Abstract
Eukaryotic RNase H2 is a heterotrimeric enzyme. Here, we show that the biochemical composition and stoichiometry of the human RNase H2 complex is consistent with the properties previously deduced from genetic studies. The catalytic subunit of eukaryotic RNase H2, RNASEH2A, is well conserved and similar to the monomeric prokaryotic RNase HII. In contrast, the RNASEH2B and RNASEH2C subunits from human and Saccharomyces cerevisiae share very little homology, although they both form soluble B/C complexes that may serve as a nucleation site for the addition of RNASEH2A to form an active RNase H2, or for interactions with other proteins to support different functions. The RNASEH2B subunit has a PIP-box and confers PCNA binding to human RNase H2. Unlike Escherichia coli RNase HII, eukaryotic RNase H2 acts processively and hydrolyzes a variety of RNA/DNA hybrids with similar efficiencies, suggesting multiple cellular substrates. Moreover, of five analyzed mutations in human RNASEH2B and RNASEH2C linked to Aicardi-Goutières Syndrome (AGS), only one, R69W in the RNASEH2C protein, exhibits a significant reduction in specific activity, revealing a role for the C subunit in enzymatic activity. Near-normal activity of four AGS-related mutant enzymes was unexpected in light of their predicted impairment causing the AGS phenotype.
INTRODUCTION
Formation and resolution of RNA/DNA hybrids created during DNA replication and repair is central to the maintenance of genome stability. RNases H are the only known enzymes that degrade the RNA strand of RNA/DNA hybrids in a sequence nonspecific manner, and therefore, are essential for DNA integrity (1). There are two types of RNases H in eukaryotes that differ by sequence, biochemical properties and substrate specificity: (i) RNase H1, which is homologous to prokaryotic RNase HI and the RNase H domain of retroviral reverse transcriptase (1); and (ii) RNase H2, which is a monomeric enzyme in prokaryotes, and is composed of three different proteins in eukaryotes (1). In Saccharomyces cerevisiae the RNase H2 heterotrimeric complex contains the catalytic subunit, similar to prokaryotic RNase HII and two other subunits that have no prokaryotic counterparts and whose functions remain unknown (2).
Crow et al. (3) defined the composition of the heterotrimeric human RNase H2 complex when they identified pathogenic mutations in the three gene orthologs of S. cerevisiae RNase H2 (Sc-RNase H2) subunits as being causative of Aicardi-Goutières Syndrome (AGS). AGS is a genetic neurological disorder with symptoms similar to those of congenital viral infection (3,4). It is characterized by loss of brain white matter, intracranial calcifications, high levels of the cytokine interferon-α in the cerebrospinal fluid and elevated number of white cells, suggesting activation of both the innate and adaptive immune responses (3). Mutations in the gene encoding the 3′-exonuclease TREX1 have also been found in AGS patients indicating nucleic acid accumulation as an inducer of AGS (5). In fact, TREX1-deficient cells accumulated single-stranded DNA molecules about 60 nt long, which induced chronic activation of the DNA damage response-signaling network (6,7).
Most of AGS-related mutations in TREX1 have been shown to abrogate its 3′ to 5′ exonuclease activity (5), although heterozygosity for certain mutations can result in other autosomal dominant disorders, such as chilblain lupus (8). A C-terminal truncation that results in mis-localization of TREX1, without affecting its enzymatic activity, is the cause of retinal vasculopathy and cerebral leukodystrophy (RVCL), an autosomal-dominant degenerative disorder (9).
Unlike for TREX1, very little is known about human RNase H2 alterations and no other disorders have been associated with defects in RNase H2. All of the AGS-related mutations found in the three subunits of RNase H2 are missense mutations (4), perhaps an indication that they are essential genes. One mutation found in RNASEH2A (G37S) has been described to decrease significantly the RNase H2 activity of the complex (3). However, the effect of other mutations in RNASEH2B and RNASEH2C subunits remains to be examined. A larger number of AGS-related mutations have been found in RNASEH2B than in other proteins. These mutations usually have a later onset and decreased phenotypic severity than mutations in other subunits (4). Because the roles of the B and C subunits are not known, it is difficult to assess the defects associated with mutations in the genes for these two proteins. Studying the functions of the RNase H2 B/C subunits and their interactions with the A subunit and other proteins would aid our understanding of how the heterotrimeric RNase H2 complex works, and how AGS is induced.
In addition to a structural role in supporting the activity of the RNASEH2A catalytic subunit, the RNASEH2B and RNASEH2C proteins may be involved in interactions with other proteins. Genetic interactions have been reported for all three genes encoding RNase H2 and SGS1 (10,11), RAD27 (12,13) and ESC2 (14,15) in yeast. SGS1p is a DNA helicase, RAD27p is the S. cerevisiae Fen1 protein involved in removal of RNA primers and ESC2p affects recombination frequencies. These studies suggest a function for RNase H2 in Okazaki fragment processing during chromosomal DNA replication/repair, although its exact role has not yet been determined. Chromosomal DNA replication in eukaryotes is orchestrated by the proliferating cell nuclear antigen (PCNA), a protein responsible for bringing to the replication fork and coordinating the activities of the elongating polymerase and other factors involved in Okazaki fragment processing (16). Most proteins that interact with PCNA share a sequence, which physically contacts PCNA (17). Recently, such a sequence has been reported in an archaeal RNase HII, and the interaction with PCNA was described as negatively affecting the enzymatic activity of RNase HII (18).
In this study we determine PCNA interaction and other contributions of the two accessory subunits, RNASEH2B and RNASEH2C, to the activity and properties of the human RNase H2 complex, and examine the effect of several AGS-related mutations, particularly those present in the B and C subunits of RNase H2.
MATERIALS AND METHODS
HeLa cells expression system and immunopurification
HeLa-XZ cells can grow both in a suspension and in an adherent state. The cell line was cultured in high glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, nonessential amino acids, 100 U/ml penicillin, 100 µg/ml streptomycin (Invitrogen) with 5% CO2 at 37°C.
Human RNASEH2A gene was cloned into XhoI/NotI site of pOPAV (19), an MMLV-derived retrovirus vector, which results in fusion of FLAG- and HA-epitopes (MDYKDDDDKLDGGYPYDVPDYAGGLE, FLAG and HA epitopes are underlined, respectively) at the N-terminus of human RNASEH2A. Establishment of stable HeLa cell line expressing the FLAG-HA-tagged human RNASEH2A (HeLa RNASEH2A cell line) was carried out as described previously (19).
Purification of RNase H2 from HeLa RNASEH2A cell was performed as follows: Cells (107) were cultured to 90% confluence and then extracted for 1 h with 500 μl lysis buffer [20 mM Tris–HCl at pH 7.9, 100 mM KCl, 5 mM MgCl2, 10% glycerol, 0.1% NP-40 and Protease inhibitor cocktail Set III EDTA-free (Calbiochem)]. The extract was incubated with 10 μl M2 anti-FLAG agarose (Sigma) for 2 h with rotation (Nutator). Beads were washed in the lysis buffer. Bound proteins were eluted by incubation for 1 h with 200 μl of lysis buffer containing 0.2 mg/ml FLAG peptide (Sigma) with rotation.
Large-scale purification of RNase H2 from HeLa cells and identification of interacting proteins with mass spectrometry was carried out as described previously (19).
Construction of plasmid for expression of human RNase H2 in Escherichia coli
To construct pET-hH2A, pET-hH2B and pET-hH2C, the DNA fragments that contain RNASEH2A, RNASEH2B and RNASEH2C were amplified from cDNAs and cloned into NdeI/XhoI site of pET15b. For coexpression of RNASEH2A and RNASEH2C, pET-hH2AC was constructed as follows: RNASEH2C cDNA fragment which is flanked by EcoRI (3′-side) and blunt-ended XbaI (5′-side) was prepared from pET-hH2C and ligated into EcoRI/blunt-ended XhoI site of pET-hH2A, which resulted in insertion of ribosomal binding site and RNASEH2C ORF at the 3′-side of RNASEH2A gene, so that expression of both RNASEH2A and RNASEH2C is under the control of single T7 promoter in pET-hH2AC. This strategy was also applied for construction of other combinations of polycistronic coexpression systems. To construct pET-hH2AB, RNASEH2B ORF fragment was inserted into 3′-side of RNASEH2A gene in pET-hH2A. Likewise, the three subunits coexpression plasmid, pET-hH2ABC, was constructed by inserting RNASEH2B ORF fragment into pET-hH2AC at the 3′-side of RNASEH2C gene. The plasmid, pET-hH2BC2, from which RNASEH2B and RNASEH2C genes are transcribed separately from different T7 promoters, was constructed as follows: RNASEH2B ORF fragment which is flanked by EcoRI (3′-side) and blunt-ended BglII (5′-side) was prepared from pET-hH2B and ligated into EcoRI/blunt-ended HindIII site of pET-hH2C. For mutational studies, the QuikChange IIXL kit (Stratagene) was used to generate all site-directed mutations and deletions, and changes were confirmed by DNA sequence determination.
