.3) | 198 (79.8) | 16 098 | 1042 | 19 895 (83.9) | 230 (92.7) | |
‘CB25’ refers to the 2002 published assembly of C. briggsae. ‘Total number of genes’ refers to the number of genes from the final (highest coverage) assembly for each species that are present in each lower coverage assembly (see Methods section). The total number of genes is listed beneath each species name, along with the estimated genome size. The ‘Mapped CEGs’ column lists numbers of the 248 CEGs that were mapped in the genome of each species. Results are shown for both contig- and scaffold-based assemblies. Figures in parentheses show values as percentages.
Do mapped CEGs faithfully represent all genes?
Eukaryotic genomes can contain many thousands of genes, though these genes might not all be present in the sequence of an incomplete genome assembly. To determine if the proportion of mapped CEGs corresponds to the proportion of all genes that can be found, we mapped the latest, complete gene catalogs of C. briggsae, H. sapiens and T. gondii against genome assemblies at various levels of sequence coverage. It is important to choose a suitable cut-off for determining whether a gene is present or not, i.e. what percentage of each gene annotation needs to be present in an individual contig or scaffold. Choosing a cut-off value that is too low makes it possible to find nearly all of the genes in most of the assemblies, because even in very low-coverage assemblies, most genes are present as fragments (Supplementary Figure S1). For instance, even in the low-coverage human 1.9× assembly, we can still find fragments of 18 528 of the 23 713 genes.
By using a cut-off of 65% (see Methods section) we find that there is a good overall correlation between the number of mapped CEGs and the total number of genes (Table 2), though there are interesting differences among the three species (Supplementary Figure S2). The results for C. briggsae show the closest agreement between the proportions of core genes and all genes that are mapped. The average discrepancy between the percentages of mapped CEGs and all genes is 0.96%. This suggests that the proportion of CEGs that can be mapped in the C. briggsae genome, regardless of the level of sequence coverage, is a good approximation for the proportion of all genes that should be present. For H. sapiens, the average discrepancy between the ability to map the two data sets is higher at 6.55%. The proportion of mapped CEGs in the human assemblies always provides an overestimation of the proportion of all genes that are present. This contrasts with T. gondii, where we appear to underestimate the proportion of all genes that are present.
The overestimation in H. sapiens is mainly caused by the high paralogy of human CEGs (32.3% of the 248 core genes had more than one orthologous protein). The other factor biasing the calculation was the length of the primary transcripts. Since the genes encoding CEGs tend to be slightly shorter than ‘normal’ genes (Supplementary Table S2), they are more likely than a normal gene to be contained completely within a short scaffold. For instance, while 19% of transcripts from all human genes are longer than 50 000 bp only 9% of human CEG transcripts exceed that length. Because of the large number of pseudogenes in the human genome, we also suspected that some pseudogenes may contribute to the overestimation, and in some of the lower-coverage simulated draft genomes a small number of pseudogenes are incorrectly classified as CEGs (Supplementary Table S3). However, in the full human genome sequence, no pseudogenes are classified as CEGs.
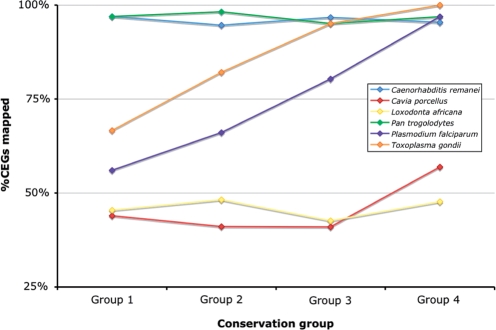
The underestimation of the number of real genes in the T. gondii assemblies is apparent at all levels of sequence coverage. Since T. gondii is an outgroup species, ∼1900 million years diverged from the higher eukaryotes used to build the CEGs (20), we sought to determine if evolutionary divergence was the source of error. We partitioned the set of 248 CEGs into four groups based on their overall degree of sequence conservation (Supplementary Table S4). Group 1 contains the most divergent CEG proteins, and Group 4 contains the most highly conserved. All of the Group 4 proteins could be mapped in the 10 × T. gondii genome, compared to about two thirds of the Group 1 proteins (Supplementary Table S5 and Figure 1). For highly diverged genomes, it is therefore necessary to use a smaller set of only the most highly conserved CEGs in order to evaluate the completeness of the gene space.
