Abstract
Regulatory influence of the G-quadruplex or G4 motif present within the nuclease hypersensitive element (NHE) in the promoter of c-MYC has been noted. On the other hand, association of NM23-H2 to the NHE leads to c-MYC activation. Therefore, NM23-H2 interaction with the G4 motif within the c-MYC NHE presents an interesting mechanistic possibility. Herein, using luciferase reporter assay and chromatin immunoprecipitation we show NM23-H2 mediated c-MYC activation involves NM23-H2-G4 motif binding within the c-MYC NHE. G4 motif complex formation with recombinant NM23-H2 was independently confirmed using fluorescence energy transfer, which also indicated that the G4 motif was resolved to an unfolded state within the protein-bound complex. Taken together, this supports transcriptional role of NM23-H2 via a G4 motif.
INTRODUCTION
Function of non B-DNA conformations in recombination, replication and particularly, regulation of gene expression has been generally appreciated in recent years, both in prokaryotes (1) and eukaryotes (2,3). Certain G-rich sequences adopt a particular type of non B-DNA structure—the G-quadruplex or G4 motif. G4 motifs are four-stranded planar array of four-hydrogen bonded guanines (G-quartets) stabilized by charge coordination with monovalent cations (especially K+) resulting from intramolecular or intermolecular association of DNA strands in parallel or antiparallel orientation (4,5). These motifs are known to form in telomere ends of eurkaryotic genomes (6,7), where they play a central role in telomere extension and are the focus of targeted anticancer drug development (6,8,9). Direct in vivo presence of G4 motifs have been observed only recently within human telomeres (10), in Escherichia Coli (11) and telomeres of Stylonychia nanochromosomes (12). In a regulatory context, emerging evidence show several important gene promoters like β-globin (13) and retinoblastoma susceptibility genes (14), the insulin gene (15), adenovirus serotype 2 (16), PDGF (17), c-KIT, hypoxia inducible factor 1-alpha, VEGF, BCL-2 (18), KRAS (19,20), c-MYB (21) and c-MYC (22,23) harbor G4 motifs with possible functional role. Furthermore, several recent genome wide studies indicate prevalence of G4 motifs within promoters of human and related species (24–26) including several bacteria (27). Several independent lines of bioinformatics analyses also suggest genome-wide regulatory function of G4 motifs (27,28).
Repression of c-MYC by G4 motif (using the G4 motif-binding ligand TMPyP4 (5,10,15,20-tetra(N-methyl-4-pyridyl) porphine chloride) and over-expression of c-MYC in case of site-specific mutation(s) that destabilized the G4 motif within the c-MYC promoter nuclease hypersensitive element (NHE IIII) indicated functional role of G4 motif in transcription (23). This was further supported by recent findings demonstrating the functional role of a G4 motif in PDGF-A expression (29). Formation of G4 motifs in promoter of three muscle-specific genes, human sarcomeric mitochondrial creatine kinase (sMtCK), mouse MCK and alpha7 integrin has also been noted where the homodimeric form of the transcription factor MyoD associated more efficiently with the tetraplex structural motifs than its target E-box (30,31). This implicates MyoD in a tetraplex-specific transcriptional role.
In this context, it is interesting to consider the metastases suppressor protein NM23-H2. Molecular mechanism of metastases suppression by NM23-H2 is not clear though for several years now the potent antimetastatic activity of NM23-H2 stands established (32). NM23-H2, its close homolog NM23-H1 along with other members belong to the NM23 family of NDP kinases (33). Earlier NDP kinases were considered essential housekeeping enzymes required for maintaining NTP pools. However, E. coli and yeast were found to be viable without NDP kinase though certain defects related to DNA repair were observed in deficient strains (34,35). Interestingly, NM23-H2 has also been reported to preferentially bind single-stranded pyrimidine-rich paranemic forms of DNA (36); the DNA binding and kinase domains appear to have independent functions though the respective catalytic residues are housed within the same site (37). Furthermore, independent studies have demonstrated that: (i) NM23-H2 binds the c-MYC promoter (NHE IIII) and transactivates c-MYC (38,39) and (ii) the NHE harbors a G4 motif that suppresses c-MYC expression (22,23).
In view of these we asked whether NM23-H2 binds the c-MYC promoter G4 motif and the role of this interaction in c-MYC promoter activation. To address this we first investigated in vivo localization of NM23-H2 on the c-MYC NHE and studied association of G4 motif with recombinant NM23-H2. In order to specifically study the role of the structural form we used promoter constructs devoid of G4 structure formation in presence of NM23-H2 in cell culture experiments. Finally NM23-H2 mutants were used to address mechanistic issues. Taken together, results support involvement of the G4 motif present within the c-MYC promoter as a structural element for association of NM23-H2 that confers regulatory control of c-MYC expression.
METHODS
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed following the protocol provided by Upstate Biotechnology with modifications as in Fast ChIP protocol (40). After 48 h of transfection of pcDNA-NM23-H2 or mutants using Lipofectamine 2000 (Invitrogen), antibody against overexpressed NM23-H2 (wild-type or H118C, K12A, H47A mutants) having myc epitope (sigma clone 9 E 10) was used to immunoprecipitate chromatin in A549 and HeLa S3 cells. For ChIP in MDA-MB-231 cells, rabbit polyclonal anti-nm 23-H antibody FL-152: sc-2882 was used to immunoprecipitate endogenous NM23-H2. Mouse IgG was used for mock immunoprecipitation in all the cell lines. Briefly, cells were fixed with 1% formaldehyde for 10 min and lysed. Chromatin was sheared to an average size of ∼1 kb using a Misonix 3000 sonicator. Lysate was precleared using protein-A sepharose beads. Twenty-five percent of lysate was used to isolate input chromatin by phenol–chloroform method and ethanol precipitation. ChIP was performed overnight at 4°C. Immune complexes were collected using herring sperm DNA saturated protein-A Sepharose in 3 h and washed extensively. Chelex-100 resin was used to extract DNA from immunoprecipitated chromatin [as described previously (40)]. To observe the effects of TMPyP4 on NM23-H2 mediated c-MYC promoter precipitation, cells were pretreated with TMPyP4 for 3 h before transfection. The following primer pair was used to amplify a 165-bp region within NHE III1 of c-MYC promoter: c-MYC_Fwd: CTACGGAGGAGCAGCAGAGAAAG, c-MYC_Rvs: GTGGGGAGGGTGGGGAAGGT. A genomic locus from human chromosome 3 was used as negative control: amplicon position: From 161189123 to 161189415 (+), size of the amplicon: 293, Fwd primer: TAGGCTGGAGGTCGTGGTTA, Rvs primer: CGGCGCTTTCGGATTAACT.
Silencing of NM23-H2 using siRNA
Commercially available synthetic siRNA duplex [Santa Cruz (sc 40774)] against NM23-H2 was used to silence NM23-H2 in A549 cells. The siRNA duplex [Santa Cruz (sc 37007] having a scrambled sequence was used as a negative control; it is known not to bind and degrade any cellular mRNA. The control siRNA duplex (sc 36869) conjugated with fluorecein was used to monitor transfection efficiency in the cultured cells. Transfection of siRNA duplexes was done using Lipofectamine 2000 as per manufacturer's protocol. After 48 h of tranfection of siRNA duplexes, total RNA from A549 cells was isolated using TRI reagent (Sigma) and cDNA synthesized using cDNA archive kit (Applied Biosystems). Primers used to probe NM23-H2 and β- actin (as control) transcript levels by semi-quantitative real-time (RT)-PCR are given as Supplementary Data.
Quantitative RT-PCR
Quantitative RT-PCR was performed using ABI 7500 and SYBR green master mix as per manufacturer's protocol. A standard curve for each gene was derived from serial dilutions with an external cDNA. Relative quantification for each of the genes was performed against a house keeping gene, β2 microglobulin (B2M), according to relative curve standard method described previously (41). Fold change was calculated relative to control siRNA treatment. Primer details are given as Supplementary Data.