Complementation assay
Escherichia coli strain MIC1066 [rnhA339::cat, recB270(Ts)] (20) was transformed with plasmids containing series of combinations of RNase H2 subunits; pET-hH2A, pET-hH2B, pET-hH2C, pET-hH2AC, pET-hH2AB, pET-hH2BC2, pET-hH2ABC. The transformants were streaked on LB-ampicillin plates, and the ability of colony formation at 32 and 42°C was examined.
Escherichia coli expression and purification of human RNase H2
For expression of human RNase H2, E. coli MIC1066 was transformed with pET-hH2ABC and cultured at 32°C overnight in MagicMediaTM (Invitrogen). Cells were harvested by centrifugation at 6000g for 10 min, suspended in 20 mM Tris–HCl pH 7.5 containing 0.5 M NaCl and 30 mM imidazole (buffer A), disrupted by sonication and centrifuged at 30 000g for 30 min. The supernatant was loaded onto a Histrap FF crude column (1 ml) (GE Healthcare) equilibrated with buffer A. The protein was eluted from the column with a step gradient of 30, 100, 300, 500 mM imidazole. The protein eluted at 300 mM imidazole was diluted by addition of 9 volumes of 20 mM Tris–HCl pH 7.5 containing 1 mM EDTA (TE buffer) and applied to Hitrap Heparin HP (GE Healthcare) equilibrated with TE buffer. The protein was eluted from the column with a step gradient of 0.1, 0.3, 0.5, 1.0, 1.5 M NaCl. The protein eluted at 1 M NaCl was dialyzed against 20 mM Tris–HCl pH 7.5 containing 1 mM EDTA and 150 mM NaCl and concentrated. The purity of the protein was confirmed by SDS–PAGE, followed by staining with Coomassie brilliant blue R250 (CBB). Expression and purification of human RNase H2 mutants were carried out as described for wild-type protein. For coexpression of RNASEH2B and RNASEH2C, E. coli MIC1066 was transformed with pET-hH2BC2 and cultured overnight in MagicMediaTM (Invitrogen) at 32°C. Two-step purification with Histrap crude FF 1 ml column (GE Healthcare) and Hitrap Heparin HP 1 ml column (GE Healthcare) was performed according to the methods described for human RNase H2 A/B/C complex. Protein concentrations were determined by measuring the absorbance at A280 of a 0.1% solution; 0.88 for human RNase H2 wild-type and AGS-related mutants (a-G37S, b-K162T, b-A177T, b-V185G and c-K143I mutants); 0.94 for c-R69W AGS-related mutant; 0.69 for RNASEH2B/C complex. These values were calculated by ɛ = 1576 M–1 for Tyr and 5225 M–1 for Trp at A280, assuming that RNase H2 A/B/C and B/C complexes have 1:1:1 and 1:1 subunit compositions, respectively (21).
Expression and purification of E. coli RNase HII
The plasmid, pET-hisEcH2, for expression of N-terminally His-tagged E. coli RNase HII was recloned into pET15b from a plasmid provided by M. Itaya, Keio University. For production of E. coli RNase HII, E. coli MIC1066 was transformed with pET-hisEcH2 and grown in LB-media at 32°C. When the absorbance at 600 nm reached around 0.5, 1 mM isopropyl β-d-thiogalactopyranoside (IPTG) was added to the culture medium and cultivation was continued for an additional 3 h. Two-step purification with Histrap crude FF 1 ml column (GE Healthcare) and Hitrap Heparin HP 1 ml column (GE Healthcare) was carried out as described for human RNase H2 purification. The purified protein was further applied to a gel filtration column (20 × 900 mm) packed with Sephacryl S-200 equilibrated with 40 mM Tris–HCl pH 7.5 containing 1 mM EDTA and 150 mM NaCl. Fractions containing the protein were collected and used for further analyses. The protein concentration was determined from the UV absorption using the A280 value of 0.55 for 0.1% solution, which was calculated as described for human RNase H2.
Expression and purification of human PCNA
The plasmid, pAVR38 (22), for production of His-tagged human PCNA was a gift from R. Woodgate, NICHD/NIH. Escherichia coli MIC1066 was transformed with pAVR38 and cultured overnight in MagicMediaTM (Invitrogen) at 32°C. Purification with Histrap crude FF 1 ml column (GE Healthcare) was carried out as described for human RNase H2 purification. The fraction eluted from Histrap crude FF was further purified with a gel filtration column as described in the methods for E. coli RNase HII purification. Fractions containing the protein were collected and used for further analyses. The protein concentration was determined from the UV absorption using the A280 value of 0.52 for 0.1% solution, which was calculated as described for human RNase H2.
RNase H substrates
Poly-rA/poly-dT with uniformly 32P-labeled poly-rA was prepared as described previously (23). Sequences of short substrates are given in Supplementary Table S1. 5′ 32P-labeled RNA20/DNA20, DNA12–RNA1–DNA27/DNA40 and DNA39–RNA1–DNA40/DNA80 hybrids were prepared by hybridizing 5′-32P-labeled RNA20, DNA12–RNA1–DNA27 and DNA39–RNA1–DNA40 with 1.5 mole equivalent of their complementary DNA.
Gel renaturation assay
Extractions of HeLa cell lines and FLAG-tag purification were performed as described in methods for HeLa cells expression system and immunopurification. Renaturation gel assays were performed with HeLa cell extracts and FLAG-tag purified fraction from the HeLa cell lines as described previously (24). The samples were applied to 15% SDS–PAGE containing poly-rA/poly-dT with uniformly 32P-labeled poly-rA, and after electrophoresis, the gel was incubated overnight in 50 mM Tris–HCl pH 7.9 containing 10 mM MgCl2, 50 mM NaCl and 10 mM 2-mercaptoethanol at room temperature to remove the SDS and allow the protein to renature and act on the substrate. Autoradiograms were developed following exposure of the gels to films.
Specific RNase H activity
The specific RNase H activities of proteins were determined using 5′ 32P-labeled DNA12-RNA1-DNA27/DNA40 hybrid and poly-rA/poly-dT with uniformly 32P-labeled poly-rA. Enzymes were diluted in 20 mM Tris–HCl pH 7.5, 10% glycerol, 0.1 mg/ml BSA. Hydrolysis of the substrates was performed at 37°C for 15 min in 50 mM Tris–HCl pH 8.5 containing 50 mM NaCl, 5 mM MgCl2, 1 mM 2-mercaptoethanol, 50 μg/ml BSA, 1% glycerol. Products of 1 μM of 32P-labeled DNA12–RNA1–DNA27/DNA40 hydrolysis were applied to a 20% TBE-urea polyacrylamide gel. The reaction products were quantified by phosphorimaging. With the poly-rA/poly-dT substrate, RNase H activity was determined by measuring the amount of radioactivity of the acid-soluble digestion product from the substrate (1 μM) as described previously (24).
Processivity assay with poly-rA/poly-dT
Analysis of processivity with poly-rA/poly-dT was carried out as described previously (25). One micromolar poly-rA/poly-dT was digested with proteins, and products of the hydrolysis were resolved on 12% TBE-urea PAGE gel. Final products were defined as oligonucleotides 12 nt or shorter in length, and intermediate products ranged from 13 to 100 nt in lanes where <85% of the starting substrate was degraded. Processivity was calculated by dividing the final products by the intermediates.
Analysis of physical interaction of RNase H2 with PCNA by gel filtration column chromatography
Gel filtrations of RNase H2, PCNA and their complex were performed at 4°C using a column (20 × 900 mm) packed with Sephacryl S-200 (GE Healthcare). The column was equilibrated with 40 mM Tris–HCl pH 7.5 containing 1 mM EDTA and 150 mM NaCl. To constitute the complex between RNase H2 and PCNA, 12 nmol of RNase H2 and 50 nmol of PCNA were incubated for 15 min at 4°C. Then, the protein mixture was chromatographed at 0.6 ml/min flow-rate. Fractions were collected and analyzed on 10–20% gradient SDS–PAGE (Bio-Rad) and stained with CBB.
