Abstract
Uncovering the nature of communication between enhancers, promoters and insulators is important for understanding the fundamental mechanisms that ensure appropriate gene expression levels. Here we describe an approach employing transient expression of genetic luciferase reporter gene constructs with quantitative RT–PCR analysis of transcription between an enhancer and Hsp70 promoter. We tested genetic constructs containing gypsy and/or Fab7 insulators in different orientations, and an enhancer from copia LTR-retroelement [(enh)copia]. A single gypsy or Fab7 insulator inserted between the promoter and enhancer in any polarity reduced enhancer action. A pair of insulators flanking the gene in any orientation exhibited increased insulation activity. We detected promoter-independent synthesis of non-coding RNA in the intergenic region of the constructs, which was induced by the enhancer in both directions and repressed by a single insulator or a pair of insulators. These results highlight the involvement of RNA-tracking mechanisms in the communications between enhancers and promoters, which are inhibited by insulators.
INTRODUCTION
Insulators are DNA elements bound by protein complexes that subsequently interrupt inappropriate interactions between neighboring chromosomal domains (1–3). Thus, insulators are important in establishing or maintaining epigenetic structures and regulating transcription in eukaryotes. Two main classes of insulators have been described: enhancer-blocking insulators, which protect promoters from activation by inappropriate enhancers, and barrier insulators, which protect active genes from repression by spreading heterochromatin structures (3).
The gypsy insulator has 12 binding sites for the Su(Hw) protein, which in turn binds with the CP190, mod(mdg4) and Topors proteins (4,5); this binding targets the gypsy insulator-containing region to the so-called insulator bodies (6–9). Fab7 is an insulator located in the bithorax complex between the iab-6 and iab-7 enhancers that control expression of the Abdominal-B gene in parasegments PS11 and PS12 (10–12). Disruption of Fab7 leads to incorrect gene activation of the gene in inappropriate cells, and results in the homeotic phenotype in the adult fly. Several types of plentiful genomic insulators may be scattered in the Drosophila genome to determine the formation of higher-order chromatin structures (8,13), and this complex pattern of insulator distribution forms an ‘insulator code’ that shapes the pattern of independent chromosomal domains.
To understand how insulators affect enhancer function it is important to know how enhancers communicate with promoters. Two models have been suggested to explain the action of insulators. The first ‘transcriptional model’ or ‘RNA tracking model’ suggests that enhancers are the initial binding sites for transcription factors, which then directly interact with the transcription complex either by a looping mechanism (14) or by a transferring from enhancer to promoter along the chromatin fiber (1,2). According to this model, insulators function as the competing targets or traps for enhancers bound by transcriptions factors. An alternative hypothesis, the ‘structural model’, proposes that the primary function of insulators is the formation of transcriptionally independent domains in which promoters are accessible only to internal enhancers (7). The current data confirm the RNA tracking model. In the human β-globin locus, the CTDF insulator has been demonstrated to act as an enhancer blocker, inhibiting promoter remodeling and transcription activation only when inserted between the enhancer and the promoter (15). Enhancer blocking also leads to accumulation of RNA polymerase II at the HS2 enhancer and within the insulator, and to reduced detection at the promoter. A recent study reported that the human ɛ-globin locus HS2 enhancer binds with RNA polymerase II and TBP, and that this complex tracks along the intervening DNA, synthesizing intergenic RNAs (16). Thus, the enhancer delivers RNA polymerase and TBP to the promoter. The insulator inserted between the enhancer and the promoter traps the enhancer complex, blocking the facilitated tracking and transcription mechanism of the enhancer complex mid-stream.
It may follow that the study of insulators using transgenic constructs cannot account for enhancers or insulators in the genomic context that interfere with the results. At the sites where genetic constructs integrate, insulators in the sequences flanking the construct could interact with the construct's regulatory elements and disrupt its expected effect. For example, the presence of two or three copies of insulators may inhibit enhancer blocking or even strengthen activation by the enhancer (1,17–19). Moreover, germ-line transformation mediated by P-elements mainly targets constructs to open chromatin regions containing enhancers and insulators (19,20), a mechanism exploited for the so-called enhancer trap screens (21). The challenges associated with current models of insulator action may arise from these complex interactions (2). To overcome the effects of the host chromatin surrounding transgenes on transgene expression, it was suggested to flank transgenes with insulators (2,22), to use targeted integration of transgenes by homologous recombination (23), or to insert specific ‘landing sites’ for transgenes integration (24). To overcome these complexities, we simplified the approach by transfecting genetic constructs into Drosophila cultured cells.
To test the hypothesis that Drosophila enhancers communicate with promoters via intergenic transcription towards the promoter, and that insulators function in blocking this interaction (2,25), we used quantitative RT–PCR to monitor transcription in different regions of transfected genetic constructs. Our data on linear and circular constructs indicate that enhancers could induce transcription in both directions, and that this transcription is inhibited by a single insulator or a pair of insulators. The level of detected synthesis of this non-coding RNA correlates with the expression of a reporter luciferase gene. These results support the RNA-tracking model of enhancer-promoter communication.
MATERIALS AND METHODS
Basic vector and e–h construct
The basic construct was prepared by insertion of an Hsp 70 promoter into the BglII site of the pGL3-Enhancer vector (Promega, AC #U47297) digested with BglII and blunted by a Klenow fragment of DNA polymerase I. The actin 5C 3′ trailer and poly(A) were then inserted, and the vector was digested with XbaI and blunted by a Klenow fragment of DNA polymerase I. The Hsp70 promoter and the 3′ trailer were amplified from pCaSpeR-hs/act DNA (AC# U60735). Oligos used to prepare the inserts were as follows: Hsp70: 5′- cccaagcttCCAATTCCCTATTCAGAGTT-3′ and 5′-cccgaattcCAATTCCCTATTCAGAGTT–3′; and 3′act-5C: 5′ccctctagaAGCCAAGTGTGAGTGTGTGTGGG-3′ and 5′-cccggatccGACCATGAAGATCAAGATCATT-3′ (artificial restriction sites are shown in lowercase). The final basic vector and its derivatives were confirmed by sequencing, and the schematic of the basic construct is shown in Figure 1A. The sites of insulator insertion are indicated.
Figure 1.
Design of the constructs with a single or pairs of gypsy insulators and results of co-transfection experiments. (A) Schematic presentation of the constructs (not to scale). The basic construct carries an Hsp70 promoter (h), the firefly luc gene, and a terminator from the actin 5C gene (3′act-5C). Numbering in the basic construct corresponds to numbering in the original vector pGL3-enhancer (Promega). The insertion positions of the enhancer [enh(copia)] and insulators (red triangles for gypsy and blue ones for Fab 7) are indicated. (B) Schematic representation of the control construct (not to scale) containing Renilla luciferase used for normalization of the transfection data. (C) Results of co-transfection experiments with constructs 1–8 together with the control Renilla luciferase construct. The abbreviations for the constructs are indicated in (A). To visualize the lowest bar, the ordinate value starts from the ‘minus’ 1000 Firefly luminescence units (FLU).