Recombinant NM23-H2 expression, phosphotransferase assay
Recombinant NM23-H2 and mutants (K12A, H118C and H47A) were expressed in E. coli using the pRSETA-NM23-H2 clones (see Supplementary Data for plasmid details) and purified using Ni–NTA chromatography to obtain His-tagged protein, which was used for all experiments. In an independent preparation His tag was removed, resin bound protein was cleaved [(0.6 µg per 25 µg of fusion protein in reaction buffer (50 mM Tris pH 8.0, 5 mM CaCl2)] and subsequently enterokinase was removed using enterokinase removal kit. NM23-H2 without His tag gave comparable results as the His-tagged product in all assays. Phosphotransferase activity of purified NM23-H2 was determined as described earlier (42). One microgram of protein was incubated with 1 mM GDP, 10 μCi [γ-32P] ATP following initiation with 0.1 mM ATP for 10 min at room temperature; reaction stopped with SDS buffer, resolved by TLC (Thin Layer chromatography) and visualized using autoradiography.
Polyacrylamide gel electrophoresis experiments detected by fluorescence energy transfer
The doubly labeled oligonucleotide measuring 0.6 μM [FG27T; 5′-F-TGG GGA GGG TGG GGA GGG TGG GGA AGG-T-3′; 5′-fluorescein (F) and 3′-TAMRA (T)] was heated at 95°C for 10 min and then slowly cooled to room temperature in 10 mM HEPES, 2 mM MgCl2 and 20 mM KCl, pH 7.4 containing 60 μM nonspecific single strand [poly(dA)] and double-stranded (ds)DNA [poly(dI.dT)]. Following this 50 μg/ml BSA was added and treated with either recombinant NM23-H2 (6 μM), TMPyP4 or both at 37°C. NM23-H2 mutants (K12A, H118C and H47A) were incubated with FG27T following an identical protocol. FG27T–protein complexes were separated on 5% native polyacrylamide gel electrophoresis (PAGE) (90 min at 70 V) in TB buffer and bands visualized by exciting at 473 nm and emission observed at either 520 or 580 nm (emission wavelength of fluorescein and TAMRA, respectively) in Fujifilm FLA2000 using ImageReader and images processed using MultiGauge and LProcess provided by manufacturer.
Fluorescence energy transfer in solution
Fifty nanomolar of the doubly labeled oligonucleotide FG27T in 10 mM HEPES, pH 7.4 (adjusted with KOH), 20 mM KCl, 2 mM MgCl2 (for protein activity) was heated at 97°C for 5 min and then slowly cooled to 37°C. Following this FG27T was incubated (10 min at 37°C) with increasing concentration of NM23-H2 in presence of 5.0 μM nonspecific dsDNA [poly(dI.dT)] and energy transfer was measured by exciting at 473 nm. Emission was observed from 500 to 700 nm using a Fluoromax 3 (Spex) spectrofluorimeter. For determining fluorescence intensity of individual fluorophores, emission intensities between 522–528 nm and 584–590 nm was averaged for donor (fluorescein) and acceptor (TAMRA; low donor emission was neglected), respectively. To estimate change in energy transfer efficiency the ratio P (= ID/(ID+IA)) was calculated, where ID and IA are average donor and acceptor intensities, respectively.
Transfection and luciferase assays
Del-4 plasmid (with 850 bp of c-MYC promoter sequence harboring the two major promoters P1 and P2 in a luciferase reporter cassette; a gift from Bert Vogelstein) alone, in combination with pcDNA3-NM23-H2 or its mutants (see Supplementary Data for cloning details) was transfected using Lipofectamine 2000 as per manufacturer's protocol in untreated or pretreated (with TMPyP4 or TMPyP2) HeLa S3 or A549 cells. Cell viability assays (MTT) indicate that on an average about 93% and 88% cells are viable after 24 and 48 h, respectively, after treatment with 100 μM TMPyP4 or TMPyP2. Furthermore, we also noted that treatment with 100 μM TMPyP4 did not influence general cell death-related gene expression changes in genome-wide expression analyses published recently (43). After 24 or 48 h cells were lysed and assayed using luciferase reporter system from Promega. Variants of the Del-4 plasmid construct, wherein specific base substitutions were incorporated such that G4 motif formation is unlikely were also employed for luciferase reporter assays. The G4 motif sequence in Del-4 and the G4-mutant plasmids (GM1 and GM2) are the following (substitutions are underlined): Del-4, TGGGGAGGGTGGGGAGGGTGGGGAAGG; GM1, TGGGGAGGGTGAGGAGGGTGGGGAAGG; GM2, TGGGGAGGGTGAAGAGAGTGGGGAAGG. The luciferase activity was measured using a Berthold Luminometer and counts normalized using the total protein concentration.
RESULTS
In vivo association of NM23-H2 to the c-MYC NHE
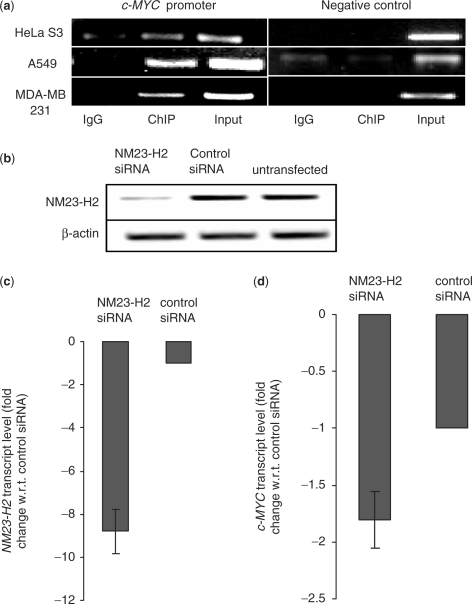
NM23-H2-mediated transactivation of the c-MYC promoter (reporter assay) via the NHE was noted several years back (38,39). Though binding of NM23-H2 to the c-MYC promoter was implicated in these studies it was not tested directly. We used ChIP to probe physical occupancy of NM23-H2 to c-MYC promoter in different human carcinoma cell lines: lung epithelial carcinoma (A549), cervical epithelial carcinoma (HeLa S3) and breast carcinoma (MDA-MB-231). Plasmid construct pCDNA3-NM23-H2 was used for expression of NM23-H2 (with Myc-epitope) in HeLa S3 and A549 cells and ChIP was performed using monoclonal anti-c-MYC antibody (from Sigma) or mouse IgG (mock precipitation). In MDA-MB-231 cells, rabbit polylclonal-nm23-H-antibody (from Santa Cruz) was used for ChIP to retrieve DNA sequences bound to endogenous NM23-H2. PCR amplification of c-MYC promoter shows significant enrichment in immunoprecipitated samples in all three cell lines compared to mock (IgG) precipitates (Figure 1a; input represents purified chromatin used as positive control). The reason for using anti-c-MYC antibody in ChIP experiments (in addition to the polyclonal nm23-H-antibody used for detecting endogenous NM23-H2 in MDA-MB-231 cells) was to immunoprecipitate specifically NM23-H2 bound chromatin fragments since the polyclonal nm23-H antibody recognizes both NM23-H2 and NM23-H1 isoforms. Furthermore, it has been demonstrated that the anti-c-MYC antibody (clone 9E 10) used by us does not immunoprecipitate endogenous c-MYC protein (44). Independently ChIP performed using polyclonal anti-nm23-H in A549 and HeLa S3 cells also gave similar results. Genomic loci taken as control showed no relative enrichment in ChIP (Figure 1a), suggesting enrichment for the c-MYC promoter was specific.
Figure 1.
NM23-H2 binds c-MYC promoter in vivo (a) ChIP assays using antibodies against NM23-H2 in multiple cell lines. Immunoprecipitated DNA samples were PCR amplified to show NM23-H2 occupancy of c-MYC promoter and negative control locus. Primer and locus information are given with ChIP methods; PCR cycles were maintained in the exponential phase of amplification. (b) Semi-quantitative RT-PCR analysis of NM23-H2 mRNA expression in A549 cells treated with siRNA duplex against NM23-H2 compared to scrambled (or control) siRNA and untransfected samples. β-actin mRNA levels remain unchanged. (c and d) Quantitative real-time PCR analysis. NM23-H2 (c) and c-MYC (d) transcript levels (fold change is relative to respective transcript levels determined in cells transfected with control siRNA) in A549 cells treated with NM23-H2 siRNA or control siRNA duplex. Experiments were performed in triplicate to analyze standard deviations in all cases.