Pull-down assay
The plasmid, pET17b-hPCNA, was constructed by subcloning the NdeI/BamHI fragment containing human PCNA gene from pAVR38 to the NdeI/BamHI site of pET17b. For production of untagged human PCNA, E. coli MIC1066 was transformed with pET17b-hPCNA and grown overnight in MagicMediaTM (Invitrogen) at 32°C. Escherichia coli MIC1066 harboring pET-hH2BC2, pET-hH2ABC and pET-hH2ABC AGS-related derivatives were grown and induced as described above. For E. coli cell lysates 2 ml of MagicMediaTM overnight cultures were harvested and resuspended in 500 μl of 40 mM Tris–HCl pH 7.5 containing 25% sucrose, 10 mg/ml lysozyme, 10 mM EDTA and 1% Triton X-100 and incubated for 30 min on ice. Next, 15 μl of 0.5 M MgCl2, 1 μl of 50 mg/ml RNase A and 10 μl of 10 U/ml DNase I were added, mixed and incubated on ice for 10 min. The samples were centrifuged and the supernatant was taken as the crude lysate. To facilitate the protein–protein interaction, the lysates containing RNase H2 and PCNA were incubated at 4°C for 1 h with rotation. His-tagged RNase H2 was pulled down by 100 μl of Talon superflow metal affinity resin (Clontech). The resin was washed with 40 mM Tris–HCl pH 7.5 containing 500 mM NaCl and 5 mM imidazole. Proteins were eluted with 40 mM Tris–HCl pH 7.5 containing 500 mM NaCl and 500 mM imidazole. The samples were analyzed by SDS–PAGE stained with CBB and western blotting with anti-PCNA antibody (Sigma).
RESULTS
Purification of human RNase H2 from HeLa cell line
To facilitate purification of human RNase H2 from human cells, we constructed a HeLa cell line that stably expresses N-terminally FLAG- and HA-tagged human RNASEH2A (HeLa RNASEH2A cell) using an MMLV-based retrovirus vector. Expression of FLAG-HA-tagged RNASEH2A in the HeLa RNASEH2A cell line was confirmed by western blotting with anti-FLAG antibody (data not shown).
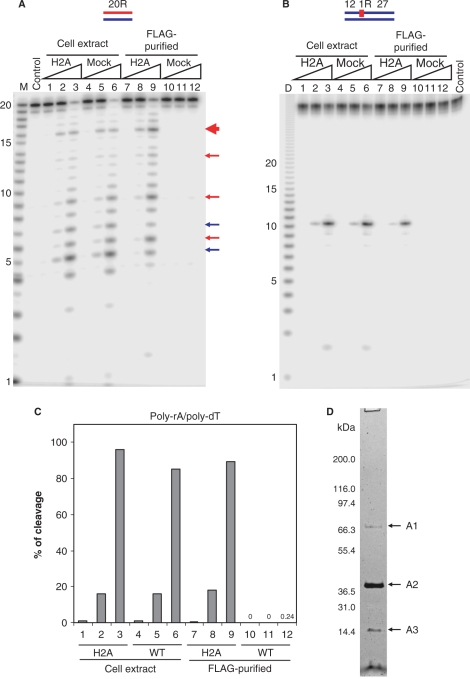
Cell extracts were prepared from HeLa RNASEH2A cells and untransduced HeLa cells, and the RNase H activity of these samples was analyzed using 5′-end labeled 32P-RNA20/DNA20 hybrid, 32P-DNA12-RNA1-DNA27/DNA40 hybrid and uniformly labeled 32P-poly-rA/poly-dT as substrates (Figure 1). The substrates with a single ribonucleotide have been reported to be cleaved by RNase H2 but not by RNase H1 (26). Pattern and efficiency of cleavage were very similar for extracts from HeLa RNASEH2A cells and untransduced HeLa cells (Figure 1), suggesting that overexpression of only the RNASEH2A subunit does not affect total RNase H activity in the HeLa cell line. Purification of human RNase H2 from the HeLa RNASEH2A cells was carried out with anti-FLAG antibody-conjugated resin, and the RNase H activity of purified fractions from HeLa RNASEH2A cells and mock cells was analyzed (Figure 1). The FLAG-tag purified fraction from HeLa RNASEH2A cells showed significantly higher RNase H2 activity than the mock-cell-purified fraction, which showed only negligible hydrolysis of the 32P-poly-rA/poly-dT substrate (Figure 1C). This one-step purification recovered 20–40% of total cell extract RNase H activity.
Figure 1.
RNase H activity in cell extracts and FLAG-tag purified fractions from RNASEH2A expressing and untransduced HeLa cell lines. The 5′-end 32P-labeled 20-bp RNA/DNA hybrid (A) and DNA12–RNA1–DNA27/DNA40 hybrid (B) were cleaved with increasing amounts of the total cell extracts and FLAG-tag purified fractions from RNASEH2A expressing cells (H2A) and untransduced cells (mock) at 37°C for 15 min. The 500 μl cell extracts from 107 cells yielded 200 μl of FLAG-purified samples as described in Materials and methods section. Ten picomoles of substrates were treated with 1 μl of the samples in 10 μl of reaction mixtures. Protein samples were diluted in Dilution Buffer. Lanes 1, 4, 7 and 10 contained 0.002 μl equivalents of the undiluted sample, lanes 2, 5, 8 and 11 contained 0.02 μl equivalents and lanes 3, 6, 9 and 12 contained 0.2 μl equivalents. After digestion the reactions were electrophoresed in a 20% TBE-urea PAGE and the gel analyzed on a phosphoimager. Note (B) the mobilities of the DNA12 product of DNA12–RNA1–DNA27/DNA40 migrates faster than the RNA size markers due to inherent differences in migration in the gels between RNA and DNA. In (A), major cleavage sites of 20-bp RNA/DNA hybrid with RNase H1 and RNase H2 are indicated with blue and red arrows, respectively. The main cleavage product of RNase H2 is indicated by a thick red arrow. Molecular size markers are indicated as M (products of digestion of 32P-labeled 20-mer RNA by Phosphodiesterase I) (measuring the sites of cleavage from the 5′- label of the 20-mer RNA) and D (products of digestion of 32P uniformly labeled poly-rA/poly-dT by mouse RNase H1) (measuring the sizes of products that have uniform sequences). (C) Uniformly 32P-labeled poly-rA/poly-dT (1 μM) was cleaved with increasing amount of the total cell extracts and FLAG-tag purified fractions. Amounts of samples in lanes 1–12 are equivalent to those of (A). The ratios of cleavage products were determined by measuring the acid-soluble radioactivity. (D) SDS–PAGE of the purified RNase H2 from HeLa RNASEH2A cells with the two-step affinity immunopurification. HeLa RNASEH2A cells were extracted and subjected to anti-FLAG and anti-HA two-step purification. The purified sample was analyzed by SDS–PAGE stained with silver staining. The fragment indicated with A1, A2 and A3 were identified by mass spectrometry, as described in text.
Total cell extracts and the FLAG-purified fraction from HeLa RNASEH2A cells produced very similar cleavage patterns (Figure 1A) of a 20-bp RNA/DNA hybrid, for which RNase H1 and RNase H2 showed different cleavage preferences (27), suggesting that RNase H2 provides most of the RNase H activity detected in this assay. Depletion of RNase H1, the other cellular RNase H, in the FLAG-purified sample resulted in only a modest decrease of cleavage products at 5u–6g and 7a–8a (Figure 1A, blue arrows), known preferred sites of RNase H1 (27). The gel renaturation assay confirmed that RNase H1 activity was depleted in the FLAG-tag purified fractions (Supplementary Figure S1). These results demonstrate that functional RNase H2 complex was purified from HeLa cells by the FLAG-tag purification.
To identify proteins interacting with RNASEH2A, two step FLAG- and HA-epitopes immunopurification was conducted from ∼1 l culture (19). Proteins that co-purified with RNASEH2A were visualized by silver staining after SDS–PAGE (Figure 1D). The major proteins (indicated by A1, A2 and A3 in Figure 1D) were identified by mass spectrometry. The identified proteins were heat-shock 70-kDa protein in A1, RNASEH2A and RNASEH2B (FLJ11712) in A2 and RNASEH2C (AYP1) in A3 (Supplementary Table S2). This result indicates that human RNase H2 forms a stable heterotrimer as does yeast RNase H2 (2). The heat-shock 70-kDa protein, an abundant protein in the HeLa cells, may or may not specifically associate with the RNase H2 complex, but it is neither present in stoichiometric amounts nor required for activity (see below). Crow et al. (3) identified the same three proteins as components of the RNase H2 complex and showed that mutations in the genes encoding any of the three subunits can cause AGS, a human neurological disorder.
Expression of the three subunits in E. coli complements an rnhA-null defect
To demonstrate that the three RNase H2 subunits can form an active enzyme in E. coli, we cloned all possible combinations of the RNase H2 genes into plasmids under the control of the T7 promoter and transformed E. coli MIC1066, a strain that is temperature-sensitive for growth due to an rnhA-null mutation in combination with a recB(ts) mutation. Without induction, transcription from the T7 promoter is often sufficient to supply enough RNase H activity for cells to grow at the restrictive temperature (20). Escherichia coli MIC1066 cells were transformed with the plasmids and streaked on LB-ampicillin plates. The plates were incubated at 32 and 42°C. Only the MIC1066 transformant harboring pET-hH2ABC could grow at 42°C, suggesting that functional human RNase H2 is reconstituted in E. coli only when the three subunits are coexpressed (data not shown).