The enhancer of copia, [enh(copia)], was amplified from genomic DNA using the oligos 5′-cccggatccAGTCCATGCCTAATAAACAATT-3′ and 5′-cccggatccGAATTCTTTTCACTCAAATTCTGAGAAGG-3′; the fragment, corresponding to a 288–431 bp region in the copia element (AC# M11240), was digested and inserted into the BamHI site of the basic construct. The obtained construct is indicated in Figure 1A as construct 2 or e–h.
Genetic constructs containing a single or a pair of gypsy insulators
A single gypsy insulator was inserted into the KpnI site of the basic construct (see above). A 431 bp region in the gypsy element (AC# M12927) was amplified from genomic DNA with the oligos 5′-cccggtaccTGGCCACGTAATAAGTGTGCG-3′ and 5′-cccggtaccGTTGTTGGTTGGCACACCACAA-3′. The obtained constructs containing the insulator either in direct or reverse orientation are indicated in Figure 1A as constructs 3 and 4, respectively. A second copy of the gypsy insulator was inserted into the HpaI site (Figure 1A, constructs 5–8).
Genetic constructs containing a single Fab7 insulator or a pair of gypsy/Fab7 insulators
A single Fab7 insulator, a 865 bp region in the Fab7 element (AC#X78983), was inserted into the KpnI site of the basic construct (see above). The region was amplified from genomic DNA using the oligos 5′-cccggtaccCAAGATTTCAAGCTGTGTGGCG-3′ and 5′-cccggtaccTTGCGACGTGAGCGACCGAAA-3′. The constructs containing the insulator either in direct or in reverse orientation are indicated in Figure 2A as constructs 9 and 10, respectively.
Figure 2.
Experiments with the constructs carrying a single Fab7 insulator or pairs of gypsy and Fab7 insulators. (A) Schematic representation of the constructs (not to scale). (B) Results of co-transfection experiments with constructs 9–14 together with the control Renilla luciferase construct. Indications are as in Figure 1.
A copy of the Fab 7 insulator was also inserted into the HpaI site of constructs 3 and 4 possessing a single gypsy insulator, as shown in Figure 1A. The obtained constructs containing the Fab 7 insulator either in direct or reverse orientation are indicated in Figure 2A as constructs 11 and 14, respectively.
Control Renilla construct
For the control luc-reporter vectors, the Hsp70 promoter was inserted into a phRL-null plasmid (Promega, AC#AF362546) via the HindIII-EcoRI sites. Then, a 3′ trailer from the actin 5C gene was inserted into the blunted XbaI site. The obtained construct was denoted as p-hsp-hRL-3′act-5C. Oligos used to prepare the insert by PCR amplification are indicated above. Amplification was performed with pCaSpeR-hs/act DNA (AC# U60735). To use smaller amounts of the control DNA in transfection experiments, amplified [enh(copia)] (as described above) was inserted into the BamHI site of p-hsp-hRL-3′act-5C to generate the control p-enh-hsp-hRL-act3′5C construct (Figure 1B).
Transfection assays
Drosophila Schneider 2 culture cells were plated one day prior to transfection at a density of 2 × 106 cells/ml into 85 mm culture dishes. To prepare liposomes, 5 ng of the experimental DNA construct was mixed with 1 µg of p-enh-hsp-hRL-act3′5C control DNA in 50 µl of serum-free medium. Then, 1 µl of TransFast reagent (Promega) diluted according to the manufacturer's recommendations was added, and the mixture was incubated at room temperature for 15 min. The cell suspension (0.5 ml) was centrifuged at 2000 rpm/min in 1.5 ml Eppendorf tubes for 5 min, and the precipitate was mixed with 58 µl of the liposome-containing sample. After incubation for 1 h at 25°C, the transfected cells were transferred into a 24-well plate containing 0.5 ml per well of the cell culture medium containing serum, and cells were incubated for 48 h. Firefly and Renilla luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega) using the Reporter Microplate Luminometer (Turner BioSystems). The firefly luciferase data were normalized to Renilla luciferase data. Excel and Origin software were used for data analysis and graphing.
Normalization of DNA content
DNA content of the constructs used for transfection assays were approximated by digestion of the aliquots with BamHI endonuclease and separation on agarose gels. Quantity ONE software (Bio-Rad) was used to normalize the DNA amounts.
RT–PCR analysis
Forty-eight hours after transfection, RNA was isolated from approximately 2 × 106 transfected Schneider 2 cells using Trizol reagent (Invitrogen) according to the manufacturer's instructions. Samples were treated with DNase using a DNA-free kit (Ambion) and ∼2 μg of total RNA, specific primers and M-MLV reverse transcriptase (Promega) were used for synthesis of cDNA and according to the manufacturer's instructions. For each PCR, the cDNA template (RT+) and the same RNA probe without addition of reverse transcriptase (RT−) were used (see Supplementary Figure 2). The number of PCR cycles varied from 28 to 45. Primers for RT–PCR were selected using the Primer Selection Tool program (http://biotools.umassmed.edu/).
The following primers were used for cDNA synthesis on the transcripts synthesized in the clockwise direction: 5′-GCTCCGTAGACGAAGCGCTCT-3′ (the region located upstream of the Hsp70 core promoter); 5′-ACACGGCGGATCTTTCCGCCCT-3′ (the 3′ region of the luc gene); 5′-TGTTTATTGCAGCTTATAATGGTT-3′ (the region downstream of the gene); 5′-CGCTGTGGAATGTGTGTCAGTTA-3′ (the region upstream of the enhancer); and 5′-GAGCTATAGGAAAGCGCCACGCTT-3′ (the region downstream of the enhancer).
For cDNA synthesis of the putative transcript synthesized in the counter-clockwise direction (reverse transcription) from the enhancer, the following primer was used: 5′-CAACAATTGCATTCATTTTATG-3′.
The following primers were used to assess transcription in a clockwise direction, as shown in Figure 3A: 5′-AAATTTCTCTGGCCGTTATTCGTT-3′ and 5′-GAGAGCAGTATGCCGTTTACTGT-3′ (the region upstream of the Hsp70 core promoter, region 1); 5′-GGTGGCTCCCGCTGAATTGG-3′ and 5′- GGCCTTTATGAGGATCTCTCTGA-3′ (the 3′ portion of the luc gene, region 2); 5′-GGCCGCTTCGAGCAGACATG-3′ and 5′-GCAGCTTATAATGGTTACAAATAA-3′ (the region just downstream of the actin 5C 3′ trailer, region 3); 5′-CAACAATTGCATTCATTTTATGTTTC-3′ and 5′-GCTCCCCAGCAGGCAGAAGTA-3′ (the region upstream of enh(copia), region 4); and 5′-GCGAGCGGTATCAGCTCACT-3′ and 5′-GGGAGAAAGGCGGACAGGTA-3′ (the region downstream of [enh(copia)], region 5).