Physical mapping of NM23-H2 to c-MYC promoter questions the regulatory aspects of this interaction. We asked whether perturbing levels of NM23-H2 would affect c-MYC expression. Small interfering RNA (siRNA from Santa Cruz) was used to silence NM23-H2 in A549 cells. Silencing was confirmed by semi-quantitative RT-PCR (Figure 1b) and quantitative RT-PCR (Figure 1c). Analysis of c-MYC transcript levels by quantitative RT-PCR after silencing of NM23-H2 revealed about 1.8-fold decrease in c-MYC levels (Figure 1d).
NM23-H2 binds G4 motif in vitro
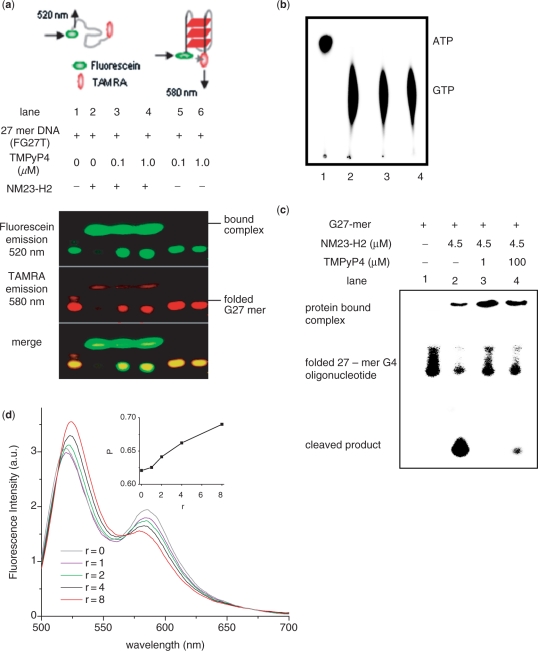
Prompted by our findings showing NM23-H2 localization on the c-MYC NHE we asked whether NM23-H2 binds to the G4 motif present within the c-MYC NHE. We checked for complex formation using recombinant NM23-H2 and the 27-mer oligonucleotide from c-MYC promoter NHE, which controls c-MYC expression and is known to adopt G4 motif in vitro (23). Fluorescence resonance energy transfer (FRET) was used to distinguish between the folded and unfolded state of the oligonucleotide (FG27T), labeled with fluorescein (donor) on one end and TAMRA (acceptor) on the other end, as described earlier (42). Reaction products were separated by native PAGE and detected by fluorescence to test whether the oligonucleotide in complex with protein was folded or unfolded. Experiments were done in the presence of 100-fold excess of nonspecific single- and dsDNA to minimize the chances of NM23-H2 binding nonspecifically. We observed primarily the folded form in the bound state on native PAGE (Figure 2a, lane 2). Interestingly, loss in energy transfer in the protein-bound complex indicated that the oligonucleotide was unfolded suggesting a G4 motif-resolving function of NM23-H2. Binding of the ligand TMPyP4 to the c-MYC promoter G4 motif has been reported earlier (23). We tested the effect of TMPyP4 on NM23-H2–G4 motif interaction; as expected, complex formation was inhibited by TMPyP4 (Figure 2a, lanes 3 and 4). Increase in energy transfer due to folding of the FG27T was independently observed in PAGE on increasing K+ ion concentration however, the effect in presence of Na+ was contrasting showing small or no change in FRET (Supplementary Figure 1).
Figure 2.
NM23-H2 binds to G4 motif in vitro. (a) Electrophoretic mobility shift of 27-mer oligonucleotide (FG27T, see Methods) from c-MYC promoter NHE in presence or absence of NM23-H2 and/or TMPyP4 in 5% native PAGE; protein bands were independently stained by Coomassie (data not shown). Top panel: schematic representation of energy transfer in unfolded and folded (parallel G4 motif) along with respective predominant emission wavelengths. (b) Protein function (kinase) activity of NM23-H2 was not affected in presence of TMPyP4. NM23-H2-mediated phosphorylation of GTP using [γ-32P]ATP was assayed in presence of TMPyP4. Lane 1: [γ-32P]ATP alone; lane 2: with NM23-H2; lanes 3 and 4: with NM23-H2 and 60 μM (lane 3) or 120 μM (lane 4) TMPyP4. (c) Nuclease activity of NM23-H2 is inhibited in presence of TMPyP4. Ten-nanomolar radiolabeled (along with 0.1 μM unlabeled) 27-mer oligonucleotide sequence (5′-TGG GGAGGGTGGGGAGGGTGG GGAAGG-3′) that forms G4 motif (23) in the c-MYC promoter NHE was incubated with 4.5 μM NM23-H2, either alone (lane 2), or with 1 μM (lane 3) or 100 μM (lane 4) TMPyP4 in presence of NM23-H2 in buffer [10 mM HEPES, 2 mM MgCl2, 20 mM KCl, 50 µg/ml BSA, 0.5 mM DTT (pH 7.4)]; lane 1: oligonucleotide only as control. All reactions in (b) and (c) had 60 μM single-strand poly(dA) as nonspecific control and TMPyP4 treatments were for 45 min followed by 3 h incubation with protein at 37°C. (d) FRET (detected in solution) of FG27T (50 nM) as a function of increasing concentration of NM23-H2 (r = NM23-H2/FG27T) in 20 mM K+; spectra were recorded following a 10-min incubation after each addition. Inset: Change in energy transfer (represented as P, determined from experimental FRET data) as a function of increasing (r); P = ID/(ID+IA), where ID and IA are average donor (fluorescein) and acceptor (TAMRA) intensities, respectively. Experiments were done in triplicate and for each value of P error of ± 0.07 arbitrary units was observed.
The inherent kinase activity of NM23-H2 was independently confirmed and found to be intact in presence of TMPyP4 (Figure 2b, lanes 3 and 4). NM23-H2 interaction with the c-MYC promoter G4 motif was independently checked using radiolabeled EMSA. Figure 2c, lane 1 shows the reported nuclease activity of the wild-type protein where protein–DNA complex formation was also detectable. In presence of TMPyP4, DNA cleavage was inhibited resulting in increased amount of complex (Figure 2c, lanes 3 and 4). In order to check whether the interaction of NM23-H2 was specific to the G4 motif formed by the single strand from the c-MYC NHE vis-à-vis the NHE duplex we looked at the NM23-H2-G4 motif interaction in presence of increasing amount of the complementary strand. This showed that on formation of duplex DNA the cleavage product mediated by NM23-H2 decreased indicating preferential binding of NM23-H2 with the single-stranded form (Supplementary Figure 2). Independent experiments were done with the FG27T oligonucleotide using CD and fluorescence emission in solution to establish loss in energy transfer once the motif is in the unfolded state (data not shown). We did not observe significant cleavage of the FRET labeled oligonucleotide in presence of NM23-H2 within the incubation times used for the PAGE experiments. Cleavage was observed when the duration was increased, this may be due to the end labeling of the oligonucleotide used for the PAGE experiments.
NM23-H2-G4 motif binding was also independently checked in solution by incubating FG27T with recombinant NM23-H2. FRET of FG27T in presence of NM23-H2 was detected in solution by monitoring change in donor emission intensity relative to total donor and acceptor emission, which was expressed as the ratio P (see ‘Methods’ section). Increase in P as a function of increasing NM23-H2/FG27T (r, Figure 2d) indicated decrease in energy transfer, supporting unfolding of the G4 motif in presence of NM23-H2. Conversely, as expected, increasing K+ ion concentration decreased P demonstrating the effect of K+ in stabilizing the folded G4 motif (Supplementary Figure 3). Taken together, this supports binding and also unfolding of the c-MYC promoter G4 motif by NM23-H2. In Figure 2d we noted that loss in energy transfer of the folded G4 motif was not complete [as expected from a fully open FG27T at r = 8; for reference see FRET of FG27T after melting (Supplementary Figure 4)] indicating only partial opening of the G4 motif in the protein-bound complex. An alternative explanation could be that the 5′ and 3′ ends of FG27T are in close proximity after unfolding and/or cleavage.