Expression and purification of human RNase H2 from E. coli
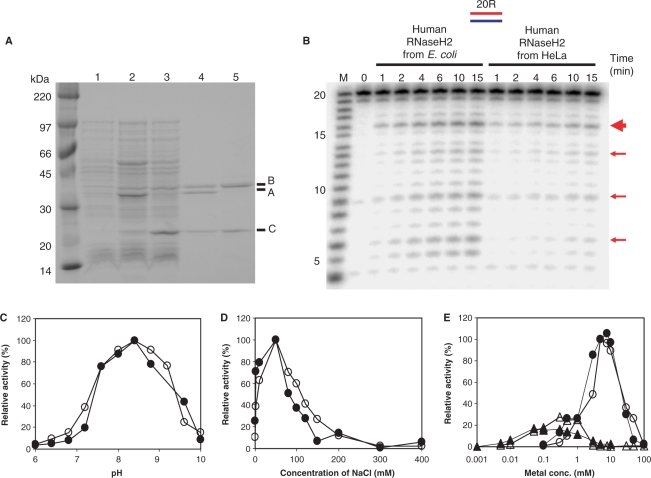
To check the properties of the recombinant human RNase H2, we expressed and purified the three subunits of the complex using E. coli MIC1066 transformed with pET-hH2ABC. In this expression system, all three subunits are N-terminally His-tagged. The expression level of RNASEH2A was estimated to be 5- to 10-fold higher than those of RNASEH2B and RNASEH2C subunits (Figure 2A). However, the three subunits were present in the soluble fraction in a roughly 1:1:1 ratio, with most of the excess RNASEH2A present in the insoluble fraction (data not shown), indicating that the A subunit is stable only as part of the trimeric complex. Coexpression of RNASEH2B and RNASEH2C subunits also yielded soluble forms of both proteins. In this case, production level of the RNASEH2C subunit was roughly 10-fold higher than that of RNASEH2B subunit (Figure 2A). These subunits were present in the soluble fraction in roughly a 1:1 ratio, and the excess of RNASEH2C accumulated in the insoluble fraction (data not shown), suggesting that the two proteins may form a complex that confers stability to the two subunits. Other combinations of expression resulted in accumulation of insoluble proteins (data not shown).
Figure 2.
Comparison of enzymatic characteristics of human RNase H2 purified from HeLa cells and E. coli. (A) SDS–PAGE of human RNase H2 expressed in E. coli. Whole-cell extracts of overnight culture of E. coli MIC1066 transformed with pET15b (lane 1), pET-hH2ABC (lane 2) and pET-hH2BC2 (lane 3), and purified human RNase H2 A/B/C complex (lane 4) and B/C complex (lane 5) were analyzed by 10–20% gradient SDS–PAGE (Bio-Rad) and the gel was stained with CBB. The molecular weight marker is Rainbow Marker from Amersham Bioscience. The proteins corresponding to RNASEH2A, RNASEH2B and RNASEH2C are indicated as A, B and C, respectively. (B) Cleavage of 20-bp substrate with human RNase H2 purified from HeLa cells and E. coli. The 5′-end 32P-labeled 20-bp RNA/DNA hybrid (10 pmol) was hydrolyzed with human RNase H2 purified from HeLa cells (as in lane 8 of Figure 1A) and E. coli (12 fmol) at 37°C for the time (min) indicated above each lane in 10 μl reaction mixtures. Molecular size marker is indicated with M (products of digestion of 32P-labeled 20-mer RNA by Phosphodiesterase I). The digested products were analyzed by 20% TBE-urea PAGE. Prominent cleavage sites were indicated with red arrows. The thick red arrow indicates the main cleavage site. Dependence of pH (C), salt concentration (D) and Mg2+ and Mn2+ ions concentration (E) of human RNase H2 purified from HeLa cells (filled) and E. coli (open) were analyzed using uniformly 32P-labeled poly-rA/poly-dT substrate. MgCl2 and MnSO4 concentrations were indicated by circle and triangle in (E), respectively.
Following coexpression of RNASEH2A/B/C or RNASEH2B/C, two-step column chromatography was used to purify the soluble proteins, and the purified samples were analyzed by SDS–PAGE (Figure 2A). After gelfiltration column chromatography, only single peaks were obtained, showing that the three subunits of human RNase H2 can form A/B/C and B/C complexes in E. coli. The elution positions of human A/B/C and B/C complexes in the gel filtration column chromatography corresponded to a subunit composition of 1:1:1 (for A/B/C complex) and 1:1 (for B/C complex). For Sc-RNase H2 A/B/C and B/C complexes were also purified with subunit composition of 1:1:1 and 1:1, respectively (28).
Enzymatic characteristics of RNase H2 purified from HeLa cells and E. coli
We compared the properties of the human RNase H2 purified from HeLa cells with those of the recombinant enzyme purified from E. coli by checking the enzymatic activity dependence on pH, salt concentration and metal ion concentration of both enzymes using as a substrate poly-rA/poly-dT with uniformly 32P-labeled poly-rA (Figure 2C–E). In all the conditions tested the enzymatic properties were equivalent between human RNase H2 purified from HeLa cells and the recombinant protein purified from E. coli.
The optimum pH for human RNase H2 activity was 8.2–8.8 and the activity was greatly decreased above the optimum pH (Figure 2C). The reported pH-dependence of Sc-RNase H2 activity is similar to that of human RNase H2, while E. coli RNase HII was shown to increase its activity as the pH increase from pH 7.0 to 9.8 (29). NaCl was used to examine the effects of salt on RNase H2 activity. Human RNase H2 exhibited the highest activity in the presence of 50 mM NaCl (Figure 2D), like Sc-RNase H2 (28) and E. coli RNase HII (29). The dependence of human RNase H2 activity on the metal ion concentration was analyzed in the presence of various concentrations of MgCl2 and MnSO4 (Figure 2E). Human RNase H2 exhibited the highest activity in the presence 5–10 mM MgCl2 and 20–30% of the maximal activity in the presence of 0.1–1.0 mM MnSO4. It has been reported that Mn2+-dependent activity of E. coli RNase HII is over 10-fold higher than that of Mg2+-dependent activity for the cleavage of either M13 RNA/DNA hybrid (29) or 12-bp RNA/DNA hybrid (30), while for the cleavage of 12-bp RNA–DNA/DNA Okazaki fragment model substrate, E. coli RNase HII shows 10-fold higher activity in the presence of MgCl2 than MnCl2. On the other hand, Sc-RNase H2 exhibited roughly 2-fold higher activity in the presence of MgCl2 than MnCl2 for the cleavage of DNA15–RNA1–DNA13/DNA29 hybrid (28). Enzymatic activity of human RNase H2 was greatly attenuated as the concentration of metal ions exceeds the optimum concentrations as reported for E. coli and Sc-RNase HII/2 (28,30). These biochemical properties are consistent with those previously described (26,31).
Cleavage pattern of 20-bp RNA/DNA hybrid was also equivalent between human RNase H2 purified from HeLa cells and the recombinant protein purified from E. coli (Figure 2B). The major cleavage site, indicated by the thicker red arrow in Figures 1A and 2B, was between 16c and 17g, 1 nt different than that reported by Pileur et al. (27). This discrepancy most likely results from the different migration of size markers with 3′-phosphate by Pileur et al. and our markers, which have 5′-phosphate moieties as do the products generated by RNases H2.
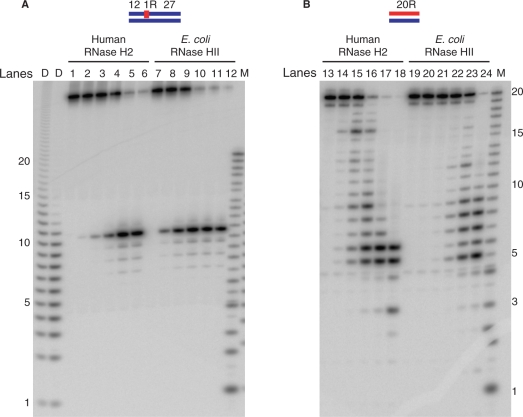
Cleavage of short hybrid substrates with human RNase H2 and E. coli RNase HII
Since the HeLa cell-derived RNase H2 and that expressed in E. coli have such similar biochemical properties, all subsequent analyses of human RNase H2 were performed using the recombinant protein purified from E. coli. To analyze the effect of RNASEH2B and RNASEH2C on the cleavage specificity of RNase H2, we compared the activity of the heterotrimeric human RNase H2 with that of the monomeric RNase HII from E. coli using two different substrates. The 5′-end labeled 32P-DNA12–RNA1–DNA27/DNA40 hybrid and 5′-end labeled 32P-RNA20/DNA20 hybrid were digested with human RNase H2 and E. coli RNase HII in the presence of optimum concentration of MgCl2 (Figure 3). Each enzyme cleaved the RNA20/DNA20 hybrid at multiple sites between 3u and 18a, but the major cleavage sites were different for each enzyme (for example, compare lanes 15 and 23 in Figure 3B). The cleavage pattern of human RNase H2 on 32P-RNA20/DNA20 hybrid was very similar to that of Sc-RNase H2 (2). 32P-DNA12–RNA1–DNA27/DNA40 hybrid was cleaved by human RNase H2 and E. coli RNase HII at the same position (Figure 3A), although E. coli RNase HII cleaved this substrate more efficiently than human RNase H2. Human RNase H2 cleaved both substrates with similar efficiencies, as seen before with the enzyme purified from HeLa cells (Figure 1A and B). However, the E. coli RNase HII cleaved more efficiently the single-ribonucleotide embedded substrate, in accordance with a recent report describing a preference of the E. coli enzyme for cleaving RNAs when part of a junction substrate (30).