Figure 3.
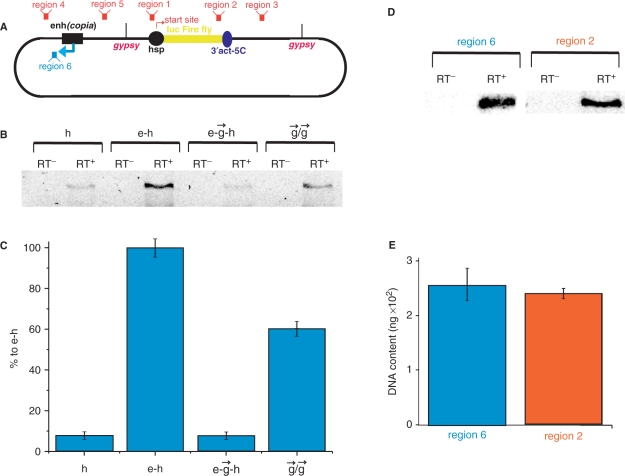
Analysis of RNA synthesized in a counter-clockwise direction on different constructs. (A) Schematic representation of the regions tested using quantitative RT–PCR. Regions 1–5 correspond to the direct transcription by a non-induced Hsp70 promoter. The blue arrow indicates the reversed putative transcription from the enhancer (region 6). (B) Results of RT–PCR (40 cycles) for the reverse promoter-independent transcription downstream from the enhancer in the reversed polarity (region 6). Constructs designated as indicated in Figure 1A. Lanes containing amplified DNA on total RNA preparations without reverse transcriptase, control on the remaining DNA (RT–), and with added reverse transcriptase (RT+), containing synthesized cDNA, are shown. (C) RT–PCR data normalized to 5.8S RNA content is shown (see Materials and Methods section). (D) Comparison of transcription activities in the regions 2 and 6 in the e–h construct. Results of RT–PCR are presented. (E) A diagrammed presentation of the RT–PCR data shown in panel D (five independent experiments).
For testing reverse transcription (Figure 3A, shown by the blue arrow), the following primers were used: 5′-TTTCAGGTTCAGGGGGAGGT-3′ and 5′-CCCCAGCAGGCAGAAGTATG-3′.
The conditions for linear PCR were selected in preliminary experiments using Mastercycler® personal (Eppendorf). The final PCR products were separated in mixed 1% agarose–2% Nu-Sieve agarose gels, and the separation data were evaluated using ‘Quantity One’ quantitation software (Bio-Rad). Statistical analysis of the fractionated DNA fragments obtained in five independent experiments was performed using Origin software. The identity of amplified DNA fragments was confirmed by sequencing. In preliminary experiments, we performed PCR using a radioactive label in duplicate and obtained similar results.
The RT–PCR data were normalized to ribosomal 5.8S RNA as follows. Two microliter aliquots from the final 40 μl cDNA probes synthesized using different specific primers on total RNA preparations (see above) were used for new cDNA synthesis with the 5.8S RNA specific primer: 5′-CAGCATGGACTGCGATATGCGTTG-3′ (26). One microliter of sample was then used for PCR using 5.8S gene-specific primers: 5′-AACTCTAAGCGGTGGATCACTC-3′ and 5′-AAAATGTCGATGTTCATGTGTCCT-3′. The data from RNA preparations corresponding to different constructs were used for normalization using ‘Quantity One’ quantitation software (Bio-Rad). The same results were obtained using rp49 as an internal reference for normalization.
RESULTS
Effects of enhancer of copia [enh(copia)] and gypsy insulator on luciferase reporter gene expression
We generated a basic construct with the firefly luciferase gene reporter (luc) driven by the Hsp70 promoter (h) and containing a 3′ maturation region from the actin 5C gene (3′act-5C) (Figure 1A). This basic construct was then used to design new constructs containing a single insulator or homologous or heterologous insulator pairs. The first insulator was inserted between [enh(copia)] (e) and the Hsp70 promoter. We selected this enhancer because it is active in Drosophila cultured cells and is promoter independent (27). Constructs 3 and 4 (Figure 1A) had a single gypsy insulator placed in direct or reverse polarity, respectively. In another set of constructs, the second gypsy insulator was inserted downstream from the gene at the HpaI site. Constructs 5–8 carried two gypsy insulators in four location combinations around the reporter gene. The construct expressing the Renilla luc gene was used for normalization of transfection data (Figure 1B).
Transfection experiments showed that the presence of the enhancer increased the expression of the luc gene by approximately 200-fold (Figure 1C). A single gypsy insulator placed between the enhancer and the Hsp70 promoter in direct or reverse polarity reduced the expression level of the reporter gene by approximately 45% compared with expression from the e–h construct (Table 1). A pair of gypsy insulators flanking the luc gene resulted in a stronger decrease in enhancer-related activation. Experiments with four constructs containing two gypsy insulators revealed an up to four-fold decrease in expression compared with constructs containing a single gypsy insulator (Figure 1C and Table 1). Insulator polarity was not critical for interruption of enhancer–promoter communication in these genetic constructs.
Table 1.
Insulation efficiency of the circular constructs
| Construct number | Construct designation | luc expression (%) |
|---|---|---|
| 1 | h | 0.2 |
| 2 | e–h | 100 |
| 3 | e– –h –h |
55 |
| 4 | e– –h –h |
57 |
| 5 | e–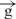 –h– –h–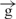
|
13 |
| 6 | e– –h– –h–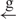
|
16 |
| 7 | e–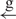 –h– –h–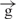
|
14 |
| 8 | e–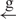 –h– –h–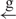
|
15 |
| 9 | e–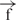 –h –h |
62 |
| 10 | e–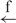 –h –h |
65 |
| 11 | e–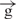 –h– –h–
|
26 |
| 12 | e– –h– –h–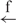
|
27 |
| 13 | e– –h– –h–
|
44 |
| 14 | e–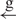 –h– –h–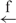
|
18 |
The effect of insulators was measured at 48 h after transfection. After 24 h, the insulation effect was less prominent (Supplementary Figure 1). The assembly of DNA into chromatin is thought to strongly increase the rate of communication between enhancer and promoter (28). Thus, the effects of the regulatory elements studied in transfection experiments are not expected to be immediate and complete. Our data implied that after a longer period of time during development, a pair of gypsy insulators could almost completely interrupt the communications between an enhancer and a promoter in Drosophila cells.
Fab7 and gypsy insulators can work together
Constructs 9 and 10 contain a single Fab7 insulator, while constructs 11–14 contain a gypsy insulator upstream of the luc gene and a Fab7 insulator downstream of the gene in different polarity (Figure 2A). It is currently unknown whether gypsy and Fab7 insulators function independently or whether they share the same proteins. We designed the latter constructs to test whether these insulators could efficiently work together.
