Abstract
Over the last 15 years, investigators have identified small noncoding RNAs as regulators of gene expression. One type of noncoding RNAs are termed microRNAs (miRNAs). miRNAs are evolutionary conserved, approximately 22-nucleotide single-stranded RNAs that target genes by inducing mRNA degradation or by inhibiting translation. miRNAs are implicated in many critical cellular processes, including apoptosis, proliferation, and differentiation. Furthermore, it is estimated that miRNAs may be responsible for regulating the expression of nearly one-third of the human genome. Despite the identification of greater than 500 mature miRNAs, very little is known about their biological functions and functional targets. In the last 5 years, researchers have increasingly focused on the functional relevance and role that miRNAs play in the pathogenesis of human disease. miRNAs are known to be important in solid organ and hematological malignancies, heart disease, as potential modulators of the immune response, and organ development. It is anticipated that miRNA analysis will emerge as an important complement to proteomic and genomic studies to further our understanding of disease pathogenesis. Despite the application of genomics and proteomics to the study of human lung disease, few studies have examined miRNA expression. This perspective is not meant to be an exhaustive review of miRNA biology but will provide an overview of both miRNA biogenesis and our current understanding of the role of miRNAs in lung disease as well as a perspective on the importance of integrating this analysis as a tool for identifying and understanding the biological pathways in lung-disease pathogenesis.
Keywords: microRNA, epigenetics, genomics
In 1993, investigators first identified a class of noncoding RNA that regulates gene expression (1) of a target gene. Later termed “microRNAs” (miRNAs), these evolutionary conserved, noncoding RNAs represent one of several recently identified small noncoding RNAs. The family of small noncoding RNAs, including small interfering RNAs (siRNA), transacting siRNAs (tasiRNAs), small scan RNAs (scnRNAS), repeat associated siRNAs (rasiRNAs), and Piwi-interacting RNAs (piRNAs) are still poorly understood but may all have the capacity for epigenetic regulation via different mechanisms (2). miRNAs are approximately 19- to 25-nucleotide (nt) single-stranded, noncoding RNAs that exist in both animals and plants and regulate gene/protein expression through direct complementarity between their 5′ region (termed “seed sequence” or “region”) and the 3′ untranslated region (UTR) of target mRNAs. Direct binding of miRNAs to a target mRNA may result in either mRNA degradation or inhibition of protein translation (3) miRNAs may indirectly alter gene expression through global effects on methylation and targeting of transcription factors essential to gene expression (4, 5). miRNAs have the capacity to bind and regulate hundreds of genes simultaneously. In fact, computational analyses suggest that miRNAs may regulate up to one-third of all protein-coding genes in human genome (6). To date, over 500 miRNAs have been identified in mammals, yet their biological relevance and functional targets remain largely unknown. The global changes in miRNA expression in several malignancies and their location in fragile chromosome loci suggest their relevance to the development and progression of malignancy (7) and other important developmentally related pathologies.
miRNAs are of importance in several biological processes including cellular differentiation, apoptosis, and proliferation. miRNAs have also been implicated in nonmalignant diseases involving the cardiovascular, endocrine, neurological systems, and immune response (8–10).
The recognition that both cells and organs have specific patterns of miRNA expression, and the novel characteristic of miRNAs targeting several genes, makes them potentially attractive candidates as therapeutic targets and proximal regulators of gene network expression (11–13). Several examples of targeting tissue-specific miRNAs exist, including miR-122 (liver) in hepatitis C, miR-1 and 133 (skeletal muscle and cardiac myocytes) in cardiac hypertrophy, and miR-375 (islet cell) in diabetes (8, 9, 14). Few studies, however, have investigated the relevance of miRNAs in diseases of the lung. The inherent complexity in simultaneous targeting of multiple genes and gene pathways by a single miRNA combined with the paucity of data on functional impact of specific miRNA makes the analysis of miRNA particularly difficult. Such complexity raises important questions for those applying miRNA analysis to the study of lung disease. Are there lung-specific miRNAs? Which miRNAs are relevant to human lung disease and how will this be determined? How can miRNA analysis best complement previously established high-throughput platforms that examine the proteome and transcriptome? Recent studies have identified distinct patterns of miRNA expression in processes such as lung development and immune response. The relevance of miRNAs to lung disease likely deserves further investigation and may emerge as being important in lung development, lung health, and lung disease.