Investigating gene space in 25 species
Table 3 shows the results of mapping the 248 CEGs into 25 species from diverse phylogenetic groups (results from additional species are included in the supplemental spreadsheet file ‘other_genomes.xls’). In most genomes (18 of 25), we were able to map >90% of the full-length CEGs. Some of the genomes with the fewest mapped proteins were guinea pig (Cavia porcellus), elephant (L. africana) and domestic cat (F. catus) with 46.0, 46.0 and 58.1% of CEGs mapped, respectively. These values are slightly inflated due to high levels of paralogy and low sequence coverage in these species, but given that these genomes were sequenced at only ∼2× coverage, it is somewhat surprising that their gene space is represented as well as it is. Some of the missing CEGs are present as fragmentary matches, and this is more pronounced in low coverage genomes (the above figures for mapped CEGs in the guinea pig, elephant, and cat rise to 68.1%, 68.5% and 75.8% when partial matches are also included).
Table 3.
Results of mapping CEGs against the genomes of various eukaryotes
| Species | Genome size (Gb) | Coverage | Full-length mapped CEGs (%) | CEGs in annotations (%) | Full-length + partially mapped CEGs (%) | Paralogy index (%) | G1 Map (%) | G4 Map (%) | G1 Identity (%) | G4 Identity (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| Mammals (placental) | ||||||||||
| Canis familiaris | 2.532 | 7.5× | 243 (98.0) | 241 (97.2) | 247 (99.6) | 37.4 | 100 | 96.9 | 38.2 | 65.5 |
| Bos taurus | 3.247 | 7.1× | 244 (98.4) | 243 (98.0) | 246 (99.2) | 33.3 | 98.5 | 95.4 | 37.8 | 65.1 |
| Pan troglodytes | 3.350 | 6.6× | 240 (96.8) | 241 (97.2) | 247 (99.6) | 39.6 | 100 | 96.9 | 38.1 | 65.1 |
| Macaca mulatta | 3.097 | 5.3× | 238 (96.0) | 237 (95.5) | 248 (100) | 36.6 | 100 | 100 | 38.0 | 65.1 |
| Felis catus | 3.000 | 2× | 144 (58.1) | – | 188 (75.8) | 17.4 | 69.7 | 75.4 | 36.3 | 61.1 |
| Loxodonta africana | 3.718 | 2× | 114 (46.0) | – | 170 (68.5) | 15.8 | 65.2 | 64.6 | 34.3 | 59.0 |
| Cavia Porcellus | 3.414 | 1.9× | 114 (46.0) | – | 169 (68.1) | 17.5 | 65.2 | 67.7 | 33.7 | 58.7 |
| Vertebrates | ||||||||||
| Ornithorynchus anatinus | 2.073 | 6× | 185 (74.6) | 175 (70.6) | 210 (84.7) | 27.1 | 75.7 | 86.1 | 35.7 | 63.9 |
| Gallus gallus | 1.100 | 6.6× | 208 (83.9) | 204 (82.3) | 212 (85.4) | 13.0 | 83.1 | 87.7 | 38.0 | 64.6 |
| Xenopus tropicalis | 1.511 | 7.7× | 237 (95.6) | 217 (87.5) | 243 (98.0) | 24.6 | 98.5 | 96.9 | 38.7 | 65.0 |
| Takifugu rubripes | 0.393 | 8.7× | 243 (98.0) | 235 (94.7) | 248 (100) | 20.6 | 98.5 | 100 | 38.4 | 65.4 |
| Insects | ||||||||||
| Anopheles gambiae | 0.278 | 10.2× | 245 (98.8) | 243 (98.0) | 247 (99.6) | 9.4 | 100 | 98.4 | 37.6 | 66.1 |