A single Fab7 insulator was as efficient as a single gypsy insulator at enhancer blocking (Figure 2B and Table 1). Again, we observed a polarity-independent 40% decrease in luc gene expression compared with the expression of the e–h construct. The four different combinations of gypsy/Fab7 insulators generated a more efficient insulation effect, up to 80%; gypsy/gypsy pairs, however, were more efficient (Table 1).
Transcription inside the intergenic region between enhancer and promoter on the circular constructs
To understand the nature of the communication between the enhancer and promoter that can be blocked by insulators, we used quantitative RT–PCR to analyze RNA synthesized from different regions of various constructs and normalized the data using 5.8S ribosomal RNA content. In separate experiments, we confirmed the absence of potentially contaminating DNA levels in the RNA probes used in our RT–PCR experiments (Supplementary Figure 2). Figure 3A shows six regions in the constructs selected for these experiments. We detected transcription in all five regions corresponding to transcription in a clockwise direction (data not shown). This transcription likely arose from the passing of the RNA polymerase II from the Hsp70 promoter along the circular constructs in a clockwise direction several kilo base pairs downstream of the actin 5C terminator. Thus, it was difficult to search for any promoter-independent transcription events in this background. For this reason, we focused our study on reverse transcription in region 6 (Figure 3A) located just downstream of the enhancer. Our experiments with the linear and circular constructs (see below, Figure 4) strongly suggest that the activation signal passes from the enhancer to the promoter in this direction. Surprisingly, we detected weak transcription in this region, even in the h construct Figure 3B). This amplified sequence corresponds to a SV40 enhancer inserted in the original pGL3-Enhancer vector used for preparation of the constructs (see Materials and Methods section). Transcription of this non-coding RNA drastically increased in the presence of the [enh(copia)] (construct e–h, Figure 3B and C), and the level of transcription was comparable with that of Hsp70-dependent transcription of the reporter luc gene in the same construct (Figure 3D and E). Interestingly, insertion of a single gypsy insulator before the promoter (construct e– –h) completely abolished this enhancer effect, while a pair of gypsy insulators (construct
–h) completely abolished this enhancer effect, while a pair of gypsy insulators (construct  /
/ ) partially restored the level of this promoter-independent transcription (Figure 3C). Clearly, strong transcription in the region 6 is induced by [enh(copia)]. To analyze the antisense transcription, we also used RT–PCR in the region complementary to region 3 and observed the transcription (data not shown). The finding that an enhancer could prompt transcription led us to speculate that weak transcription detected in these experiments only after 40 cycles of RT–PCR was due to recognition of the heterologous SV40 enhancer by the Drosophila transcription machinery. This hypothesis was directly confirmed (Supplementary Figure 3).
) partially restored the level of this promoter-independent transcription (Figure 3C). Clearly, strong transcription in the region 6 is induced by [enh(copia)]. To analyze the antisense transcription, we also used RT–PCR in the region complementary to region 3 and observed the transcription (data not shown). The finding that an enhancer could prompt transcription led us to speculate that weak transcription detected in these experiments only after 40 cycles of RT–PCR was due to recognition of the heterologous SV40 enhancer by the Drosophila transcription machinery. This hypothesis was directly confirmed (Supplementary Figure 3).
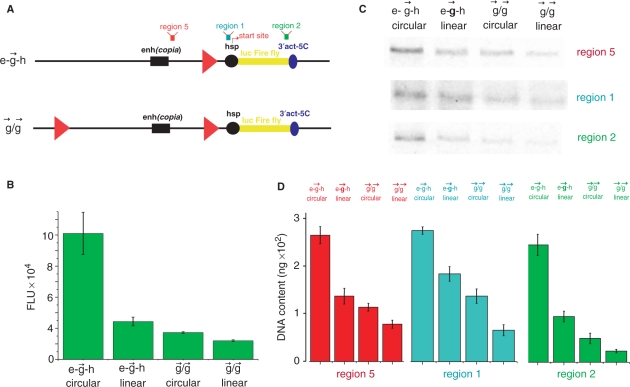
Figure 4.
Analysis of RNA synthesized on linear constructs containing gypsy insulators. (A) Schematic representation of the e–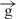 –h and
–h and 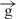 /
/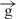 constructs and the regions tested by quantitative RT–PCR. Regions 5, 1 and 2 correspond to the sequences located just downstream of the enhancer, the upstream regulatory region of the Hsp70 promoter, and the 3′ end of the luc gene, respectively. (B) Results of co-transfection experiments with the circular and linear constructs. (C) RT–PCR for the regions 5, 1 and 2. The constructs are designated as indicated in Figure 1A. (D) A diagrammed presentation of the RT–PCR data shown in panel C (five independent experiments).
constructs and the regions tested by quantitative RT–PCR. Regions 5, 1 and 2 correspond to the sequences located just downstream of the enhancer, the upstream regulatory region of the Hsp70 promoter, and the 3′ end of the luc gene, respectively. (B) Results of co-transfection experiments with the circular and linear constructs. (C) RT–PCR for the regions 5, 1 and 2. The constructs are designated as indicated in Figure 1A. (D) A diagrammed presentation of the RT–PCR data shown in panel C (five independent experiments).
A single gypsy insulator in linear constructs is as efficient as two gypsy insulators in circular constructs
We next used the linear constructs for transcriptional analysis and tested the expression of the reporter gene on the linear e– –h and
–h and 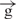 /
/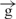 constructs. In our experiments with circular constructs, a single gypsy insulator reduced reporter gene expression only up to 45% relative to expression of the initial e–h construct (Figure 1). This difference is likely due to the communication between [enh(copia)] in these constructs with the promoter from the opposite direction, where no insulator was inserted. To assess the real effect coming from a single gypsy insulator placed between the enhancer and promoter, we digested two constructs using the FseI endonuclease and assessed activity in transfection experiments. Figure 4B shows that the linear
constructs. In our experiments with circular constructs, a single gypsy insulator reduced reporter gene expression only up to 45% relative to expression of the initial e–h construct (Figure 1). This difference is likely due to the communication between [enh(copia)] in these constructs with the promoter from the opposite direction, where no insulator was inserted. To assess the real effect coming from a single gypsy insulator placed between the enhancer and promoter, we digested two constructs using the FseI endonuclease and assessed activity in transfection experiments. Figure 4B shows that the linear 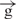 construct exhibited only 30% luc activity compared to its circular form. Linearization of the
construct exhibited only 30% luc activity compared to its circular form. Linearization of the 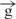 /
/ construct only slightly reduced reporter gene expression. These data strongly suggested that physical cutting of the e–
construct only slightly reduced reporter gene expression. These data strongly suggested that physical cutting of the e– –h construct was as efficient in blocking enhancer–promoter communication as was the insertion of the second insulator. In fact, the linear e–
–h construct was as efficient in blocking enhancer–promoter communication as was the insertion of the second insulator. In fact, the linear e–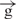 –h construct exhibited approximately the same expression level as the
–h construct exhibited approximately the same expression level as the  /
/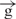 circular construct.
circular construct.