Figure 3.
Comparison of cleavage pattern of short substrates with human RNase H2 and E. coli RNase HII. The 5′-end 32P-labeled DNA12–RNA1–DNA27/DNA40 hybrid (A) and 20-bp RNA/DNA hybrid (B) were cleaved with human RNase H2 and E. coli RNase HII at 37°C for 15 min. The reaction volume was 10 μl and the substrate concentration was 1 μM. Amounts of proteins for human RNase H2 were 1.2 fmol (lanes 2 and 14), 12 fmol (lanes 3 and 15), 120 fmol (lanes 4 and 16), 1.2 pmol (lanes 5 and 17), 12 pmol (lanes 6 and 18). The amounts of protein for E. coli RNase HII were 7.6 fmol (lanes 8 and 20), 76 fmol (lanes 9 and 21), 760 fmol (lanes 10 and 22), 7.6 pmol (lanes 11 and 23), 76 pmol (lanes 12 and 24). Lanes 1, 7, 13 and 19 contained no enzymes. The digested products were analyzed by 20% TBE-urea PAGE. Molecular size markers are indicated as M (products of digestion of 32P-labeled 20-mer RNA by Phosphodiesterase I) and D (products of digestion of 32P-labeled poly-rA/poly-dT by mouse RNase H1).
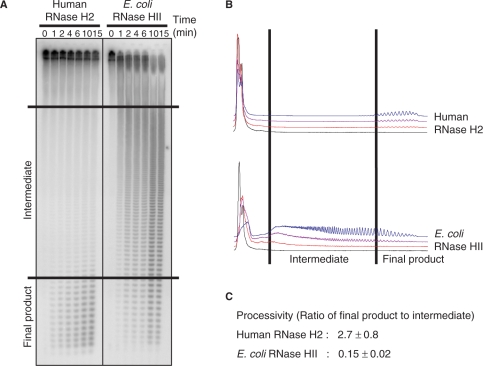
Human RNase H2 cleaves poly-rA/poly-dT in a processive manner
To determine whether the RNASEH2B and RNASEH2C subunits of human RNase H2 contribute to substrate interactions, we examined the mode of cleavage of human RNase H2 and E. coli RNase HII on uniformly labeled 32P-poly-rA/poly-dT (Figure 4). Short oligonucleotides with very few intermediate-sized products appeared throughout the reaction using human RNase H2. In contrast, E. coli RNase HII initially generated a broad size range of cleavage products that were eventually converted to short oligonucleotides. Relative quantities of small and intermediate products were determined for samples in which <15% of the initial substrate had been degraded. Processivity, defined as the ratio of the amounts of small- to intermediate-sized products as described previously (25), showed a more than a 10-fold difference between the human RNase H2 and E. coli RNase HII (Figures 4 and 5).
Figure 4.
Cleavage of poly-rA/poly-dT substrate with human RNase H2 and E. coli RNase HII. (A) The uniformly 32P-labeled poly-rA/poly-dT (1 μM) was digested with human RNase H2 (0.25 fmol) and E. coli RNase HII (76 fmol) at 37°C for the times indicated for each lane. Samples were analyzed by 20% TBE-urea PAGE as described in Materials and methods section. (B) Graphical representations of degradation of the substrate at the time points 0 (black), 2 (red), 6 (purple) and 15 min (blue) are shown. (C) Processivity values are from at least three independent experiments.
Figure 5.
Relative specific activity and processivity. (A) Schematic representation of the recombinant human RNase H2 three subunits. Numbers represent the position of the amino acid residues relative to the N-terminal methionine after the pET15b-derived His-tag. AGS-related mutations examined in this study are shown above each subunit. Conserved catalytic residues (D34, E35, D141 and D169) are shown below RNASEH2A. The region shown in black (K294 to F301) in RNASEH2B represents the PIP-box. (B, C) A bar graph of relative specific activity and processivity: (B) Relative specific activity (red) and processivity (green) of human RNase H2 wild type and AGS-related mutants and E. coli RNase HII (EcHII) was analyzed using uniformly 32P-labeled poly-rA/poly-dT as a substrate. (C) Relative specific activity of human RNase H2 wild-type and AGS-related mutants was analyzed by 32P-labeled DNA12–RNA1–DNA27/DNA40 hybrid. The relative values were normalized to wild type RNase H2 (100%). The error bars represent the standard deviation of at least three independent measurements.
We previously reported that eukaryotic RNase H1 cleaves the poly-rA/poly-dT substrate in a processive manner that requires the N-terminal hybrid binding domain (HBD) and that E. coli RNase HI, which does not possess an HBD, cleaves the substrate in a distributive manner (25). We compared human RNase H2 and mouse RNase H1 and found that processivity of these proteins was very similar but RNase H1 of mouse had a 4-fold higher specific activity (data not shown).
Effect of AGS-related mutation on activity and processivity
Mutations in RNASEH2B and RNASEH2C have been described to cause AGS (3,4), although their effect on RNase H activity is not known. Here, we analyzed the enzymatic activity of five mutations, three in RNASEH2B: Lys162 to Thr (RNase H2b-K162T), Ala177 to Thr (RNase H2B-A177T), Val185 to Gly (RNase H2b-G185V) and two in RNASEH2C: Arg69 to Trp (RNase H2C-R69W) and Lys143 to Ile (RNase H2C-K143I). We also re-examined the activity of a previously described mutation in RNASEH2A: Gly37 to Ser (RNase H2A-G37S). We produced the AGS-related mutant proteins in E. coli and purified them as described for wild-type protein. Production level and solubility of the AGS mutant proteins were similar to those of wild-type protein (data not shown). Specific activity and processivity of the mutant proteins relative to the activity and processivity of the wild-type enzyme are shown in Figure 5. RNase H2A-G37S showed greatly reduced activity as reported previously (3). Processivity was also reduced in the G37S mutation. RNase H2C-R69W retained 30–40% of RNase H activity, while the processivity of the mutant protein was not significantly different from that of wild-type protein. All other mutant RNases H2 have activity and processivity values similar to wild type.
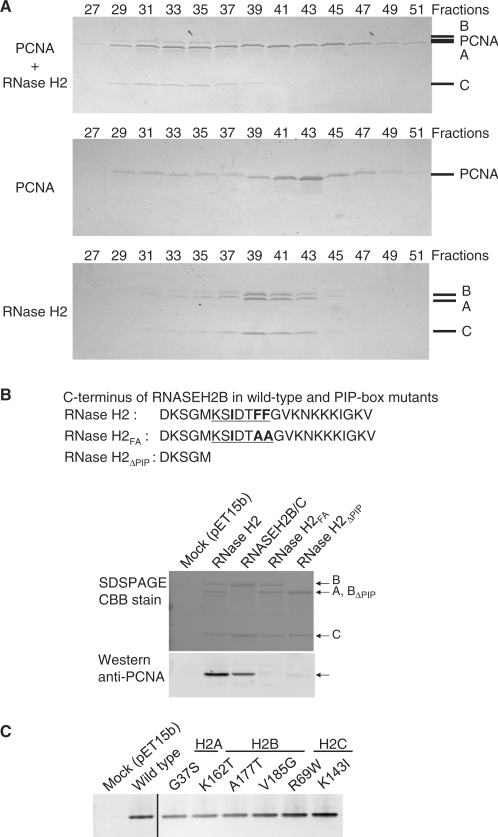
Conservation of PCNA-interacting motif at the C-terminus of RNASEH2B
We found that a putative PCNA interacting protein-box sequence (PIP-box) is present at the C-terminus of human RNASEH2B at residues 294–301 (Figures 5 and 6). The PIP-box (32) is one of the few regions of RNASEH2B that is highly conserved among species (3). Many PCNA interacting proteins contain the typical PIP-box (16), QxxmxxΦΦ (m, aliphatic hydrophobic residues; Φ, aromatic residues; x, any residue). Eukaryotic RNASEH2B conserves the three hydrophobic residues motif, mxxΦΦ, while the first Gln is not conserved in the putative PIP-box of RNASEH2B.
Figure 6.