miRNA Biogenesis and Regulation
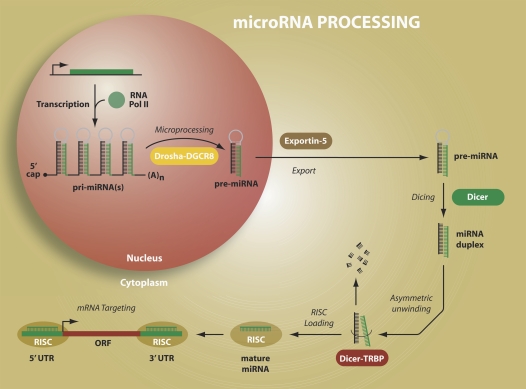
miRNAs are located in several different genomic locations (including within introns of protein- coding genes, within introns or exons of noncoding RNAs), as independent transcription units, or clustered as a group or family (“polycistronic”) sharing similar functions (15). miRNA processing requires a series of steps, each of which are quite complex and the focus of intense investigation. Within the nucleus, miRNAs are first transcribed as long primary transcripts by RNA polymerase II into primary miRNAs (pri-miRNAs), which range from hundreds to thousands of nucleotides in length (Figure 1). While in the nucleus, an RNase III termed “Drosha” cleaves both strands of the pri-miRNA to release a 70- to 100-nt stem loop, which is termed the precursor miRNA (pre-miRNA) (16). The pre-miRNA is exported from the nucleus to the cytoplasm by Exportin5/RanGTP. Once in the cytoplasm, a second RNase III, termed “Dicer” in conjunction with a double-stranded RNA binding domain (dsRBD) cleaves the pre-miRNA, releasing a 19- to 25-nt RNA duplex (mature miRNA and its complement miRNA*) (17). A single strand of the miRNA/miRNA* duplex is released and incorporated into the miRNA-induced silencing complex (miRISC) while the other strand is degraded. The miRISC represents a regulatory complex consisting of several protein factors, including the Argonaut protein, and is the focus of extensive study (18, 19). The miRISC guides miRNAs to the target mRNA to affect either mRNA degradation or translational inhibition (17). This is partially accomplished through core base-pair complementarity between the miRNA 5′ region (termed the “seed sequence”) and target mRNA 3′ UTR. The mechanisms by which miRNAs regulate gene and protein expression remain poorly understood, and investigators recognize that several factors beyond the seed sequence may affect targeting, including proximity of other miRNAs and distance of the miRNA/mRNA pairing from the stop codon (3). miRNA may also bind to the 5′ UTR and suppress (20) or enhance translation following binding to adenine-uracil (AU)-rich elements (21). Despite these complicated scenarios, most studies of miRNA target identification have focused on the ability of miRNA to suppress translation following binding to the 3′ UTR of the mRNA.
Figure 1.
miRNA processing. miRNAs are first transcribed as long primary transcripts by RNA polymerase II into primary miRNAs (pri-miRNAs). While in the nucleus, an RNase termed “Drosha” cleaves both strands of the pri-miRNA to release a 70- to 100-nucleotide stem loop termed “precursor miRNA” (pre-miRNA). The pre-miRNA is exported from the nucleus to the cytoplasm by Exportin5/RanGTP. Once in the cytoplasm, a second RNase III, termed “Dicer,” in conjunction with a double-stranded RNA binding domain cleaves the pre-miRNA, releasing an approximately 22-nucleotide RNA. A single strand of the duplex is released and incorporated into the miRNA-induced silencing complex (miRISC) while the other strand is degraded. MiRISCs guide miRNAs to the target mRNA to affect either mRNA degradation or translational inhibition. This is partially accomplished through core base-pair complementarity between the miRNA 5′region (termed the “seed sequence”) and target mRNA 3′ UTR. miRNA may also bind to the 5′ UTR and suppress or enhance translation following binding to AU-rich elements.