These data demonstrate that a single insulator located between an enhancer and a promoter can essentially reduce the effect of the enhancer. Up to 50% of insulation was observed in our experiments with circular plasmids carrying a single gypsy or Fab7 insulator inserted in any polarity (Table 1). This effect was more apparent with linear DNAs: up to 85% of insulation was observed 48 h after transfection.
Analysis of transcription in linear and circular constructs containing gypsy insulators
The above results demonstrating the various enhancer-promoter interactions on linear and circular constructs containing a single or pair of gypsy insulators rendered these constructs a powerful model system for the quantitative analysis of this communication. Two main regions were selected for such analyses: the intergenic region located just downstream of [enh(copia)] and the upstream regulation region of the Hsp70 promoter located just upstream of its transcription start site (Supplementary Figure 5). The aim was to check the delivery putative RNA signal from the enhancer inside the promoter. We compared the transcriptional activity in these regions (regions 5 and 1 on Figure 4A) with the transcription inside the luc reporter gene (region 2 on Figure 4A). We detected a rather weak transcription in both region 5 and region 1 in the linear e– –h construct (Figure 4C and D). Previously we detected a strong transcription on the circular e–h construct in the reversed polarity coming from the enhancer. Now, using the constructs possessing gypsy insulator, we observed direct transcription towards the promoter in region 5 during not complete insulation of the enhancer signal. Linearization of the constructs containing a single gypsy insulator or the pair of insulators decreased this transcription, likely due to the interruption of transcription inside the circular constructs in the clockwise direction from the Hsp70 promoter. Interestingly, transcription was also detected in the upstream regulatory Hsp70 promoter region. It follows that the signal from the enhancer reaches the promoter region and the nature of this signal is RNA being synthesized along the intergenic region. Our results suggest a correlation between the strength of this transcription and the expression of the luc gene detected by both luminescence measurements and by quantitative RT–PCR (Figure 4B and C). Similar data were obtained from analysis of linear and circular e–
–h construct (Figure 4C and D). Previously we detected a strong transcription on the circular e–h construct in the reversed polarity coming from the enhancer. Now, using the constructs possessing gypsy insulator, we observed direct transcription towards the promoter in region 5 during not complete insulation of the enhancer signal. Linearization of the constructs containing a single gypsy insulator or the pair of insulators decreased this transcription, likely due to the interruption of transcription inside the circular constructs in the clockwise direction from the Hsp70 promoter. Interestingly, transcription was also detected in the upstream regulatory Hsp70 promoter region. It follows that the signal from the enhancer reaches the promoter region and the nature of this signal is RNA being synthesized along the intergenic region. Our results suggest a correlation between the strength of this transcription and the expression of the luc gene detected by both luminescence measurements and by quantitative RT–PCR (Figure 4B and C). Similar data were obtained from analysis of linear and circular e– –h and
–h and 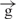 /
/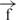 constructs (Supplementary Figure 4).
constructs (Supplementary Figure 4).
Enhancer induces the promoter-independent transcription towards the promoter
Using four circular constructs, we assessed the transcription in a counter-clockwise direction the region located before the enhancer (Figure 3). Detection of a clockwise transcription inside the intergenic region in two linear constructs containing insulators (Figure 4) prompted us to perform similar analysis of direct transcription from the enhancer towards the promoter in the linear h, e–h and e–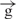 –h constructs. The aim of this experiment was to compare the transcription levels in three regions of the linear constructs: region 5, located just downstream from the enhancer; region 1, corresponding to the upstream regulatory region of the Hsp70 promoter; and inside the luc gene (region 2). Figure 5B shows the expected levels of the luc expression in the constructs tested by luminescence. Using the h construct, no transcription was detected in regions 5 and 1 under the conditions used, whereas in region 2, a weak transcription was clearly observed (Figure 5C). Interestingly, all three regions were actively transcribed in the e–h construct. These results of active promoter independent transcription induced by enhancer are in agreement with the above data demonstrating the same level of counter-clockwise and direct luc transcription in the e–h construct (Figure 3). It follows that active transcription induced by enhancer in the intergenic region 5 and the upstream regulatory region of the promoter leads to the active transcription of the reporter gene. Insertion of the gypsy insulator leads to simultaneous inhibition of transcription in regions 5 and 1 and in the luc gene (Figure 5C and D). The level of transcription detected in the e–
–h constructs. The aim of this experiment was to compare the transcription levels in three regions of the linear constructs: region 5, located just downstream from the enhancer; region 1, corresponding to the upstream regulatory region of the Hsp70 promoter; and inside the luc gene (region 2). Figure 5B shows the expected levels of the luc expression in the constructs tested by luminescence. Using the h construct, no transcription was detected in regions 5 and 1 under the conditions used, whereas in region 2, a weak transcription was clearly observed (Figure 5C). Interestingly, all three regions were actively transcribed in the e–h construct. These results of active promoter independent transcription induced by enhancer are in agreement with the above data demonstrating the same level of counter-clockwise and direct luc transcription in the e–h construct (Figure 3). It follows that active transcription induced by enhancer in the intergenic region 5 and the upstream regulatory region of the promoter leads to the active transcription of the reporter gene. Insertion of the gypsy insulator leads to simultaneous inhibition of transcription in regions 5 and 1 and in the luc gene (Figure 5C and D). The level of transcription detected in the e–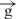 –h construct is low in all three regions analyzed.
–h construct is low in all three regions analyzed.
Figure 5.
Analysis of RNA synthesized on linear h, e–h and e–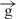 –h constructs. (A) Schematic representation of the constructs and the regions tested using quantitative RT–PCR. (B) Results of co-transfection experiments with the linear constructs. (C) Results of RT–PCR for the regions 5, 1 and 2. Designation of the constructs as indicated in Figure 1A. To visualize the lowest bar the ordinate value starts from the ‘minus’ 2000 FLU. (D) A diagrammed presentation of the RT–PCR data shown in (C) (five independent experiments).
–h constructs. (A) Schematic representation of the constructs and the regions tested using quantitative RT–PCR. (B) Results of co-transfection experiments with the linear constructs. (C) Results of RT–PCR for the regions 5, 1 and 2. Designation of the constructs as indicated in Figure 1A. To visualize the lowest bar the ordinate value starts from the ‘minus’ 2000 FLU. (D) A diagrammed presentation of the RT–PCR data shown in (C) (five independent experiments).
DISCUSSION
Quantitative analysis of gypsy and Fab7 insulators by transfection experiments in Drosophila cells
The transgenic approach is a method commonly employed in mammalian cells for the study of enhancers and insulators (29–31). Two approaches have been used in Drosophila for the identification and characterization of enhancer-blocking insulators. The first is a position effect assay assessing the activation of transgenic promoters by enhancers located at the site of integration of a transgene without enhancers (32). The second approach also uses transgenic genetic constructs, but possessing all studied enhancers and insulators (4,32). Nevertheless, the latter approach cannot exclude the communications of functional elements in the site of integration with the elements of the construct.