NM23-H2 interaction with c-MYC promoter G4 motif induces c-MYC expression
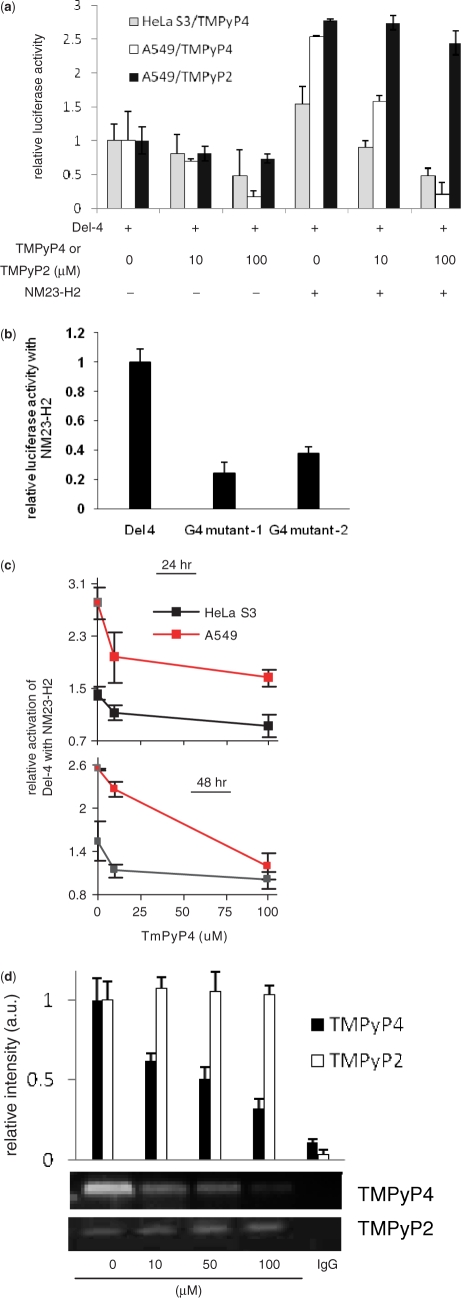
Using a luciferase reporter construct harboring the c-MYC promoter (Del-4) we first confirmed (under our experimental conditions) transactivation of the c-MYC promoter when transfected with NM23-H2 in HeLa S3 and A549 cells (Figure 3a). As noted before in A31 murine cells (39) increased c-MYC promoter activity was found in presence of NM23-H2 in both HeLa S3 and A549 cells (Figure 3a). Next, we asked whether formation of the reported G4 motif within the c-MYC promoter (NHE) had any role in NM23-H2-mediated promoter activity. To test this we used Del-4 based reporter constructs [G4-mutants 1 and 2 (GM1 and GM2); see ‘Methods’ section] that are unlikely to adopt the G4 motif. In an earlier study it was demonstrated that the GM1 construct harboring a single G to A substitution that destabilizes the G4 motif gives ∼3-fold over-expression of the c-MYC promoter in contrast to the repressor-like activity observed otherwise (23). In addition, we used the second plasmid construct GM2, where two G-repeats within the sequence were disrupted thereby making G4 motif formation unlikely (this was confirmed by CD, data not shown). As expected GM2 had ∼2.5-fold higher promoter activity compared to Del-4 (Supplementary Figure 5). Luciferase activity was measured for Del-4, GM1 and GM2 in presence and absence of NM23-H2. NM23-H2-mediated transactivation in case of GM1 and GM2 mutants were only ∼30% and 40%, respectively, of that noted for Del-4 (compared to untransfected control in each case; Figure 3b). Though it is difficult to rule out the possibility that influence of NM23-H2 is limited when the background reporter activity is inherently high (2- to 3-fold in case of the G4 mutants), taken together with other observations reported here these results support the role of NM23-H2-G4 motif interaction in the transactivation process.
Figure 3.
G4 motif is required for NM23-H2-mediated transactivation of c-MYC. (a) NM23-H2-mediated transactivation of c-MYC is inhibited in presence of TMPyP4, but not TMPyP2. Cells were transfected with Del-4 (reporter plasmid with c-MYC promoter) and treated with TMPyP4, TMPyP2, NM23-H2 expressing pCDNA3 plasmid or in combination as indicated (TMPyP4/TMPyP2 treatment was for 3 h, prior to transfection). Luciferase activity is after 48 h, normalized to Del-4 transfected untreated cells (not treated with TMPyP4/TMPyP2). (b) Disruption of G4 motif leads to decreased transactivation. NM23-H2-induced promoter activity of the reporter construct Del-4 and G4-mutant constructs (where specific substitutions that destabilize G4 motif formation were introduced within Del-4); luciferase activity is relative to NM23-H2 untransfected cells in each case. (c) Loss in transactivation in presence of TMPyP4, normalized to cells treated with same concentration of TMPyP4 but without NM23-H2. (d) ChIP with NM23-H2-specific antibody is affected in presence of TMPyP4 but not TMPyP2. Dose-dependent ChIP signal of the 165-bp c-MYC promoter NHE harboring the G4 motif sequence in HeLa S3 cells when treated with TMPyP4 or TMPyP2. IgG: ChIP using mouse IgG as nonspecific negative control.
Role of NM23-H2-G4 motif interaction in transcription of c-MYC was also tested using a second approach—by using a G4 motif-binding ligand. Suppression of c-MYC promoter activity in the presence of TMPyP4 has been observed before (23). We independently observed this in HeLa S3 and A549 cells where increasing amount of TMPyP4 led to decreased promoter activity (Figure 3a). Having established the independent effect of TMPyP4, we checked for NM23-H2-mediated transactivation in the presence of TMPyP4 with the reasoning that TMPyP4 binding to the folded G4 motif may inhibit interaction with NM23-H2. On the other hand, if the transactivation mechanism does not involve the G4 motif then effect of TMPyP4 is unlikely. Decrease in the transactivation potential of NM23-H2 with increasing TMPyP4 concentration was clearly observed in both, HeLa S3 and A549 cells at both 24 and 48 h after NM23-H2 transfection (Figure 3a and c). This supports the model of NM23-H2 interacting with G4 motif. Biochemical effects of TMPyP4 that underlie its binding to G4 motif (9,45), like inhibition of telomerase (46) and resulting biological effects of anaphase bridge formation and in vivo antitumor activity (47) are reasonably established. These effects were observed to be prominent with TMPyP4 and not its positional-isomer TMPyP2, which was deficient in binding to G4 motif but not dsDNA (23). Therefore, to further validate the specificity of above results observed in presence of TMPyP4, we used TMPyP2 as a negative control. We first observed that TMPyP2, in contrast to TMPyP4, had limited effect on suppression of c-MYC promoter activity in A549 cells in absence of NM23-H2 (Figure 3a). This was consistent with previous experiments demonstrating specific effect of TMPyP4 and not TMPyP2 in G4-motif-mediated suppression of c-MYC expression (48). In the presence of NM23-H2, we noted with interest that effect of TMPyP2 on c-MYC transactivation was marginal relative to that observed for TMPyP4 (Figure 3a). Taken together, these observations support a regulatory model suggesting binding of NM23-H2 specifically to the G4 motif intracellularly.