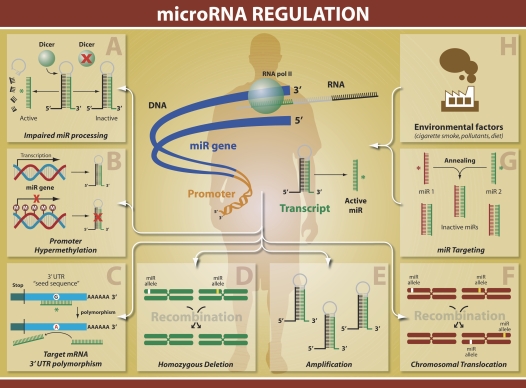
The determinants of miRNA regulation are multifactorial (Figure 2). The observation that regulation can be dependant on chromosomal location is based on several studies in both solid and hematologic malignancies. miRNAs are often located at chromosomal regions susceptible to deletions, translocations, and amplifications (7). In addition, miRNA/miRNA interactions, epigenetic regulation, and possibly environmental factors have all been implicated in miRNA regulation (22, 23). Recent studies also demonstrate that the presence of polymorphisms in the 3′ UTR of a target gene may also compromise miRNA binding and regulation. One such example is the recent identification of single nucleotide polymorphisms (SNP) in the 3′UTR of HLA-G, which is an asthma susceptibility gene (24).
Figure 2.
miRNA regulation. MiRNA regulation is multifactorial including (A) impaired processing (B) methylation (C) target 3′UTR polymorphism (D) homozygous deletion, (E) amplification, (F) translocation, (G) miRNA/miRNA targeting and (H) environmental factors.
Tools for miRNA and Target Gene Identification
miRNAs are structurally different from traditional RNAs; therefore, methods to profile, quantify, and visualize RNAs need to be adapted for miRNA. Several methodologies exist, including Northern blotting, complimentary DNA (cDNA) arrays, bead-based flow cytometry, and quantitative reverse transcriptase–polymerase chain reaction (qRT-PCR). Northern blotting is currently the gold standard of miRNA detection. Protocols for Northern blotting using DNA oligo probes have been modified to detect miRNA (25). The sensitivity of the assay may be improved up to 10-fold using locked nucleic acids probes (LNA) (26). The advantage of Northern blotting is the ability to visualize both the mature and precursor miRNA. Disadvantages include the large amount of RNA required.
cDNA arrays, modified to detect miRNA, have become the most widely used method for profiling miRNA expression (27, 28). Most miRNA discovery protocols begin with profiling the expression of several hundreds of miRNAs using cDNA arrays. The sheer number of miRNAs that are placed on the array is an advantage of this approach. However, there are factors that must be considered in the development of an miRNA array including probe thermal stability (which can be achieved through the use of LNA or locked nucleic acid modificiation), miRNA specificity, and adequate controls (29). Another method to rapidly profile miRNA expression is a bead-based/flow cytometry method (30). In this method, each latex bead is assigned to a unique miRNA, and the miRNA expression is determined following detection by flow cytometry.
Qualitative RT-PCR techniques have been modified to quantify both the mature (31) and precursor miRNA (32). Validation of miRNA profiling studies is typically performed by qRT-PCR. Current technology enables direct profiling of miRNA expression using qRT-PCR. An advantage of profiling miRNA expression with sensitive qRT-PCR is the ability to profile very small amounts of RNA and to quantify miRNA expression levels. For example, the expression of 220 miRNAs was profiled in a single embryonic stem cell using qRT-PCR (33). Despite the advent of different methodologies, there is no clear consensus on the ideal high throughput platform for miRNA profiling.
miRNA target gene prediction is based on searching established databases that list potential miRNA target genes including PicTar, TargetScan, miRanda, miRBase (http://www.sanger.ac.uk/Software/Rfam/mirna/) and miRGen (http://www.diana.pcbi.upenn.edu/miRGen.html), all of which are freely accessible. Once a target gene or genes are identified, the mRNA and protein levels of the target are measured after in vitro or in vivo manipulation of miRNA expression levels. For example, the addition of a synthetic miRNA oligonucleotide should reduce the levels of mRNA and protein expression (34). The next step is to transfect the cell of interest with luciferase reporter constructs with the predicted miRNA target site cloned into the 3′ UTR of the reporter gene (35). Once this is achieved, deletional or mutational analysis of the luciferase reporter constructs can be performed to identify putative miRNA regulatory elements. This approach relies upon the ability of the miRNA to bind to the 3′ UTR. Co-labeling cells with in situ hybridization probe for a given miRNA and its putative mRNA target is another method to predict a functional target.
miRNAs in Lung Disease
miRNAs and lung development.