We developed a system for the quantitative analysis of effects of enhancers and insulators on the expression of a reporter luc gene in Drosophila cultured cells. This simple system has another important advantage, in that it is well suited for very sensitive transcriptional analysis, especially for transcription of non-Drosophila sequences that are present only in transfected constructs. The system allows for more rapid testing of genetic constructs than the transgenic approach and, more importantly, it excludes effects from the surrounding host chromatin on the expression of the constructs. The possible disadvantage of the described system is that it may not be useable for study of tissue-specific functional genomic elements that require nuclear proteins not expressed in cultured cells.
Transfected plasmids that are maintained for few months have been shown to form episomally maintained concatameric structures (33). Our experiments using 48 h transfection assays did not reveal the formation of concatamers, as tested by analysis of DNA isolated from the transfected cells (data not shown). Thus we conclude that the observed effects (luciferase expression and intergenic transcription) originate from single plasmids.
Two insulators are required to protect a promoter from non bona fide enhancers
Our results demonstrate that all studied pairs of gypsy/gypsy or gypsy/Fab7 insulators are active. In the corresponding constructs, the counter-clockwise communication between enhancer and promoter was interrupted either by gypsy or by Fab7 insulators (constructs 5–8 and 11–14, Table 1). Only in one combination with gypsy, with Fab7 inserted in a reversed polarity (construct 13), we observed a weak efficiency of this insulator. The different insulators form active pairs in the remaining three combinations. The data led us to conclude that both insulators could share in at least some overlapping proteins required for their enhancer blocking activity. Our data also indicate that gypsy or Fab7 insulators could block the communications between an enhancer and a promoter in any orientation when placed between them. Interestingly, a single Fab7 insulator, or together with gypsy, is weaker than a single gypsy or gypsy/gypsy pair. This ‘weak insulating activity’ of Fab7 is consistent with the recent data demonstrating that in the Drosophila embryo, the Fab7 insulator could be bypassed by the promoter targeting sequence from the Abd-B locus (34).
The results of our experiments with the linear constructs show that a single insulator could protect a promoter from an enhancer. This is true only for artificial constructs possessing one enhancer. In the genomic context, however, each gene is located among of various enhancers (8,9). To protect promoters from the ‘foreign’ enhancers, insulators could interact with one another or with SARs/MARs and form chromatin loops (1,6,35). A gene within the loop is isolated from interference of non bona fide enhancers. Our data on the circular constructs demonstrate that a single insulator cannot protect a promoter from an enhancer communicating with a promoter from a counter-clockwise direction. Thus, a gene should be protected from incorrect enhancers by insulators from both sides, and another insulator is required. This conclusion is in agreement with recent data highlighting the involvement of insulators in stabilizing the contacts between distant genomic regulatory regions leading to the formation of chromatin loops (25).
Enhancers communicate with promoters by inducing transcription in both directions and insulators inhibit this transcription
In this study, we presented evidence that transcripts are coming from enhancer to promoter. In the circular constructs, this transcription occurs in both directions (Figures 3 and 5). We tested the transcripts from the sequences located immediately around [enh(copia)] and corresponding to non-Drosophila sequences in the constructs, and concluded that this transcription is induced inside the enhancer. This transcription flow reaches the upstream regulatory region of the Hsp70 promoter located upstream of its transcription start site (Supplementary Figure 5 shows the position of the primers used for cDNA synthesis and for PCR amplification of Hsp70 promoter sequences). This region contains the TATA box and sequences where GAGA factor and HSF bind (36–38). GAGA has been suggested to facilitate long-range activation that mediates enhancer–promoter communications (39). Our data support the view that GAGA could interact with enhancers. RNA tracking could be considered a mechanism of enhancement of transcription by delivery of transcriptional complex components from enhancers to upstream regulatory regions of promoters. Studies have demonstrated that the human HS2 enhancer of the ɛ-globin gene initiates synthesis of non-coding poly(A) RNAs and delivers RNA polymerase II and TBP to the promoter (16,40). Our data in Drosophila support this view on the mechanisms of enhancer action.
The data presented here clearly support the RNA tracking mechanism model of communication between an enhancer and promoter. Our data argues that enhancers are not simply the binding sites for transcriptions factors, but are sites that also likely bind transcription complexes. In this respect, enhancers could be considered relatives or homologues of promoters. The observation that the SV40 enhancer is slightly active in Drosophila cells (Supplementary Figure 3) might indicate that these elements are conserved in evolution.
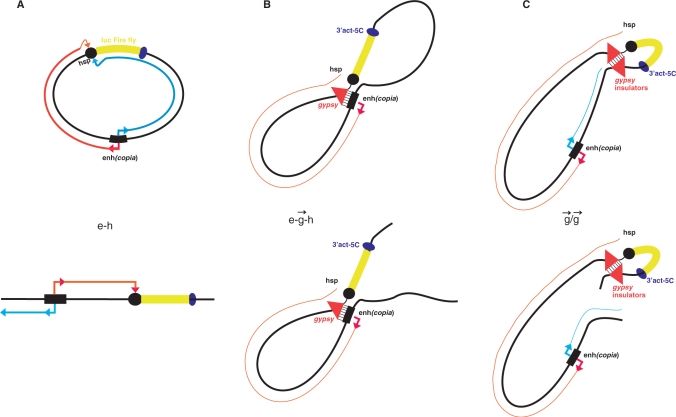
Our data demonstrate that an insulator inhibits both the transcription in the intergenic region located downstream from an enhancer and the transcription within the Hsp70 promoter upstream of its transcription start site (Figure 4C and D). It follows that an insulator does not interrupt the RNA synthesis coming to it, but rather reduces the level of transcription by interfering with the initiation of RNA synthesis inside an enhancer. The decrease of transcription in the intergenic region 5 (before the insulator) and in the upstream regulatory region of the Hsp70 promoter (after the insulator) was similar as that observed in the linear construct e–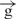 –h (Figure 5). Thus, we conclude that the insulator does not work as an obstacle in transcriptional communication between an insulator and a promoter. If this was the case, we should observe high levels of transcription in region 5 and low levels in region 1 (Figure 5). Our data argues in favor of direct contacts between the enhancer and gypsy insulator that suppress transcription in the entire region between enhancer and promoter (Figure 6B). We speculate that direct binding of insulators with enhancers is one possible mechanism leading to the inhibition of transcription induced by enhancers (Figure 6B). Another mechanism is the formation of topologically and transcriptionally isolated loops formed by a pair of insulators (Figure 6C). We speculate that in our transfection experiments with constructs containing a single or a pair of insulators, the formation of chromatin structures characteristic to chromosomal regions containing enhancers and insulators was not complete over 48 h, providing a possible explanation as to why we did not observe complete stop of transcription induced by insulators. The interposed insulator in integrated constructs has been reported to reduce enhancer activity 4–8-fold downstream of the insulator, while only about 20% of inhibition of transcription was observed in the downstream region (16). Our experiments clearly indicated stronger inhibition of transcription (up to 80%) in regions both upstream and downstream of the insulator.