In this context, we further reasoned that G4 motif binding of NM23-H2 may be impeded by TMPyP4 in vivo and tested ChIP signal in presence of increasing TMPyP4. Diminished ChIP signals indicating decreased NM23-H2 occupancy of the c-MYC NHE in the presence of TMPyP4 (Figure 3d) was in line with this hypothesis, lending support to the transcriptional activation model that involves G4 motif binding of NM23-H2. Inhibition of NM23-H2-G4 motif interaction by TMPyP4 is interesting considering that TMPyP4-mediated stabilization of the G4 motif could, in effect, present a well-formed G4 structure for more efficient NM23-H2 binding. However, based on inhibition of NM23-H2-induced activation of c-MYC in presence of TMPyP4, it appears more likely that TMPyP4 distorts the G4 motif and/or occupies the binding site required for NM23-H2 binding. We used TMPyP2 as a negative control and observed that NM23-H2-ChIP was not dependent on TMPyP2 (Figure 3d). This supported specific G4 motif binding of TMPyP4 in vivo and also indicated that non-specific binding of TMPyP4 to dsDNA was unlikely. As a second control we used ChIP with NFkB (p65) antibody. ChIP, in this case, was not dependent on TMPyP4 (data not shown) indicating that nonspecific binding of TMPyP4 to dsDNA was unlikely.
NM23-H2 mutants lacking phosphotransferase or nuclease function retain c-MYC transactivation
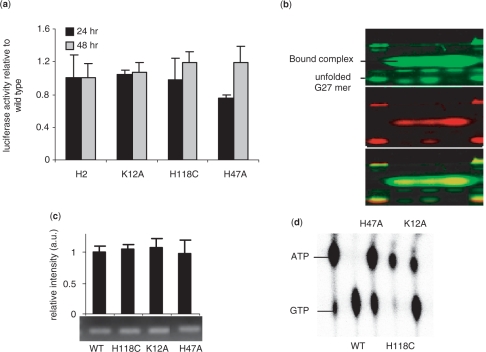
In addition to transcription, NM23-H2 has been attributed various other functions including suppression of metastases (49), reactivation of CD 4+T cells (50) and regulation of endocytosis of a G-protein-coupled receptor (51). Taken together, this makes NM23-H2 an intriguing multifunctional protein where interplay or ‘crosstalk’ between functions is an interesting mechanistic possibility. In this context, the connection between enzymatic phosphotransferase activity and transcription has been addressed before (52). We explored the interplay between transcription and endonuclease functions by expressing the endonuclease mutant NM23-H2K12A [characterized before (53)] in HeLa S3 cells and checked its potential to transactivate the c-MYC promoter. Transactivation of c-MYC promoter by NM23-H2K12A was comparable to that of wild-type NM23-H2 at both 24 and 48 h after transfection (Figure 4a). Next, we probed NM23-H2K12A for its ability to bind the G4 motif and observed significant loss of FRET in the DNA-bound complex (Figure 4b, lane 2) indicating a G4 motif-resolving function like the wild-type protein (shown in Figure 2a). We further confirmed in vivo occupancy of NM23-H2K12A on the c-MYC promoter. ChIP assay in HeLa S3 cells revealed that in vivo c-MYC promoter occupancy by NM23-H2K12A was similar to that of NM23-H2WT (Figure 4c). The mutant NM23-H2H118C which lacks phosphotransferase activity is known to retain its transcription and DNA-binding activity (54). Likewise, our luciferase results using the Del-4 construct in HeLa S3 cells also showed c-MYC promoter transactivation that is similar to NM23-H2WT (Figure 4a). We further probed the interaction of NM23-H2H118C interaction with the G4 motif. Figure 4b (lane 3) shows complex formation of NM23-H2H118C with the G4 motif was similar to the wild-type and K12A mutant. This indicated that a form of NM23-H2 devoid of phosphorylation resolved the G4 motif like the wild-type factor. In this case also ChIP indicated promoter occupancy by NM23-H2H118C was similar to that of the wild-type protein (Figure 4c). Based on these, we conclude that the endonuclease and phosphotransferase activities of NM23-H2 are independent of its transcriptional regulatory activity. NM23-H2 mutants, H118C and K12A have been well characterized (52,53), we confirmed the reported phosphotransferase activities under our experimental conditions for both mutants (Figure 4d). The DNA cleavage activity of the mutants was also independently confirmed to observe complete abrogation in case of K12A mutant, while H118C mutant showed no appreciable change in DNA cleavage relative to wildtype (data not shown).
Figure 4.
Effect of NM23-H2 mutants on c-MYC transactivation. (a and b) Luciferase reporter activity of the Del-4 plasmid on co-transfection with plasmids expressing NM23-H2 (labeled as H2) and respective mutants in HeLa S3 cells (a); PAGE of complex formation of FG27T with NM23-H2 mutants detected by fluorescence (b) (lanes 1 and 5: FG27T alone; lane 2: K12A; lane 3: H118C; lane 4: H47A); (c) Enrichment of the 165-bp c-MYC promoter fragment in ChIP with NM23-H2-specific antibody for NM23-H2 wild-type (WT) and each mutant. (d) Phosphotransferase assay of mutants showing decreased phosphotransfer relative to WT activity in case of H47A; activities as shown for H118C and K12A mutants were reported earlier (52,53).
Phosphorylated NM23-H2 is deficient in c-MYC transactivation
We noted that though the un-phosphorylated form (NM23-H2H118C) had been studied extensively, role of a phosphorylated form of NM23-H2 had not been investigated. To test the role of phosphorylated NM23-H2 in transactivation we developed the mutant NM23-H2H47A based on the mutant homolog reported from Mycobacterium tuberculosis, which was deficient in transphosphorylation while autophosphorylation was intact (55). This results in a phosphorylated protein that is trapped with phosphate on the histidine-118 residue. Limited phosphate transfer resulting in relatively lower amount of GTP in case of human NM23-H2H47A with respect to the wild type activity was confirmed (Figure 4d, lane 3). NM23-H2H47A was transfected into HeLa S3 cells to investigate the role of phosphorylated form in transactivation. Interestingly, NM23-H2H47A showed decreased transactivation of the c-MYC promoter at 24 h (Figure 4a), which was not due to decreased in vitro DNA binding (Figure 4b, lane 4 shows substantial amount of retarded band) or in vivo promoter occupancy (ChIP, Figure 4c). Wild-type-like transactivation observed at 48 h (Figure 4a) may be due to slow loss of the protein-bound phosphate as the recombinant NM23-H2H47A showed loss of phosphate with time under in vitro conditions (data not shown). We also noted that the DNA complex with the phosphorylated form appeared to show more energy transfer (retarded band is predominantly green in Figure 4b, lane 4) relative to wild-type and other mutants. Though further experiments will be required to understand the role of phosphorylation in this respect, it is interesting to note that phosphorylated NM23-H2, in principle, mechanistically connects the enzymatic phosphotransferase function of multifunctional NM23-H2 with its transcriptional role.
DISCUSSION
Results presented here show transactivation of c-MYC induced by NM23-H2 could be due to binding of NM23-H2 to the G4 motif within c-MYC promoter. This is supported by in vitro and in vivo (ChIP) interaction of NM23-H2-G4 motif and decreased transactivation observed on specifically destabilizing the G4 motif. In addition, reduced transactivation and intracellular DNA binding (ChIP) in presence of TMPyP4, and not TMPyP2, argues for G4 motif-specific binding by NM23-H2 as a plausible mode of c-MYC activation. Though several proteins are known to interact with G4 motifs (56) in vivo interaction of the G4 motif with a regulatory factor and the role of this interaction in transcription was not reported earlier. Recent biochemical results showing binding of homodimeric MyoD to tetraplex motifs present in promoter sequences of the muscle-specific genes alpha7 integrin and sarcomeric Mitochondrial Creatine Kinase (sMtCK) implicates a similar model in relation to regulation of muscle-specific genes (57). Selection of NM23-H2 as the putative factor to test G4 motif mediated transcription needs mention as NM23-H2 has been attributed with several diverse functionalities including kinase activity, promoter-binding and transcription, DNA cleavage and DNA repair (58). Several previous models have implicated the possibility of G4 motif binding to NM23-H2, wherein NM23-H2 binding to paranemic forms of DNA (G4 and i-motif) present in the c-MYC promoter has been postulated (23,59). This, in addition to the observation that NM23-H2 binds the c-MYC promoter NHE, and mutations within the NHE alter NM23-H2 mediated regulation of c-MYC in Burkitt's Lymphoma (60), make NM23-H2 an interesting first candidate in the context of our study though several other factors (like SP1, CNBP, hnRNP K) that bind G-rich sequences would be equally interesting to study.