| Apis mellifera | 0.231 | 7.5× | 228 (91.9) | 173 (69.7) | 243 (98.0) | 6.1 | 98.5 | 98.4 | 38.7 | 65.9 |
| Nematodes | ||||||||||
| Caenorhabditisbriggsae | 0.108 | 12× | 246 (99.2) | 242 (97.6) | 247 (99.6) | 8.1 | 100 | 98.4 | 35.0 | 62.9 |
| Caenorhabditis brenneri | 0.150 | 9.5× | 245 (98.8) | – | 248 (100) | 53.5 | 98.5 | 98.4 | 34.6 | 62.1 |
| Caenorhabditis remanei | 0.152 | 9× | 238 (96.0) | – | 245 (98.8) | 15.5 | 98.5 | 100 | 34.9 | 62.9 |
| Trichinella spiralis | 0.065 | >30× | 233 (94.0) | – | 238 (96.0) | 7.7 | 97.0 | 98.4 | 34.8 | 61.5 |
| Chordates | ||||||||||
| Ciona intestinalis | 0.173 | 11× | 239 (96.4) | 203 (81.8) | 243 (98.0) | 6.3 | 95.5 | 100 | 37.5 | 64.8 |
| Plants | ||||||||||
| Populus trichocarpa | 0.480 | 7.5× | 244 (98.4) | 246 (99.2) | 248 (99.6) | 71.3 | 100 | 100 | 35.0 | 62.1 |
| Oryza sativa | 0.430 | – | 244 (98.4) | 185 (74.6) | 246 (99.2) | 51.6 | 98.5 | 98.4 | 34.2 | 61.4 |
| Chlamydomonas reinhardtii | 0.120 | 12.8× | 231 (93.1) | 221 (89.1) | 233 (94.0) | 6.9 | 87.9 | 98.4 | 31.7 | 59.7 |
| Fungi | ||||||||||
| Neurospora crassa | 0.039 | >10× | 245 (98.8) | 236 (95.1) | 245 (98.8) | 3.7 | 97.0 | 100 | 33.3 | 58.8 |
| Magnoporthe grisea | 0.040 | 7× | 243 (97.9) | 237 (95.5) | 246 (99.6) | 4.1 | 98.5 | 98.4 | 33.0 | 59.3 |
| Protozoan | ||||||||||
| Plasmodium falciparum | 0.023 | – | 186 (75.0) | 204 (82.2) | 187 (75.4) | 4.3 | 56.1 | 96.9 | 25.6 | 52.4 |
| Giardia lamblia | 0.011 | 11× | 115 (46.4) | 135 (54.4) | 115 (46.4) | 3.4 | 18.2 | 67.7 | 26.7 | 44.7 |
Genome sizes are estimates from experimental data. Coverage refers to approximate values of sequence coverage for WGS genomes only. The ‘Full-length mapped CEGs’ column lists numbers and percentages (in parentheses) of the 248 CEGs that were mapped in the genome of each species. ‘CEGs in annotations’ refers to the number of CEGs found in the current set of gene annotations (when available) for each genome. The ‘Full-length + partially mapped CEGs’ column corresponds to the number of full-length CEGs that were mapped (column 4) plus the numbers of CEG fragments that were mapped. The ‘Paralogy index’ indicates the fraction of mapped CEGs for which we detected at least one potential paralog. G1 and G4 mapped percentage corresponds to the number of CEGs from the conservation groups (in Table 3) that have been partially mapped. G1 and G4 identity percent corresponds to the average percentage identity of the global pairwise alignment of the predicted CEGs against the CEGs of the six original species. The latest available versions of genomes were used for this analysis (see Supplementary Table S6 for more details) apart from C. intestinalis for which the v1.95 assembly was used. Genome sizes are estimates. Coverage refers to approximate values of sequence coverage for WGS genomes only.