–h (Figure 5). Thus, we conclude that the insulator does not work as an obstacle in transcriptional communication between an insulator and a promoter. If this was the case, we should observe high levels of transcription in region 5 and low levels in region 1 (Figure 5). Our data argues in favor of direct contacts between the enhancer and gypsy insulator that suppress transcription in the entire region between enhancer and promoter (Figure 6B). We speculate that direct binding of insulators with enhancers is one possible mechanism leading to the inhibition of transcription induced by enhancers (Figure 6B). Another mechanism is the formation of topologically and transcriptionally isolated loops formed by a pair of insulators (Figure 6C). We speculate that in our transfection experiments with constructs containing a single or a pair of insulators, the formation of chromatin structures characteristic to chromosomal regions containing enhancers and insulators was not complete over 48 h, providing a possible explanation as to why we did not observe complete stop of transcription induced by insulators. The interposed insulator in integrated constructs has been reported to reduce enhancer activity 4–8-fold downstream of the insulator, while only about 20% of inhibition of transcription was observed in the downstream region (16). Our experiments clearly indicated stronger inhibition of transcription (up to 80%) in regions both upstream and downstream of the insulator.
Figure 6.
Models illustrating mechanisms of enhancer and insulator action. (A) Promoter-independent RNA synthesis driven by an enhancer recruits RNA polymerase II (pol II) to the promoter. (B) A single insulator interacts with an enhancer and blocks the reverse promoter-independent transcription directed by an enhancer and/or the tunnel delivering RNA polymerase II. (C) A pair of insulators forms more a stable complex, shapes loops and isolates RNA tracking directed to a promoter.
For many years the effects of insulators on enhancers were explained by two alternative models: the tracking or transcriptional model, and the structural or direct contact model (1,2). Based on the data reported here, we consider these models not as alternatives, but as supplementing each other: RNA synthesis is a mechanism of communications between enhancers and promoters, while formation of structures in which insulators are involved is a protection mechanism from the influence of the ‘wrong’ enhancers on promoters. Another part of this protection mechanism is the ability of an insulator to inhibit the transcription induced by a ‘foreign’ enhancer, likely by direct interaction with such an enhancer. Our data on insulator pairs agree with the looping model of insulator action (1,2). A pair of insulators can form a looped domain fixed in an insulator body located at the nuclear periphery. In these constructs, the loop efficiently cuts off the enhancer and its communications with the promoter. We also suggest that interactions between insulators are more stable than between an enhancer and an insulator, and a pair of insulators might form a separate gene-containing loop that is physically isolated from communications via an RNA tracking mechanism acting in another loop (Figure 6C). We observed a decrease of delivery of RNA signal to the Hsp70 upstream region in the 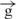 /
/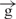 construct from the enhancer (Figure 4C and D). We speculate that in the e–
construct from the enhancer (Figure 4C and D). We speculate that in the e–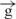 –h construct, a physical interaction between the enhancer and insulator might occur, leading to a blockage of transcription initiation in a counter-clockwise direction inside the enhancer (Figure 6B) and to inhibition of transcription in direct polarity (Figure 5C and D). Our data suggest that enhancers communicate with promoters by RNA tracking. We speculate that insulators interrupt this communication either by inhibition of RNA synthesis by direct contact with an enhancer, or by formation of a transcriptionally independent loop by a pair of insulators (Figure 6). Our ongoing detailed analysis of start sites of promoter-independent transcription in these constructs may address some questions these results raise.
–h construct, a physical interaction between the enhancer and insulator might occur, leading to a blockage of transcription initiation in a counter-clockwise direction inside the enhancer (Figure 6B) and to inhibition of transcription in direct polarity (Figure 5C and D). Our data suggest that enhancers communicate with promoters by RNA tracking. We speculate that insulators interrupt this communication either by inhibition of RNA synthesis by direct contact with an enhancer, or by formation of a transcriptionally independent loop by a pair of insulators (Figure 6). Our ongoing detailed analysis of start sites of promoter-independent transcription in these constructs may address some questions these results raise.
SUPPLEMENRATY DATA
Supplementary Data are available at NAR Online.
FUNDING
Molecular and Cellular Biology Program of the Russian Academy of Sciences, Russian Ministry of Education and Science (02.512.11.2259); Russian Foundation for Basic Research (08-04-00058-a). Funding to pay the Open Access publication charges for this article was provided by Russian Foundation for Basic Research (08-04-00058-a).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Yu.N. Toropchina for technical assistance.
REFERENCES
- 1.Mongelard F, Corces VG. Two insulators are not better than one. Nature Struct. Biol. 2001;8:192–194. doi: 10.1038/84905. [DOI] [PubMed] [Google Scholar]
- 2.Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 3.Sun FL, Elgin SC. Putting boundaries on silence. Cell. 1999;99:459–462. doi: 10.1016/s0092-8674(00)81534-2. [DOI] [PubMed] [Google Scholar]
- 4.Geyer PK, Corces VG. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 1992;6:1865–1873. doi: 10.1101/gad.6.10.1865. [DOI] [PubMed] [Google Scholar]
- 5.Capelson M, Corces VG. The ubiquitin ligase dTopors directs the nuclear organization of a chromatin insulator. Mol. Cell. 2005;20:105–116. doi: 10.1016/j.molcel.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 6.Gerasimova TI, Corces VG. Polycomb and trithorax group proteins mediate the function of a chromatin insulator. Cell, 1998;92:511–521. doi: 10.1016/s0092-8674(00)80944-7. [DOI] [PubMed] [Google Scholar]
- 7.Gerasimova TI, Byrd K, Corces VG. A chromatin insulator determines the nuclear localization of DNA. Mol. Cell. 2000;6:1025–1035. doi: 10.1016/s1097-2765(00)00101-5. [DOI] [PubMed] [Google Scholar]