In order to discern that transcriptional activation was due to NM23-H2 binding to G4 motif and not dsDNA several complementary approaches were considered. First, base substitutions that specifically disrupt the G4 motif showed much decreased NM23-H2-mediated transactivation in two independent cases. Though possible, it is unlikely that NM23-H2 binding to dsDNA would be significantly perturbed due to these substitutions. Second, we used a G4 motif-binding ligand TMPyP4 as a probe for the folded motif. Several studies have established intracellular G4 motif-binding of TMPyP4 in telomerase inhibition and as a repressor of c-MYC (23,61). It was demonstrated that at 100 μM TMPyP4, effect of the ligand on c-MYC repression was abrogated when specific base substitutions that disrupted G4 motif structure were incorporated within the promoter—any effect due to dsDNA binding of TMPyP4 is expected to be retained, particularly at such high concentrations (23). We observed loss in NM23-H2-induced transactivation of c-MYC in presence of TMPyP4. This indicated inhibition of NM23-H2 binding by TMPyP4 suggesting a G4 motif-NM23-H2 interaction. Third, we employed TMPyP2, which has significantly lower affinity for the G4 motif compared to TMPyP4; however, both ligands have similar affinity for dsDNA. Unlike TMPyP4, loss in transactivation of c-MYC was not evident in presence of TMPyP2. In addition, ChIP experiments in presence of TMPyP2 showed unperturbed binding of NM23-H2 to the c-MYC NHE, in contrast to that noted for TMPyP4. These results argue against dsDNA binding of NM23-H2 inside cells, which was expected to show similar effects in presence of both ligands. However, we found that TMPyP4/TMPyP2 inhibited nuclease activity of NM23-H2 in a similar fashion in vitro. A possible reason may be nonspecific/low affinity binding of TMPyP2 to G4 DNA; an interaction that is expected to be limited inside cells due to the presence of excess dsDNA. Finally, we observed specific NM23-H2-G4 motif binding in vitro, in presence of excess non-specific single strand and dsDNA. In further support of our findings, it has been noted by others that recombinant NM23-H2 binds to single-stranded but not the double stranded form of the telomeric repeat sequence (62).
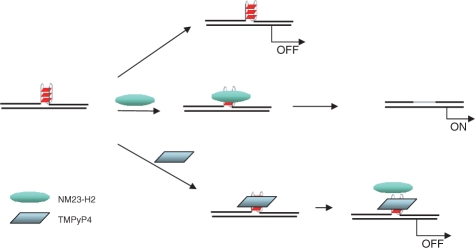
Together with other observations (including inhibitory effect of TMPyP4 and not TMPyP2 on NM23-H2-mediated activation of c-MYC), reduced transactivation of c-MYC by NM23-H2 on disruption of the G4 motif presents the G4 motif as a structural element necessary for NM23-H2-mediated activation of c-MYC. In contrast, earlier studies (23) and also observations made by us (for GM2) show that disruption of the promoter G4 motif leads to increased expression of c-MYC, in absence of NM23-H2. This is supported by TMPyP4-induced stabilization of the G4 motif resulting in reduced promoter activity, thereby establishing the G4 motif as a suppressor of c-MYC expression. This makes the contrasting role of the c-MYC-promoter G4 motif in presence of NM23-H2 evident. Findings reported here support a model implicating the structural G4 motif (which is a repressor) as a binding site for NM23-H2. Binding of NM23-H2 may remove (possibly by unwinding) the G4 motif leading to c-MYC activation. In Figure 5 we propose a model summarizing above aspects, which builds on the previously reported ‘repressor-like’ influence of the G4 motif and takes into account observations made in this study showing the negative influence of both, G4 motif-disruption and TMPyP4-binding, on NM23-H2-mediated activation of c-MYC expression. Direct interaction of NM23-H2 with the folded G4 motif noted in vitro help in further validating this regulatory model. It is also possible that in addition to removal of the G4 motif (repressor) NM23-H2 binding recruits other ancillary factors for transcriptional activation.
Figure 5.
Proposed regulatory model for NM23-H2-G4 motif interaction in c-MYC transcription activation. Repressive influence of the G4 motif (noted earlier) is transformed in to activation of c-MYC expression on NM23-H2 binding to the G4 motif, which could be due to removal of the repressor (G4 motif) and/or recruitment of ancillary transcriptional activators. TMPyP4-binding to the G4 motif inhibits NM23-H2-G4 motif interaction possibly by disrupting the G4 motif structure.
Our observations suggest NM23-H2 as an activator of c-MYC expression whereby we note decreased levels of NM23-H2 give lower c-MYC levels. On the other hand, decreased expression of NM23-H2 has been associated with enhanced metastatic potential (63,64). In this context, it is interesting to consider the effect of NM23-H2 on c-MYC expression. Decreased expression of c-MYC induces cell cycle arrest and apoptosis in many cell types including the lung epithelial carcinoma, A549 cells, where cells appear to halt at G1/S transition and adopt characteristics of apoptosis (65,66). It is also known that activation of apoptosis significantly reduces the outcome of metastatic spread (67). Therefore, decreased NM23-H2 levels in cancer cells, which in turn would result in reduced c-MYC expression is expected to decrease apoptosis of cancer cells endowing them with enhanced metastatic potential. Our observations are preliminary in this context and warrant detailed experiments to investigate the role of NM23-H2 in apoptosis mediated metastases suppression.
In conclusion, based on observations presented herein it is tempting to speculate the functional role of G4 motifs as structural recognition elements for NM23-H2 or other similar factors. Alteration in chromatin leading to formation/deformation of G4 motifs may lead to transcriptional regulation thereby directly linking chromosomal structure to specific gene regulatory events. Notably, supercoiling-dependent single-stranded non-B-DNA forms have been demonstrated to regulate several operons in E. coli (1). Further support for structure-mediated regulatory control comes from recent findings showing formation of non-B-DNA structures could directly result from supercoiling induced during transcription where the non-B structural forms, in turn, recruit structure-specific regulatory factors for specific gene regulatory control (68). Genome-wide promoter occupancy of NM23-H2 and its role in gene expression in tandem with other intracellular [several proteins have been implicated as G4 motif-binding (56)] and extracellular G4 motif-binding molecules (9) will be required to better understand the regulatory implications of G4 motifs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Fellowships from CSIR (to P.K., K.H., A.K., and A.V); UGC (to R.K.T.); DBT (to An.K.); and research grants: CSIR (IAP 001) and DBT (PR6752) to S.C. Funding for Open Access Publication charges were waived by Oxford University Press.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Chowdhury lab and Munia Ganguli for discussion and comments on the manuscript. The authors wish to thank anonymous referees for comments/suggestions that helped in improving the manuscript.