In platypus (O. anatinus), we mapped only 74.6% of the CEGs despite the genome having 6× coverage. The N50 scaffold length of the genome is high (531 kb) but the average contig size is only 7232 bp with 28% of the genome sequence in fragments smaller than 20 000 bp. Since the average platypus transcript is 22 000 bp, many genes are not fully contained in a scaffold sequence and this leads to fragmentation of the gene space and a high number of partial predictions (10%).
The chicken (G. gallus) genome assembly is derived from 6.6× sequence coverage, yet we were only able to map 83.9% of the CEGs. The chicken sequencing consortium produced a comprehensive EST collection, so we mapped the ESTs against the 36 missing CEGs to determine if the CEGs were missing from the genome assembly or from the organism. Of the 36 missing genes, 29 have at least one matching EST, indicating that the missing genes are not missing from the organism. When the ESTs of these missing genes are aligned back to the genome, 45% could not be mapped to the genome sequence at all. Of the 55% of ESTs that did match the genome, about half matched unanchored sequences that are not integrated into the main genome assembly, and the remaining half had only partial matches to the main genome sequence (on average, matches occurred across just 40% of the length of the EST sequence).
Another species with a low fraction of mapped CEGs was T. spiralis. Because of errors in the initial estimate of genome size, the sequence coverage for this species may be as high as 30×. This is partially supported by contigs and scaffold sequences having much higher N50 sizes than the other nematodes in this study (data not shown). However, we still fail to map 15 CEGs in this species, although at least five of these missing genes are present as fragments. For seven cases we find the candidate locus but the coding sequence contains frame-shifts. Given that there are few paralogs and the genome is compact, it is unlikely that these are pseudogenes. Whether the frame-shifts are intrinsic properties of the genome, sequencing errors, or assembly artifacts is currently unknown.
Other species with lower than expected numbers of mapped CEGs include P. falciparum (75.0%), G. lamblia (46.4%), and to a lesser extent, Chlamydomonas reinhardtii (93.1%). These divergent genomes follow a similar pattern as T. gondii where highly conserved proteins are mapped more frequently than poorly conserved ones (Figure 1). Consequently, the fraction of mapped CEGs is an underestimate of the completeness of the gene space. Considering only the most highly conserved Group 4 proteins, the genomes of P. falciparum and C. reinhardtii appear mostly complete with 98.4% and 96.9% of the CEGs represented, but G. lamblia has only 67.7%. Since G. lamblia is an outgroup to the higher eukaryotes used to build the CEGs, the low fraction of mapped CEGs may not be an accurate reflection of the state of the gene space.
Figure 1.
Mapping results for six selected species in four subsets of core genes. Group 1 represents the least conserved of all CEGs and Group 4 the most conserved.
For the 19 species with available gene annotations, we find that that there is a good overlap between the proteins predicted by the CEGMA mapping protocol and those provided by the relevant genome consortia. However, in most cases (14 of 19) CEGMA predicts some core genes that are not present in the current annotations, and for a few species many CEGs appear missing from the annotations (e.g. CEGMA finds 228 CEGs in the honey bee genome, though only 173 appear in the available annotations). In a few cases (4 of 19) the consortium annotations include CEGs that are not mapped by the CEGMA protocol. This is more pronounced in the two protozoan species P. falciparum and G. lamblia and their evolutionary divergence would again be most likely to account for this.