Whereas few studies have explored the role for miRNAs in lung disease, several have suggested their importance in lung development and lung homeostasis. Researchers recently demonstrated that a global reduction in miRNA processing, through lung-specific, targeted deletion of Dicer, results in abnormal embryonic lung development manifested by apoptosis and abnormal airway branching (36). Both individual miRNAs and clusters may be important in normal lung development. Although Dicer affects miRNA maturation, the miRNA cluster, miR-17–92, is important in lung development and homeostasis. MiR 17–92 cluster expression is high in embryonic development and steadily declines through development and into adulthood (37). Ventura and colleagues demonstrated that mice deficient in the miR-17–92 cluster exhibit an altered phenotype characterized by smaller size, hypoplasia of the lung, ventricular septal defects, and deficiencies in normal B-cell development (38). Conversely, murine models of lung-specific miR-17–92 cluster overexpression results in an abnormal phenotype manifested by absence of terminal air sacs, which are replaced with highly proliferative, undifferentiated epithelium (37). Interestingly, a recent study demonstrated a lack of age-related changes in miRNA expression within murine lungs. Such a finding may suggest that miRNAs are important to maintaining homeostasis in gene and protein expression within the lung (39). The observation that overexpression of the miR-17–92 cluster results in the overpopulation of an undifferentiated cell population strongly suggests its importance in lung disease. The link between normal cellular growth and differentiation and disease development is not new. miRNAs such as the miR-17–92 cluster appears critical in establishing normal cellular growth and differentiation. This same cluster may ultimately prove relevant to lung diseases that partially rely on normal cellular repair and regeneration.
miRNAs as modulators of the immune response.
The inflammatory response to either the innate or acquired immune system involves the activation of numerous genes. In response to endotoxin, miR-132, miR-146a, and miR-155 are up-regulated in macrophage cell lines (40), while miR-125b is decreased (41). Toll-like receptor (TLR) -2, -4, or -5 activation and proinflammatory cytokines, tumor necrosis factor (TNF)-α and IL-1β, rapidly induce the expression of mir-146a, whereas viral-associated TLR-3 and TLR-9 activation do not (40). Endotoxin-induced nuclear factor-κB (NF-κB) activity in immune cells is important in inducing gene expression that facilitates the resolution of inflammation. Of the numerous genes activated by NF-κB, one would expect that miRNAs are also targeted. Indeed, miR-146a expression is directly regulated by NF-κB activation (40). Expression of miR-146a targets several downstream signaling molecules of TLRs and IL-1 receptor (40), suggesting that this miRNA may temporally limit the immune response. In contrast to miR-146a, miR-155 is induced by both innate and viral immune responses by macrophages (42). Notably, miR-155 is increased in macrophages stimulated with viral or bacterial/viral host cytokines, IFN-β or IFN-γ, respectively. Interestingly, TLRs that respond to either bacterial or viral insult also induce miR-155.
Whereas the above studies are based on in vitro observations, miRNA expression from the lungs of mice exposed to aerosolized endotoxin reveals a very different expression pattern (43). Only miR-146a is among the top 46 significantly up-regulated miRNAs from the mouse lungs (43), suggesting that many different lung cells respond to bacterial challenge. miRNA expression in the lung coincides with a reduction of TNF-α, macrophage inflammatory protein, and keratinocyte-derived chemokine in bronchoalveolar fluid from these mice (43).
A comparison of miRNA expression in activated T cells to that of endotoxin-treated mice or macrophages indicates commonality between both the innate and acquired immune responses. Shared between the two immune responses are miR-21, -103, -155, and -204 (43, 44). Of note, all down-regulated miRNAs in the activated T cells (miR-16, -26, -30b, -30c, -150, -181, and let-7 family members) are among the significantly expressed miRNAs in endotoxin-treated mouse lungs, indicating that these may be specific for the innate immune responses.
In addition to T-cell and macrophage responses, miR-155 is also important in B-cell activation events. Vigorito and colleagues reported that the absence of miR-155 in B cells results in an attenuated production of IgG1 antibodies (45). Of its many potential targets in B cells, miR-155 appears to regulate the transcription factor PU.1 to influence antibody production. These observations are consistent with the phenotype of mice lacking miR-155. In addition to a decrease in antibody production, these mice also displayed reduced IL-2 and IFN-γ responses, indicating that both T and B cells depend on miR-155 for immune challenges (46). In addition, dendritic cells from these mice fail to present antigen to T cells upon endotoxin challenge. Although there is no defect in the development of mononuclear phagocytes in miR-155 null mice, there is extensive remodeling of the lung that is associated with an apparent accumulation of these cells.