- 8.Ramos E, Ghosh D, Baxter E, Corces VG. Genomic organization of gypsy chromatin insulators in Drosophila melanogaster. Genetics. 2006;172:2337–2349. doi: 10.1534/genetics.105.054742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parnell TJ, Kuhn EJ, Gilmore BL, Helou C, Wold MS, Geyer PK. Identification of genomic sites that bind the Drosophila suppressor of Hairy-wing insulator protein. Mol. Cell. Biol. 2006;26:5983–5993. doi: 10.1128/MCB.00698-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mihaly J, Hogga I, Barges S, Galloni M, Mishra RK, Hagstrom K, Muller M, Schedl P, Gausz J, Gyukovics H, et al. Chromatin domain boundaries in the bithorax complex. Cell. Mol. Life Sci. 1998;54:60–70. doi: 10.1007/s000180050125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou J, Barolo S, Szymansky P, Levin M. The Fab-7 element of the bithorax complex attenuates enhancer-promoter interactions in the Drosophila embryo. Genes Dev. 1996;10:3195–3201. doi: 10.1101/gad.10.24.3195. [DOI] [PubMed] [Google Scholar]
- 12.Hagstrom K, Muller M, Schedl P. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev. 1996;10:3202–3215. doi: 10.1101/gad.10.24.3202. [DOI] [PubMed] [Google Scholar]
- 13.Gerasimova TI, Lei EP, Bushey AM, Corces VG. Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila. Mol. Cell. 2007;14:761–772. doi: 10.1016/j.molcel.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hampsey M, Reinberd D. RNA polymerase II as a control panel for multiple coactivator complexes. Curr. Opin. Genet. Dev. 1999;9:132–139. doi: 10.1016/S0959-437X(99)80020-3. [DOI] [PubMed] [Google Scholar]
- 15.Zhao H, Dean A. An insulator blocks spreading of histone acetylation and interferes with RNA polymerase II transfer between an enhancer and gene. Nucleic Acids Res. 2004;32:4903–4914. doi: 10.1093/nar/gkh832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu X, Ling J, Zhang L, Pi W, Wu M, Tuan D. A facilitated tracking and transcription mechanism of long-range enhancer function. Nucleic Acids Res. 2007;35:5532–5544. doi: 10.1093/nar/gkm595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai HN, Shen P. Effects of cis arrangement of chromatin insulator on enhancer-blocking insulator activity. Science. 2001;291:493–495. doi: 10.1126/science.291.5503.493. [DOI] [PubMed] [Google Scholar]
- 18.Muraviova E, Golovin A, Gracheva E, Parshikov A, Belenjaya T, Pirotta V, Georgiev P. Loss of insulator activity by paired Su(Hw) chromatin insulators. Science. 2001;291:495–498. doi: 10.1126/science.291.5503.495. [DOI] [PubMed] [Google Scholar]
- 19.West AG, Gaszner M, Felsenfeld G. Insulators: many functions, many mechanisms. Genes Dev. 2006;16:271–288. doi: 10.1101/gad.954702. [DOI] [PubMed] [Google Scholar]
- 20.Bownes M. Preferential insertion of P elements into genes expressed in the germ-line of Drosophila melanogaster. Mol. Gen. Genet. 1990;222:457–460. doi: 10.1007/BF00633856. [DOI] [PubMed] [Google Scholar]
- 21.O’Kane CJ, Gering W. Detection in situ of genomic regulatory element in Drosophila. Proc. Natl Acad. Sci. USA. 1987;84:9123–9127. doi: 10.1073/pnas.84.24.9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roseman RR, Swan JM, Geyer PK. A Drosophila insulator protein facilitates dosage compensation of the X chromosome min-white gene located at autosomal insertion sites. Development. 1995;121:3573–3582. doi: 10.1242/dev.121.11.3573. [DOI] [PubMed] [Google Scholar]
- 23.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 24.Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 2008;40:476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wellace JA, Felsenfeld G. We gather together: insulators and genome organization. Curr. Opin. Genet. Dev. 2007;17:400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tchurikov NA, Kretova OV. Suffix-specific RNAi leads to silencing of F element in Drosophila melanogaster. PLoS ONE. 2007;2:e476. doi: 10.1371/journal.pone.0000476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cavarec L, Jensen S, Heidmann T. Identification of a strong transcriptional activator for the copia retrotransposon responsible for its differential expression in Drosophila hydei and melanogaster cell lines. Biochem. Biophys. Res. Commun. 1984;203:392–399. doi: 10.1006/bbrc.1994.2195. [DOI] [PubMed] [Google Scholar]
- 28.Rubtsov MA, Polikov YS, Bondarenko VA, Wang YH, Studitsky VM. Chromatin structure can strongly facilitate enhancer action over a distance. Proc. Natl Acad. Sci. USA. 2006;103:17690–17695. doi: 10.1073/pnas.0603819103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Recillas-Targa F, Bell AC, Felsenfeld G. Positional enhancer-blocking activity of the chicken beta-globin insulator in transiently transfected cells. Proc. Natl. Acad. Sci. USA. 1999;96:14354–14359. doi: 10.1073/pnas.96.25.14354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bell AC, West AG, Felsenfeld G. Insulators and boundaries: versatile regulatory elements in the eukaryotic genome. Science. 2001;291:447–450. doi: 10.1126/science.291.5503.447. [DOI] [PubMed] [Google Scholar]
- 31.Farrell CM, West AG, Felsenfeld G. Conserved CTCF insulator elements flank the mouse and human beta-globin loci. Mol. Cell. Biol. 2002;11:3820–3831. doi: 10.1128/MCB.22.11.3820-3831.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kellum R, Schedl P. A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991;64:941–950. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- 33.O'Donnell RA, Preiser PR, Williamson DH, Moore PW, Cowman AF, Crabb BS. An alteration in concatameric structure is associated with efficient segregation of plasmids in transfected Plasmodium falciparum parasites. Nucleic Acids Res. 2001;29:716–724. doi: 10.1093/nar/29.3.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin Q, Chen Q, Lin L, Smith S, Zhou J. Promoter targeting sequence mediates enhancer interference in the Drosophila embryo. Proc. Natl Acad. Sci. USA. 2007;104:3237–3242. doi: 10.1073/pnas.0605730104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Majumder P, Cai HN. The functional analysis of insulator interactions in the Drosophila embryo. Proc. Natl Acad. Sci. USA. 2003;100:5223–5228. doi: 10.1073/pnas.0830190100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gilmour DS, Thomas GH, Elgin SC. Drosophila nuclear proteins bind to regions of alternating C and T residues in gene promoters. Science. 1989;245:1487–1490. doi: 10.1126/science.2781290. [DOI] [PubMed] [Google Scholar]
- 37.Weber JA, Taxman DJ, Lu Q, Gilmour DS. Molecular architecture of the hsp70 promoter after deletion of the TATA box or the upstream regulation region. Mol. Cell. Biol. 1997;17:3799–3808. doi: 10.1128/mcb.17.7.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Georgel PT. Chromatin potentiation of the Hsp70 promoter is linked to GAGA-factor recruitment. Biochem. Cell. Biol. 2005;83:555–565. doi: 10.1139/o05-060. [DOI] [PubMed] [Google Scholar]
- 39.Mahmoudi T, Katsani KR, Verrijzer CP. GAGA can mediate enhancer function in trans by linking two separate DNA molecules. EMBO J. 2002;21:1775–1781. doi: 10.1093/emboj/21.7.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ling J, Baibakov B, Pi W, Emerson BM, Tuan D. The HS2 enhancer of the β-globin locus control region initiates synthesis of non-coding, polyadenylated RNAs independent of cis-linked globin promoter. J. Mol. Biol. 2005;350:883–896. doi: 10.1016/j.jmb.2005.05.039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.