REFERENCES
- 1.Hatfield GW, Benham CJ. DNA topology-mediated control of global gene expression in Escherichia coli. Annu. Rev. Genet. 2002;36:175–203. doi: 10.1146/annurev.genet.36.032902.111815. [DOI] [PubMed] [Google Scholar]
- 2.Bacolla A, Wells RD. Non-B DNA conformations, genomic rearrangements, and human disease. J. Biol. Chem. 2004;279:47411–47414. doi: 10.1074/jbc.R400028200. [DOI] [PubMed] [Google Scholar]
- 3.Rich A, Zhang S. Timeline: Z-DNA: the long road to biological function. Nat. Rev. Genet. 2003;4:566–572. doi: 10.1038/nrg1115. [DOI] [PubMed] [Google Scholar]
- 4.Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 5.Balagurumoorthy P, Brahmachari SK. Structure and stability of human telomeric sequence. J. Biol. Chem. 1994;269:21858–21869. [PubMed] [Google Scholar]
- 6.Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 7.Parkinson GN, Lee MP, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 8.Incles CM, Schultes CM, Kempski H, Koehler H, Kelland LR, Neidle S. A G-quadruplex telomere targeting agent produces p16-associated senescence and chromosomal fusions in human prostate cancer cells. Mol. Cancer Ther. 2004;3:1201–1206. [PubMed] [Google Scholar]
- 9.Neidle S, Read MA. G-quadruplexes as therapeutic targets. Biopolymers. 2000;56:195–208. doi: 10.1002/1097-0282(2000)56:3<195::AID-BIP10009>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 10.Chang CC, Chu JF, Kao FJ, Chiu YC, Lou PJ, Chen HC, Chang TC. Verification of antiparallel G-quadruplex structure in human telomeres by using two-photon excitation fluorescence lifetime imaging microscopy of the 3,6-Bis(1-methyl-4-vinylpyridinium)carbazole diiodide molecule. Anal. Chem. 2006;78:2810–2815. doi: 10.1021/ac052218f. [DOI] [PubMed] [Google Scholar]
- 11.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 13.Howell RM, Woodford KJ, Weitzmann MN, Usdin K. The chicken beta-globin gene promoter forms a novel ‘cinched’ tetrahelical structure. J. Biol. Chem. 1996;271:5208–5214. doi: 10.1074/jbc.271.9.5208. [DOI] [PubMed] [Google Scholar]
- 14.Murchie AI, Lilley DM. Retinoblastoma susceptibility genes contain 5′ sequences with a high propensity to form guanine-tetrad structures. Nucleic Acids Res. 1992;20:49–53. doi: 10.1093/nar/20.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Catasti P, Chen X, Moyzis RK, Bradbury EM, Gupta G. Structure-function correlations of the insulin-linked polymorphic region. J. Mol. Biol. 1996;264:534–545. doi: 10.1006/jmbi.1996.0659. [DOI] [PubMed] [Google Scholar]
- 16.Kilpatrick MW, Torri A, Kang DS, Engler JA, Wells RD. Unusual DNA structures in the adenovirus genome. J. Biol. Chem. 1986;261:11350–11354. [PubMed] [Google Scholar]
- 17.Ma D, Xing Z, Liu B, Pedigo NG, Zimmer SG, Bai Z, Postel EH, Kaetzel DM. NM23-H1 and NM23-H2 repress transcriptional activities of nuclease-hypersensitive elements in the platelet-derived growth factor-A promoter. J. Biol. Chem. 2002;277:1560–1567. doi: 10.1074/jbc.M108359200. [DOI] [PubMed] [Google Scholar]
- 18.Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cogoi S, Xodo LE. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006;34:2536–2549. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cogoi S, Paramasivam M, Spolaore B, Xodo LE. Structural polymorphism within a regulatory element of the human KRAS promoter: formation of G4-DNA recognized by nuclear proteins. Nucleic Acids Res. 2008;36:3765–3780. doi: 10.1093/nar/gkn120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palumbo SL, Memmott RM, Uribe DJ, Krotova-Khan Y, Hurley LH, Ebbinghaus SW. A novel G-quadruplex-forming GGA repeat region in the c-myb promoter is a critical regulator of promoter activity. Nucleic Acids Res. 2008;36:1755–1769. doi: 10.1093/nar/gkm1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simonsson T, Pecinka P, Kubista M. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998;26:1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao Y, Du Z, Li N. Extensive selection for the enrichment of G4 DNA motifs in transcriptional regulatory regions of warm blooded animals. FEBS Lett. 2007;581:1951–1956. doi: 10.1016/j.febslet.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 26.Yadav VK, Abraham JK, Mani P, Kulshrestha R, Chowdhury S. QuadBase: genome-wide database of G4 DNA—occurrence and conservation in human, chimpanzee, mouse and rat promoters and 146 microbes. Nucleic Acids Res. 2008;36:D381–D385. doi: 10.1093/nar/gkm781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rawal P, Kummarasetti VB, Ravindran J, Kumar N, Halder K, Sharma R, Mukerji M, Das SK, Chowdhury S. Genome-wide prediction of G4 DNA as regulatory motifs: role in Escherichia coli global regulation. Genome Res. 2006;16:644–655. doi: 10.1101/gr.4508806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du Z, Zhao Y, Li N. Genome-wide analysis reveals regulatory role of G4 DNA in gene transcription. Genome Res. 2008;18:233–241. doi: 10.1101/gr.6905408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin Y, Rezler EM, Gokhale V, Sun D, Hurley LH. Characterization of the G-quadruplexes in the duplex nuclease hypersensitive element of the PDGF-A promoter and modulation of PDGF-A promoter activity by TMPyP4. Nucleic Acids Res. 2007;35:7698–7713. doi: 10.1093/nar/gkm538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Etzioni S, Yafe A, Khateb S, Weisman-Shomer P, Bengal E, Fry M. Homodimeric MyoD preferentially binds tetraplex structures of regulatory sequences of muscle-specific genes. J. Biol. Chem. 2005;280:26805–26812. doi: 10.1074/jbc.M500820200. [DOI] [PubMed] [Google Scholar]
- 31.Yafe A, Etzioni S, Weisman-Shomer P, Fry M. Formation and properties of hairpin and tetraplex structures of guanine-rich regulatory sequences of muscle-specific genes. Nucleic Acids Res. 2005;33:2887–2900. doi: 10.1093/nar/gki606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palmieri D, Horak CE, Lee JH, Halverson DO, Steeg PS. Translational approaches using metastasis suppressor genes. J. Bioenerg. Biomembr. 2006;38:151–161. doi: 10.1007/s10863-006-9039-9. [DOI] [PubMed] [Google Scholar]
- 33.Lascu I, Gonin P. The catalytic mechanism of nucleoside diphosphate kinases. J. Bioenerg. Biomembr. 2000;32:237–246. doi: 10.1023/a:1005532912212. [DOI] [PubMed] [Google Scholar]
- 34.Lu Q, Zhang X, Almaula N, Mathews CK, Inouye M. The gene for nucleoside diphosphate kinase functions as a mutator gene in Escherichia coli. J. Mol. Biol. 1995;254:337–341. doi: 10.1006/jmbi.1995.0620. [DOI] [PubMed] [Google Scholar]
- 35.Izumiya H, Yamamoto M. Cloning and functional analysis of the ndk1 gene encoding nucleoside-diphosphate kinase in Schizosaccharomyces pombe. J. Biol. Chem. 1995;270:27859–27864. doi: 10.1074/jbc.270.46.27859. [DOI] [PubMed] [Google Scholar]
- 36.Hildebrandt M, Lacombe ML, Mesnildrey S, Veron M. A human NDP-kinase B specifically binds single-stranded poly-pyrimidine sequences. Nucleic Acids Res. 1995;23:3858–3864. doi: 10.1093/nar/23.19.3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Postel EH, Weiss VH, Beneken J, Kirtane A. Mutational analysis of NM23-H2/NDP kinase identifies the structural domains critical to recognition of a c-myc regulatory element. Proc. Natl Acad. Sci. USA. 1996;93:6892–6897. doi: 10.1073/pnas.93.14.6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Postel EH, Berberich SJ, Flint SJ, Ferrone CA. Human c-myc transcription factor PuF identified as nm23-H2 nucleoside diphosphate kinase, a candidate suppressor of tumor metastasis. Science. 1993;261:478–480. doi: 10.1126/science.8392752. [DOI] [PubMed] [Google Scholar]