Paralogy index
We define the ‘paralogy index’ as the proportion of mapped CEGs with paralogs. This figure partly depends on genome coverage since CEGMA has difficulties sorting orthologs from paralogs in incomplete genomes. The species with the highest paralogy indexes tend to be higher plants and vertebrates (Table 3). Plants are represented by rice (Oryza sativa) and the poplar tree (P. trichocarpa) and these have the highest (P. trichocarpa, 71.3%) and third-highest (O. sativa, 51.6%) proportion of paralogs in the 25 species studied. The paralogy index for most mammals is in the 30–40% range. Within the vertebrates, the species with the lowest fraction of paralogs (13.0%) is the chicken (G. gallus). This result is supported by data suggesting that the chicken genome has undergone extensive gene loss (21).
Among the Caenorhabditids, C. brenneri is an outlier. The C. elegans, C. briggsae and C. remanei genome assemblies roughly match their expected sizes (3,15), but the C. brenneri assembly is nearly 200 Mb though its expected size is 150 Mb (http://genome.wustl.edu). The paralogy index of C. brenneri is also much higher (53.5%) than the others (5.7%, 8.1% and 15.5%). One possible reason that C. brenneri is an outlier is that its genome assembly contains some heterozygosity, and this results in creating artificial paralogs and inflating the genome size.
DISCUSSION
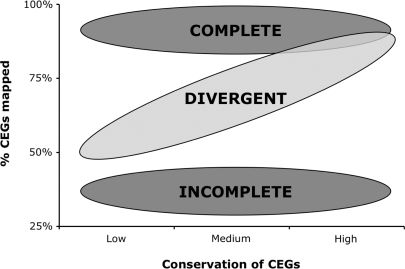
We have previously shown that CEGMA is a useful tool for predicting the orthologs of a set of core genes in newly sequenced genomes that may have little or no annotation (10). In this article, we show that with some minor adjustments to take paralogy and divergence into account, CEGMA can be used to assess the completeness of the gene space. The expected outcomes of mapping CEGs are summarized in Figure 2. In highly divergent genomes, we successfully map highly conserved proteins more frequently than poorly conserved ones, and an estimate of completeness should ideally be made by only using the most conserved (Group 4) CEGs.
Figure 2.
Summary of the three main patterns of results that can be expected when studying a new genome sequence. X-axis represents whether the mapping protocol uses subsets of CEGs that are the most or least conserved.
The gene spaces of platypus and chicken appear to be relatively incomplete given their sequence coverage. It may be useful to reassemble them with a different or updated genome assembler. Our 10× reassembly of C. briggsae is more complete (i.e. more CEGs mapped) than the original 12× assembly. Similarly, the dog genome was vastly improved with an updated assembler (9). The C. brenneri genome also appears relatively incomplete. Here, it may be that a genome assembler designed to deal with heterozygosity may improve the genome (23). Updates to a genome assembler do not always produce better sequences however. Comparing the latest (v2.0) 11× assembly of C. intestinalis to the the older v1.95 assembly (also 11×) sees a 10-fold increase in N50 length from 234 500 to 2 571 800 nt but the newer assembly contains fewer CEGs than the older one. Upon closer inspection, we found that several of the CEGs present in v1.95 were lost in v2.0, and for this reason we used v1.95 in this study. Core genes—by their very nature—are expected to be present in all complete genome sequences, though we found that most sets of gene annotations that accompany genome sequences have missed core genes that CEGMA detected.
WGS assemblies are complex entities. Assessing the completeness of a genome is not a simple task, and reliance on a single metric, such as N50 length, can be misleading. We believe that mapping highly conserved proteins provides a practical view of the gene space, and reflects on the utility of the genome assembly as a whole. The proteins we employed in this study are common to higher eukaryotes but any set of proteins that tends to be single copy and highly conserved in a particular clade could be used. It is difficult to reliably map divergent proteins, and for this reason, estimates of gene space completeness may not be very accurate in divergent genomes.