PCNA binding motif at the C-terminus of eukaryotic RNASEH2B. Amino acid sequences of C-terminus of archaeal RNase HII and eukaryotic RNASEH2B and the consensus PCNA interacting protein box (PIP-box) are aligned. m is aliphatic hydrophobic, Φ is aromatic and x is any residue (32). Conserved residues are in boldface. Hs, Homo sapiens; Mm, Mus musculus; Rn, Rattus norvegicus; Xt, Xenopus tropicalis; Gg, Gallus gallus; Tn, Tetraodon nigroviridis; Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans; Am, Apis mellifera; Dd, Dictyostelium discoideum; Os, Oryza sativa; Spo, Schizosaccharomyces pombe; Sce, Saccharomyces cerevisiae.
RNase H2 physically interacts with PCNA in vitro
To determine whether the putative PIP-box present in RNASEH2B confers binding to PCNA, we analyzed the physical interaction between RNase H2 and PCNA by gel filtration column chromatography and by co-precipitation using affinity tags. PCNA and RNase H2 were purified from E. coli and applied to a gel filtration column. PCNA and RNase H2 when applied separately eluted in fractions 39–45 and 37–43, respectively (Figure 7A). A mixture of 12 nmol of RNase H2 complex and 50 nmol of the homotrimeric PCNA was analyzed on the same gel filtration column, and we found that the elution of RNase H2 shifted to fractions 31–37, indicating that RNase H2 formed a complex with PCNA (Figure 7A). Elution of PCNA occurred over a broad range, some of which was coincident with the shifted RNase H2, with the excess PCNA eluting later.
Figure 7.
Physical interaction of RNase H2 with PCNA. (A) Physical interaction between RNase H2 and PCNA was analyzed using gel filtration column chromatography. Mixture of RNase H2 and PCNA (top), PCNA alone (middle) and RNase H2 alone (bottom) were analyzed on the gel filtration column. The indicated fractions were subjected to 10–20% gradient SDS–PAGE and the proteins were visualized by CBB. A representative result is shown in this figure. A, B, C, and PCNA mark the migration of RNASEH2A, RNASEH2B, RNASEH2C and PCNA, respectively. (B) PCNA interacts with C-terminal tail of RNASEH2B: Bacterial lysate containing untagged PCNA was incubated with bacterial lysates containing His-tagged wild-type RNase H2 A/B/C complex, B/C complex, mutant A/B/C complex with two F to A mutations in PIP-box (RNase H2FA) and mutant A/B/C complex with PIP-box deleted (RNase H2ΔPIP). Pulled-down samples were analyzed by SDS–PAGE with CBB stain and western blotting with anti-PCNA antibody. (C) AGS-related mutations do not affect physical interaction between RNase H2 and PCNA: Bacterial lysate containing untagged PCNA was incubated with bacterial lysates containing His-tagged wild-type RNase H2 and mutant RNase H2 with AGS-related mutations. Pulled-down samples were analyzed by western blotting with anti-PCNA antibody.
To demonstrate that the putative PIP-box is responsible for the RNase H2 interaction with PCNA, we constructed RNASEH2B mutations either by changing the two conserved Phe to Ala (RNase H2FA) or by deleting the C-terminal 18 amino acids that include the putative PIP-box (RNase H2ΔPIP) (Figures 6 and 7B), and the physical interaction of PCNA with wild-type human RNase H2 and the PIP-box mutants was analyzed by metal affinity pulldown assays (Figure 7B). Escherichia coli harboring pET15b was used as a negative control. PCNA was pulled down together with RNase H2 A/B/C trimeric complex and B/C dimeric complex. Alterations in the PIP-box resulted in a great decrease on the amount of PCNA interacting with RNase H2. These results suggest that the PIP-box is required for physical interaction with PCNA.
Effects of the six AGS-related mutations on the physical interaction with PCNA were also examined (Figure 7C). As expected, since the mutations did not map to the PIP-box, the amounts of PCNA pulled down with these AGS-related mutants were equivalent to that of wild-type protein.
Effect of PCNA on RNase H2 activity
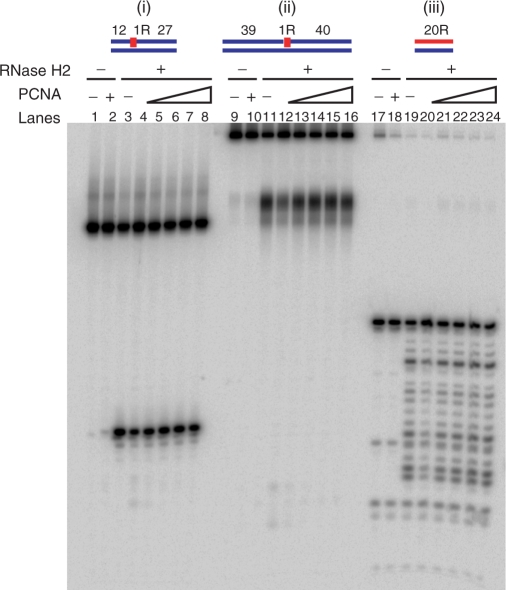
To determine whether RNase H2 activity is affected by interacting with PCNA, RNase H assays were performed with a very wide range in the ratio of RNase H2/PCNA concentrations. The reactions contained 150 fmol of 5′-end 32P-labeled DNA12–RNA1–DNA27/DNA40 hybrid, DNA39–RNA1–DNA40/DNA80 hybrid and RNA20/DNA20 hybrid. RNase H2 amounts were 3 fmol [Figure 8 (i) and (ii)] and 1.2 fmol [Figure 8 (iii)]. PCNA levels ranged from 0.3 to 30 pmol some 2–200 times that of the substrates. Representative data are shown in Figure 8. Enzymatic activity and processivity of human RNase H2 were unaffected under any of the conditions tested. Similarly, the activity of yeast RNase H2 also was unaffected by the presence of PCNA (data not shown).
Figure 8.
Cleavage of oligomeric substrates with RNase H2 in the presence of PCNA. The 5′-end 32P-labeled DNA12–RNA1–DNA27/DNA40 hybrid (i), DNA39–RNA1–DNA40/DNA80 hybrid (ii) and 20-bp RNA/DNA hybrid (ii) were cleaved with human RNase H2 (3 fmol for DNA12–RNA1–DNA27/DNA40 and DNA39–RNA1–DNA40/DNA80 hybrids, 1.2 fmol for 20-bp RNA/DNA hybird) at 37°C for 15 min. The reaction volume was 10 μl. Substrates amounts were 150 fmol. PCNA quantities were 300 fmol (lanes 4, 12 and20), 1 pmol (lanes 5, 13 and 21), 3 pmol (lanes 6, 14 and 22), 10 pmol (lanes 7, 15 and 23), 30 pmol (lanes 2, 8, 10, 15, 18 and 24). Lanes 1, 9 and 17 contained no enzyme. Reactions were analyzed by 20% TBE-urea PAGE.
DISCUSSION
Human and S. cerevisiae RNASEH2A have strong sequence similarity to prokaryotic RNases HII, which is functionally active as a single polypeptide. However, both eukaryotic RNases H2 require two additional subunits to constitute an active enzyme. In this report, we describe a role for the RNASEH2B and RNASEH2C subunits in providing a structural support for the catalytic RNASEH2A subunit to become active, and hydrolyze its RNA/DNA hybrid substrate in a processive manner. The RNASEH2B and RNASEH2C subunits also act as a platform for interactions with other proteins, such as PCNA, which in this work was found to bind human RNase H2 through the PIP-box located in the RNASEH2B subunit.
Role of RNASEH2B and RNASEH2C subunits
Tagged-RNASEH2A in stably transfected HeLa cells permitted purification of the three-subunit complex in a 1:1:1 ratio, with little or no excess RNASEH2A suggesting that any protein not interacting with the RNASEH2B and RNASEH2C proteins did not survive as a separate protein. Unless RNASEH2B and RNASEH2C were present during expression in E. coli, RNASEH2A was found only in the insoluble fraction. Thus, it seems that RNASEH2B and RNASEH2C provide a means to generate an active enzyme by forming a scaffold on which the catalytic RNASEH2A can assume its appropriate configuration.
Human RNASEH2B and RNASEH2C subunits share little amino acid sequence similarity with their S. cerevisiae homologues (3,28), however, both human and Sc-RNase H2 B and C subunits can be purified as soluble dimers after expression in E. coli [Figure 2A and (28)], suggesting that B/C complex formation may be important in both organisms. Such B/C complexes could serve as a platform that incorporates additional proteins to support functions other than RNase H2. Direct protein–protein assessments in S. cerevisiae showed that RNASEH2A interacts exclusively with RNASEH2B and RNASEH2C. In contrast, RNASEH2B has been reported to interact with BUD32 and INP52 (33) in affinity capture-MS studies. This method also uncovered the binding of RNASEH2C to KSS1 and TOP2 (33). In our assays we found that RNASEH2B interacts with PCNA when is part of a functional complex containing A/B/C subunits or as part of the inactive B/C complex.