- 39.Berberich SJ, Postel EH. PuF/NM23-H2/NDPK-B transactivates a human c-myc promoter-CAT gene via a functional nuclease hypersensitive element. Oncogene. 1995;10:2343–2347. [PubMed] [Google Scholar]
- 40.Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 2006;1:179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- 41.Larionov A, Krause A, Miller W. A standard curve based method for relative real time PCR data processing. BMC. Bioinformatics. 2005;6:62. doi: 10.1186/1471-2105-6-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar P, Verma A, Saini AK, Chopra P, Chakraborti PK, Singh Y, Chowdhury S. Nucleoside diphosphate kinase from Mycobacterium tuberculosis cleaves single strand DNA within the human c-myc promoter in an enzyme-catalyzed reaction. Nucleic Acids Res. 2005;33:2707–2714. doi: 10.1093/nar/gki568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verma A, Halder K, Halder R, Yadav VK, Rawal P, Thakur RK, Mohd F, Sharma A, Chowdhury S. Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J. Med. Chem. 2008;51:5641–5649. doi: 10.1021/jm800448a. [DOI] [PubMed] [Google Scholar]
- 44.Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phan AT, Kuryavyi V, Gaw HY, Patel DJ. Small-molecule interaction with five-guanine-tract G-quadruplex structure from the human MYC promoter. Nat. Chem. Biol. 2005;1:167–173. doi: 10.1038/nchembio723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shammas MA, Shmookler Reis RJ, Akiyama M, Koley H, Chauhan D, Hideshima T, Goyal RK, Hurley LH, Anderson KC, Munshi NC. Telomerase inhibition and cell growth arrest by G-quadruplex interactive agent in multiple myeloma. Mol. Cancer Ther. 2003;2:825–833. [PubMed] [Google Scholar]
- 47.Izbicka E, Wheelhouse RT, Raymond E, Davidson KK, Lawrence RA, Sun D, Windle BE, Hurley LH, Von Hoff DD. Effects of cationic porphyrins as G-quadruplex interactive agents in human tumor cells. Cancer Res. 1999;59:639–644. [PubMed] [Google Scholar]
- 48.Grand CL, Han H, Munoz RM, Weitman S, Von Hoff DD, Hurley LH, Bearss DJ. The cationic porphyrin TMPyP4 down-regulates c-MYC and human telomerase reverse transcriptase expression and inhibits tumor growth in vivo. Mol. Cancer Ther. 2002;1:565–573. [PubMed] [Google Scholar]
- 49.Steeg PS, Bevilacqua G, Kopper L, Thorgeirsson UP, Talmadge JE, Liotta LA, Sobel ME. Evidence for a novel gene associated with low tumor metastatic potential. J. Natl Cancer Inst. 1988;80:200–204. doi: 10.1093/jnci/80.3.200. [DOI] [PubMed] [Google Scholar]
- 50.Srivastava S, Li Z, Ko K, Choudhury P, Albaqumi M, Johnson AK, Yan Y, Backer JM, Unutmaz D, Coetzee WA, et al. Histidine phosphorylation of the potassium channel KCa3.1 by nucleoside diphosphate kinase B is required for activation of KCa3.1 and CD4 T cells. Mol. Cell. 2006;24:665–675. doi: 10.1016/j.molcel.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 51.Rochdi MD, Laroche G, Dupre E, Giguere P, Lebel A, Watier V, Hamelin E, Lepine MC, Dupuis G, Parent JL. Nm23-H2 interacts with a G protein-coupled receptor to regulate its endocytosis through an Rac1-dependent mechanism. J. Biol. Chem. 2004;279:18981–18989. doi: 10.1074/jbc.M312621200. [DOI] [PubMed] [Google Scholar]
- 52.Postel EH, Ferrone CA. Nucleoside diphosphate kinase enzyme activity of NM23-H2/PuF is not required for its DNA binding and in vitro transcriptional functions. J. Biol. Chem. 1994;269:8627–8630. [PubMed] [Google Scholar]
- 53.Postel EH, Abramczyk BM, Levit MN, Kyin S. Catalysis of DNA cleavage and nucleoside triphosphate synthesis by NM23-H2/NDP kinase share an active site that implies a DNA repair function. Proc. Natl Acad. Sci. USA. 2000;97:14194–14199. doi: 10.1073/pnas.97.26.14194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Postel EH, Abramczyk BA, Gursky SK, Xu Y. Structure-based mutational and functional analysis identify human NM23-H2 as a multifunctional enzyme. Biochemistry. 2002;41:6330–6337. doi: 10.1021/bi025606+. [DOI] [PubMed] [Google Scholar]
- 55.Tiwari S, Kishan KV, Chakrabarti T, Chakraborti PK. Amino acid residues involved in autophosphorylation and phosphotransfer activities are distinct in nucleoside diphosphate kinase from Mycobacterium tuberculosis. J. Biol. Chem. 2004;279:43595–43603. doi: 10.1074/jbc.M401704200. [DOI] [PubMed] [Google Scholar]
- 56.Fry M. Tetraplex DNA and its interacting proteins. Front. Biosci. 2007;12:4336–4351. doi: 10.2741/2391. [DOI] [PubMed] [Google Scholar]
- 57.Shklover J, Etzioni S, Weisman-Shomer P, Yafe A, Bengal E, Fry M. MyoD uses overlapping but distinct elements to bind E-box and tetraplex structures of regulatory sequences of muscle-specific genes. Nucleic Acids Res. 2007;35:7087–7095. doi: 10.1093/nar/gkm746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Postel EH. Multiple biochemical activities of NM23/NDP kinase in gene regulation. J. Bioenerg. Biomembr. 2003;35:31–40. doi: 10.1023/a:1023485505621. [DOI] [PubMed] [Google Scholar]
- 59.Postel EH, Berberich SJ, Rooney JW, Kaetzel DM. Human NM23/nucleoside diphosphate kinase regulates gene expression through DNA binding to nuclease-hypersensitive transcriptional elements. J. Bioenerg. Biomembr. 2000;32:277–284. doi: 10.1023/a:1005541114029. [DOI] [PubMed] [Google Scholar]
- 60.Ji L, Arcinas M, Boxer LM. The transcription factor, Nm23H2, binds to and activates the translocated c-myc allele in Burkitt's lymphoma. J. Biol. Chem. 1995;270:13392–13398. doi: 10.1074/jbc.270.22.13392. [DOI] [PubMed] [Google Scholar]
- 61.Cuesta J, Read MA, Neidle S. The design of G-quadruplex ligands as telomerase inhibitors. Mini. Rev. Med. Chem. 2003;3:11–21. doi: 10.2174/1389557033405502. [DOI] [PubMed] [Google Scholar]
- 62.Nosaka K, Kawahara M, Masuda M, Satomi Y, Nishino H. Association of nucleoside diphosphate kinase nm23-H2 with human telomeres. Biochem. Biophys. Res. Commun. 1998;243:342–348. doi: 10.1006/bbrc.1997.8097. [DOI] [PubMed] [Google Scholar]
- 63.Miyazaki H, Fukuda M, Ishijima Y, Takagi Y, Iimura T, Negishi A, Hirayama R, Ishikawa N, Amagasa T, Kimura N. Overexpression of nm23-H2/NDP kinase B in a human oral squamous cell carcinoma cell line results in reduced metastasis, differentiated phenotype in the metastatic site, and growth factor-independent proliferative activity in culture. Clin. Cancer Res. 1999;5:4301–4307. [PubMed] [Google Scholar]
- 64.Hartsough MT, Steeg PS. Nm23/nucleoside diphosphate kinase in human cancers. J. Bioenerg. Biomembr. 2000;32:301–308. doi: 10.1023/a:1005597231776. [DOI] [PubMed] [Google Scholar]
- 65.Zajac-Kaye M. Myc oncogene: a key component in cell cycle regulation and its implication for lung cancer. Lung Cancer. 2001;34(Suppl. 2):S43–S46. doi: 10.1016/s0169-5002(01)00343-9. [DOI] [PubMed] [Google Scholar]
- 66.Hoffman B, Liebermann DA. The proto-oncogene c-myc and apoptosis. Oncogene. 1998;17:3351–3357. doi: 10.1038/sj.onc.1202592. [DOI] [PubMed] [Google Scholar]
- 67.Mehlen P, Puisieux A. Metastasis: a question of life or death. Nat. Rev. Cancer. 2006;6:449–458. doi: 10.1038/nrc1886. [DOI] [PubMed] [Google Scholar]
- 68.Kouzine F, Sanford S, Elisha-Feil Z, Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat. Struct. Mol. Biol. 2008;15:146–154. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.