A description of the software used in this article along with the source code is available from (http://korflab.ucdavis.edu/Datasets/genome_completeness). The website will include analyses of additional genomes as they become available.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Human Genome Research Institute (HG004348 to I.K.). Funding for open access charge: National Institutes of Health (1R01HG004348).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are extremely grateful to the various sequencing centers that have freely provided the genome sequences and related data used in this study. Specifically we would like to acknowledge the following institutions: The Arabidopsis Information Resource (TAIR), Berkeley Drosophila Genome Project (BDGP), The Broad Institute, Celera Genomics, Ensembl, FlyBase, The Fugu Genome Consortium, Genoscope, Human Genome Sequencing Center at Baylor College of Medicine, The Institute of Genomic Research (TIGR), PlasmoDB, Saccharomyces Genome Database (SGD), ToxoDB, US Department of Energy Joint Genome Institute, Washington University Genome Sequencing Center, and The Wellcome Trust Sanger Institute. We would also like to thank John Spieth (WashU GSC) for useful discussions.
REFERENCES
- 1.Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 2.Liolios K, Tavernarakis N, Hugenholtz P, Kyrpides NC. The Genomes On Line Database (GOLD) v.2: a monitor of genome projects worldwide. Nucleic Acids Res. 2006;34:D332–D334. doi: 10.1093/nar/gkj145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Consortium CeS. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 4.Goffeau AR, Aert ML, Agostini-Carbone A, Ahmed M, Aigle L, Alberghina K, Albermann M, Albers M, Aldea D, Alexandraki G, et al. The Yeast Genome Directory. Nature. 1997;387(Suppl.):1–105. [Google Scholar]
- 5.Initiative TAG. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 6.Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 7.Aparicio S, Chapman J, Stupka E, Putnam N, Chia JM, Dehal P, Christoffels A, Rash S, Hoon S, Smit A, et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- 8.Batzoglou S, Jaffe DB, Stanley K, Butler J, Gnerre S, Mauceli E, Berger B, Mesirov JP, Lander ES. ARACHNE: a whole-genome shotgun assembler. Genome Res. 2002;12:177–189. doi: 10.1101/gr.208902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas E.J., III, Zody MC, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 10.Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 11.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 13.Birney E, Clamp M, Durbin R. GeneWise and genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parra G, Agarwal P, Abril JF, Wiehe T, Fickett JW, Guigo R. Comparative gene prediction in human and mouse. Genome Res. 2003;13:108–117. doi: 10.1101/gr.871403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stein LD, Bao Z, Blasiar D, Blumenthal T, Brent MR, Chen N, Chinwalla A, Clarke L, Clee C, Coghlan A, et al. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 2003;1:E45. doi: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mullikin JC, Ning Z. The phusion assembler. Genome Res. 2003;13:81–90. doi: 10.1101/gr.731003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gregory TR, Nicol JA, Tamm H, Kullman B, Kullman K, Leitch IJ, Murray BG, Kapraun DF, Greilhuber J, Bennett MD. Eukaryotic genome size databases. Nucleic Acids Res. 2007;35:D332–D338. doi: 10.1093/nar/gkl828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boardman PE, Sanz-Ezquerro J, Overton IM, Burt DW, Bosch E, Fong WT, Tickle C, Brown WR, Wilson SA, Hubbard SJ. A comprehensive collection of chicken cDNAs. Curr. Biol. 2002;12:1965–1969. doi: 10.1016/s0960-9822(02)01296-4. [DOI] [PubMed] [Google Scholar]
- 19.Consortium CS. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 20.Hedges SB, Blair JE, Venturi ML, Shoe JL. A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol. Biol. 2004;4:2. doi: 10.1186/1471-2148-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hillier LW, Miller W, Birney E, Warren W, Hardison RC, Ponting CP, Bork P, Burt DW, Groenen MA, Delany ME, et al. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 22.Vinson JP, Jaffe DB, O'Neill K, Karlsson EK, Stange-Thomann N, Anderson S, Mesirov JP, Satoh N, Satou Y, Nusbaum C, et al. Assembly of polymorphic genomes: algorithms and application to Ciona savignyi. Genome Res. 2005;15:1127–1135. doi: 10.1101/gr.3722605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.