In our human RNase H2 A/B/C coexpression system in E. coli, only the A/B/C complex was present in a soluble form, while in the case of Sc-RNase H2, coexpression of the three subunits in E. coli resulted in accumulation of both A/B/C and B/C complexes (28). Coexpression of the human proteins produced a large (∼10-fold) excess of the RNASEH2A subunit (Figure 2), while in the Sc-RNase H2 expression system the amount of RNASEHA subunit was much lower than that of RNASEH2B and RNASEH2C. These results suggest that the ratio of expression of the three subunits may be regulated to produce A/B/C or B/C complexes. The mRNAs encoding the three subunits of Sc-RNase H2 are most abundant during the S and G2/M phases of the cell cycle (34,35). Coincident changes in S. cerevisiae mRNAs could indicate that all the proteins that interact with B/C complex, including the A subunit, are involved in processes that occur in S and G2/M phases. The data of mRNAs encoding the human RNase H2 subunits are not clearly consistent but there does appear to be some change in amounts during the cell cycle (36).
Physical interaction between RNase HII/2 and PCNA is evolutionally conserved
One important contribution of RNASEH2B is to provide a PIP-box for interaction with PCNA (Figure 7). PCNA functions as a platform to recruit interacting proteins at the ‘right time and place’ during DNA replication and repair (17). In some cases, PCNA has been shown to stimulate the enzymatic activity of the interacting protein (17) most likely by facilitating substrate binding. In our assays, using linear substrates of various lengths, addition of PCNA showed no effect on the enzymatic activity of RNase H2 (Figure 8), although PCNA could be required for loading RNase H2 into certain substrates, such as circular nucleic acids, or RNA/DNA hybrids with blocked ends, substrates similar to the ones found in vivo.
Recently, it was shown that Pab-RNase HII interacts with PCNA through its C-terminal tail, which contains a PIP-box (18). We searched a NCBI database and found that many archaeal RNases HII possess putative PIP-boxes at their C-termini (Supplementary Table S3). It was reported that the enzymatic activity of Pab-RNase HII was inhibited by 80% in assays using equimolar amounts of RNase HII, a DNA7-RNA1-DNA7/DNA15 hybrid substrate and PCNA (18), a result quite different from our observation with the human enzyme. The archaeal enzyme has the PIP-box directly attached to the single protein comprising the enzyme. Perhaps the more complex human RNase H2 has adapted a different approach to interact with PCNA that does not interfere with its activity. These results suggest that physical interaction between RNase HII/2 and PCNA is conserved from archaea to eukaryotes, although the effect on enzymatic activity needs further studies.
During lagging strand synthesis, PCNA brings together and coordinates the proteins involved in DNA elongation and maturation, such as DNA polymerase δ(Pol δ), the flap cleavage endonuclease, FEN1 and the nick closure ligase, Lig1 (32). The nick translation activity of Pol δ in cooperation with FEN1 and DNA2 nucleases has been shown to be the main pathway for Okazaki fragment maturation (32). However, several yeast genetic studies have indicated that deletion of RNase H2 in combination with defects in DNA2 (37) or RAD27 (FEN1) (12,13) has synthetic lethal or sickness phenotypes, suggesting functional overlapping of RNase H2 with these Okazaki fragment maturation enzymes (32). The fact that RNase H2 binds to PCNA also suggests that RNase H2 is involved in replication and repair. Analysis on the in vivo interaction with PCNA will give new insights into the physiological function of RNase H2.
Substrate recognition by RNase H2
Human RNase H2 acts in a processive manner for which RNASEH2B and RNASEH2C may be important since the single polypeptide RNase HII enzyme from E. coli acts in a distributive manner (Figure 4). RNase H1/I, the other class of RNases H follows the same pattern: they are processive enzymes in eukaryotic cells while their prokaryotic counterparts are not. This strongly suggests the presence of long, complex hybrids in eukaryotes that require processive cleavage.
Human RNase H2 does not exhibit any significant preference for cleaving the single ribonucleotide-embedded substrate over a typical RNA/DNA hybrid (Figure 4). However, several prokaryotic RNases HII prefer hydrolyzing the embedded ribonucleotide hybrid, and in a recent report, it was described that the RNase HII homologue from a highly thermophilic bacterium, Thermus thermophilus, cleaved the embedded ribonucleotide hybrid and the ribonucleotide adjacent to the DNA in a model Okazaki fragment hybrid, but it did not hydrolyzed the usual RNA/DNA substrate (30). Moreover, bacterial RNase HIII, which is a type 2 RNase H and structurally (38) very similar to archaeal (39,40) and bacterial RNases HII (PDB:2etj), would not hydrolyze a single embedded ribonucleotide DNA substrate (41).
The ability of human RNase H2 to act in a processive manner and hydrolyze a variety of RNA/DNA hybrids with similar efficiencies suggests that it may participate in many cellular processes (e.g. as a surveillance mechanism to find and cleave single ribonucleotides in DNA, leading to their removal by repair enzymes, or acting to remove R-loops that may be generated during transcription).
Effect of AGS-related mutation
Mutations in the three subunits of RNase H2 and TREX1, a ssDNA exonuclease, have been found as causative of AGS (3,5), suggesting that accumulation of DNA and RNA by-products is a main factor that induces virus infection-like phenotypes in AGS (3).
The earliest appearance and most severely affected AGS patients have mutations in the RNASEH2A and RNASEH2C subunits (RNASEH2A G37S and RNASEH2C R69W) whereas phenotypic presentation of AGS in children with RNASEH2B mutations occurs significantly later (4). In addition, patients with RNASEH2B mutations tend to live longer than those with mutations in other genes.
Of all the AGS-related mutations examined in this study, only the RNASEH2A G37S substitution exhibited a severe decrease in the enzymatic activity of the complex, as previously reported (3). RNASEH2C R69W mutant enzyme was about 35% as active as wild-type RNase H2, which may account for the severity of AGS-phenotype in patients with this mutation. However, the other mutant proteins, RNase H2B-K162T, -A177T and -V185G and RNase H2C-K143I had almost normal levels of RNase H activity (Figure 5). A recent report (28) describes the effect of AGS-causing mutations on the activity of RNase H2 from S. cerevisiae and Thermococcus kodakaraensis. While G37 is clearly conserved in these different organisms, and consequently mutations of this amino acid have similar effects, the great divergence of the B and C subunits between human and S. cerevisiae make it difficult to assess the effects of AGS-related mutation in the yeast system (28). However, it is clear that RNase HII activity of T. kodakaraensis is impaired when the conserved Gly is changed to other amino acids with bulkier side chains, presumably limiting access to the active site.
The failure to note differences in activity in our in vitro studies does not preclude small defects in enzymatic activity that would induce the accumulation of unprocessed hybrids over long periods of time. Perhaps the mutations that give rise to later onset of AGS symptoms, such as those found in the RNASEH2B, are not related to specific activity per se but to stability or complex formation. If our suggestion that the RNASEH2B/C heterodimer is the platform on which the RNASEH2A subunit is properly assembled, some of the mutations in the RNASEH2B or RNASEH2C subunits may be defective in assembly/stability.
The 13 missense mutations present in the RNASEH2B gene are in exons 2–8 (4). Interestingly, no AGS-related mutation has been found in exons 9–11. Thus, the PIP-box of RNASEH2B is normal in all of the examined AGS-related mutant proteins, and consequently they showed normal PCNA interaction (Figure 7C). Interaction with PCNA, which is required to participate in DNA replication and repair, may be essential for RNase H2 function and therefore mutations in the PIP-box of RNASEH2B may not be viable.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health; Human Development, National Institutes of Health; and the National Institutes of Health (GM32431 to P.M.B.). Chon,H. is a Japan Society for the Promotion of Science Research Fellow in Biomedical and Behavioral Research at the National Institutes of Health. Funding for open access charge: Intramural Research Program of the National Institutes of Health, NICHD.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Shun-ichi Tanaka for help in pull-down assay and Andrew Jackson for helpful comments on the manuscript, and Roger Woodgate and Mitsuhiro Itaya for plasmids.
REFERENCES
- 1.Crouch RJ, Toulme JJ, editors. Ribonucleases H. Paris: INSERM; 1998. [Google Scholar]
- 2.Jeong HS, Backlund PS, Chen HC, Karavanov AA, Crouch RJ. RNase H2 of Saccharomyces cerevisiae is a complex of three proteins. Nucleic Acids Res. 2004;32:407–414. doi: 10.1093/nar/gkh209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crow YJ, Leitch A, Hayward BE, Garner A, Parmar R, Griffith E, Ali M, Semple C, Aicardi J, Babul-Hirji R, et al. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutières syndrome and mimic congenital viral brain infection. Nat. Genet. 2006;38:910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- 4.Rice G, Patrick T, Parmar R, Taylor CF, Aeby A, Aicardi J, Artuch R, Montalto SA, Bacino CA, Barroso B, et al. Clinical and molecular phenotype of Aicardi-Goutières syndrome. Am. J. Hum. Genet. 2007;81:713–725. doi: 10.1086/521373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crow YJ, Hayward BE, Parmar R, Robins P, Leitch A, Ali M, Black DN, van Bokhoven H, Brunner HG, Hamel BC, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutières syndrome at the AGS1 locus. Nat. Genet. 2006;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- 6.Yang YG, Lindahl T, Barnes DE. Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell. 2007;131:873–886. doi: 10.1016/j.cell.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 7.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee-Kirsch MA, Chowdhury D, Harvey S, Gong M, Senenko L, Engel K, Pfeiffer C, Hollis T, Gahr M, Perrino F, et al. A mutation in TREX1 that impairs susceptibility to granzyme A-mediated cell death underlies familial chilblain lupus. J. Mol. Med. 2007;85:531–537. doi: 10.1007/s00109-007-0199-9. [DOI] [PubMed] [Google Scholar]
- 9.Richards A, van den Maagdenberg A.M.JM, Jen JC, Kavanagh D, Bertram P, Spitzer D, Liszewski MK, Barilla-LaBarca ML, Terwindt GM, Kasai Y, et al. C-terminal truncations in human 3′-5′ DNA exonuclease TREX1 cause autosomal dominant retinal vasculopathy with cerebral leukodystrophy. Nat. Genet. 2007;39:1068–1070. doi: 10.1038/ng2082. [DOI] [PubMed] [Google Scholar]
- 10.Ooi SL, Shoemaker DD, Boeke JD. DNA helicase gene interaction network defined using synthetic lethality analyzed by microarray. Nat. Genet. 2003;35:277–286. doi: 10.1038/ng1258. [DOI] [PubMed] [Google Scholar]
- 11.Ii M, Brill SJ. Roles of SGS1, MUS81, and RAD51 in the repair of lagging-strand replication defects in Saccharomyces cerevisiae. Curr. Genet. 2005;48:213–225. doi: 10.1007/s00294-005-0014-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loeillet S, Palancade B, Cartron M, Thierry A, Richard GF, Dujon B, Doye V, Nicolas A. Genetic network interactions among replication, repair and nuclear pore deficiencies in yeast. DNA Repair. 2005;4:459–468. doi: 10.1016/j.dnarep.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 13.Pan X, Ye P, Yuan DS, Wang X, Bader JS, Boeke JD. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- 14.Tong A.HY, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 15.Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 16.Bambara RA, Murante RS, Henricksen LA. Enzymes and reactions at the eukaryotic DNA replication fork. J. Biol. Chem. 1997;272:4647–4650. doi: 10.1074/jbc.272.8.4647. [DOI] [PubMed] [Google Scholar]
- 17.Majka J, Burgers P.MJ. The PCNA-RFC families of DNA clamps and clamp loaders. In: Kivie M, editor. Progress in Nucleic Acid Research and Molecular Biology. New York: Academic Press; 2004. pp. 227–260. [DOI] [PubMed] [Google Scholar]
- 18.Meslet-Cladiére L, Norais C, Kuhn J, Briffotaux J, Sloostra JW, Ferrari E, Hübscher U, Flament D, Myllykallio H. A novel proteomic approach identifies new interaction partners for proliferating cell nuclear antigen. J. Mol. Biol. 2007;372:1137–1148. doi: 10.1016/j.jmb.2007.06.056. [DOI] [PubMed] [Google Scholar]
- 19.Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 2001;15:1229–1241. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cerritelli SM, Shin DY, Chen HC, Gonzales M, Crouch RJ. Proteolysis of Saccharomyces cerevisiae RNase H1 in E coli. Biochimie. 1993;75:107–111. doi: 10.1016/0300-9084(93)90031-m. [DOI] [PubMed] [Google Scholar]
- 21.Goodwin TW, Morton RA. The spectrophotometric determination of tyrosine and tryptophan in proteins. Biochem. J. 1946;40:628–632. doi: 10.1042/bj0400628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vidal AE, Kannouche P, Podust VN, Yang W, Lehmann AR, Woodgate R. Proliferating cell nuclear antigen-dependent coordination of the biological functions of human DNA polymerase τ. J. Biol. Chem. 2004;279:48360–48368. doi: 10.1074/jbc.M406511200. [DOI] [PubMed] [Google Scholar]
- 23.Dirksen ML, Crouch RJ. Selective inhibition of RNase H by dextran. J. Biol. Chem. 1981;256:11569–11573. [PubMed] [Google Scholar]
- 24.Carl PL, Bloom L, Crouch RJ. Isolation and mapping of a mutation in Escherichia coli with altered levels of ribonuclease H. J. Bacteriol. 1980;144:28–35. doi: 10.1128/jb.144.1.28-35.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaidamakov SA, Gorshkova II, Schuck P, Steinbach PJ, Yamada H, Crouch RJ, Cerritelli SM. Eukaryotic RNases H1 act processively by interactions through the duplex RNA-binding domain. Nucleic Acids Res. 2005;33:2166–2175. doi: 10.1093/nar/gki510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eder PS, Walder RY, Walder JA. Substrate specificity of human RNase H1 and its role in excision repair of ribose residues misincorporated in DNA. Biochimie. 1993;75:123–126. doi: 10.1016/0300-9084(93)90033-o. [DOI] [PubMed] [Google Scholar]
- 27.Pileur F, Toulme JJ, Cazenave C. Eukaryotic ribonucleases HI and HII generate characteristic hydrolytic patterns on DNA-RNA hybrids: further evidence that mitochondrial RNase H is an RNase HII. Nucleic Acids Res. 2000;28:3674–3683. doi: 10.1093/nar/28.18.3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rohman MS, Koga Y, Takano K, Chon H, Crouch RJ, Kanaya S. Effect of the disease-causing mutations identified in human RNase H2 on the activities and stabilities of yeast RNase H2 and archaeal RNase HII. FEBS J. 2008;275:4836–4849. doi: 10.1111/j.1742-4658.2008.06622.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohtani N, Haruki M, Muroya A, Morikawa M, Kanaya S. Characterization of ribonuclease HII from Escherichia coli overproduced in a soluble form. J. Biochem. 2000;127:895–899. doi: 10.1093/oxfordjournals.jbchem.a022684. [DOI] [PubMed] [Google Scholar]
- 30.Ohtani N, Tomita M, Itaya M. Junction ribonuclease: a ribonuclease HII orthologue from Thermus thermophilus HB8 prefers the RNA−DNA junction to the RNA/DNA heteroduplex. Biochem. J. 2008;412:517–526. doi: 10.1042/BJ20080140. [DOI] [PubMed] [Google Scholar]
- 31.Toulmé JJ, Frank P, Crouch RJ. Human RNases H. In: Crouch RJ, Toulmé JJ, editors. Ribonucleases H. INSERM, Paris: Les Editions; 1998. pp. 147–162. [Google Scholar]
- 32.Garg P, Burgers P.MJ. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit. Rev. Biochem. Mol. Biol. 2005;40:115–128. doi: 10.1080/10409230590935433. [DOI] [PubMed] [Google Scholar]
- 33.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 34.Arudchandran A, Cerritelli SM, Narimatsu SK, Itaya M, Shin DY, Shimada Y, Crouch RJ. The absence of ribonuclease H1 or H2 alters the sensitivity of Saccharomyces cerevisiae to hydroxyurea, caffeine and ethyl methanesulphonate: implications for roles of RNases H in DNA replication and repair. Genes Cells. 2000;5:789–802. doi: 10.1046/j.1365-2443.2000.00373.x. [DOI] [PubMed] [Google Scholar]
- 35.Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, Brown PO, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Budd ME, Tong A.HY, Polaczek P, Peng X, Boone C, Campbell JL. A network of multi-tasking proteins at the DNA replication fork preserves genome stability. PLoS Genet. 2005;1:e61. doi: 10.1371/journal.pgen.0010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chon H, Matsumura H, Koga Y, Takano K, Kanaya S. Crystal structure and structure-based mutational analyses of RNase HIII from Bacillus stearothermophilus: a new type 2 RNase H with TBP-like substrate-binding domain at the N terminus. J. Mol. Biol. 2006;356:165–178. doi: 10.1016/j.jmb.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 39.Lai L, Yokota H, Hung LW, Kim R, Kim SH. Crystal structure of archaeal RNase HII: a homologue of human major RNase H. Structure. 2000;8:897–904. doi: 10.1016/s0969-2126(00)00179-9. [DOI] [PubMed] [Google Scholar]
- 40.Muroya A, Tsuchiya D, Ishikawa M, Haruki M, Morikawa M, Kanaya S, Morikawa K. Catalytic center of an archaeal type 2 ribonuclease H as revealed by X-ray crystallographic and mutational analyses. Protein Sci. 2001;10:707–714. doi: 10.1110/ps.48001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haruki M, Tsunaka Y, Morikawa M, Kanaya S. Cleavage of a DNA−RNA–DNA/DNA chimeric substrate containing a single ribonucleotide at the DNA-RNA junction with prokaryotic RNases HII. FEBS Letts. 2002;531:204–208. doi: 10.1016/s0014-5793(02)03503-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.