Although the immune system uses miRNAs to regulate development and immune responses, viruses exploit the function of miRNAs by either changing their expression (47) or releasing viral miRNA orthologs into infected cells (48). Recently, an miRNA in Kaposi's sarcoma-associated herpes virus was identified homologous to miR-155 (48) Thus, the immune regulation imparted by miRNAs is important to maintain and limit immune responses. Dysregulation of these functions are evident in many inflammatory diseases (43, 44).
miRNAs and lung cancer.
Researchers have identified abnormal expression of miRNAs in several types of malignancies including colorectal cancer, lymphoma, glioblastoma, breast cancer, lung cancer, and hepatocellular carcinoma (49–51). miRNAs may function as either tumor suppressors or oncogenes (49). Our knowledge of miRNAs in lung cancer is just starting to emerge. One of the first miRNAs identified (let-7) appears to be important in lung cancer. Overexpression of let-7 inhibits RAS protein expression and let-7 complementary sites are present in human NRAS and KRAS 3′-UTR (52). Both in vitro and in vivo models show that let-7 is important to lung tumorigenesis (53, 54). These observations are supported clinically by the correlation between reduced let-7 expression in 143 resected lung cancer cases and poor clinical outcome (55). More fundamentally, global impairments in miRNA processing can promote lung tumorigenesis. Targeted silencing of components of the miRNA machinery including Dicer, Drosha, and DGCR8 have resulted in lung tumor development (56). Several miRNAs have also been identified as potential therapeutic targets based on their direct effects on cancer cell phenotype. As an example, miR-128B, which is predicted to target epidermal growth factor receptor, demonstrates frequent loss of heterozygosity in lung tumors (57). Furthermore, loss of herozygosity correlated with response to epidermal growth factor receptor inhibition. Other miRNAs such as miR-221,222 and 17–92 have been implicated in altering lung cancer cell phenotype and sensitizing lung cancer cells to cytotoxic agents (58–60). MiR-21 is an miRNA that is overexpressed in several solid malignancies including those of the lung, breast, and colon (61). MiR-21 alters cancer cell phenotype through targeting genes critical to cell transformation including programmed cell death and the tumor suppressor tropomysin (62, 63).
High-throughput interrogations of the proteome and genome are currently being used to identify the molecular heterogeneity and predictive signatures in lung cancer (64, 65). The first such study of miRNAs conducted by Yanaihara and colleagues examined global miRNA expression patterns using cDNA array-based technology. The authors compared miRNA profiles in tumor versus adjacent uninvolved lung in 104 cases of non–small cell lung cancer (66). The investigators identified 43 miRNAs that were differentially expressed between lung tumors and adjacent uninvolved lung. In addition, five distinct miRNAs (miR-155, 17–3p, let-7a-2, 145, and 21) predicted prognosis among patients with lung cancer. A more recent study using a RT-PCR– based platform examined miRNA expression in 112 non–small cell lung cancer tumors. They also identified a signature that predicted survival (high-risk miRNAs miR-137, miR-182*, and miR-372, low-risk miRNAs miR-221 and let-7a) (67) The observed difference in signatures between the two studies were likely related to study design and the platform selected for analysis.
Incorporating miRNAs into a Systems Biology Approach to Human Disease
How do we fit miRNAs expression into a diagnostic framework to determine individualized molecular networks determining health and disease? The answer may reside in the emerging field of systems biology. Classical reductionist approaches to identifying single gene targets for diagnosis and therapy of complex human lung disease have been largely unproductive. As an alternative, the field of systems biology seeks to understand the regulation and dynamics of gene and protein network activation by integrating available information with human phenotypes. MiRNAs may turn out to be proximal regulators of gene transcription and protein production, which makes them exciting targets to control protein network activation. Moreover, an understanding of which protein networks are activated in individuals in health and disease should enable the opportunity to diagnose disease before it is clinically manifest. Once defined, these miRNAs can be targeted to regulate gene and protein network activity to promote health.
Other applications of these tools are to explore miRNA, gene and protein expression in healthy and diseased lung tissue to define molecular underpinnings. For example, these tools can determine if a single pathologic lung phenotype arises from a single pathological mechanism or represents separate molecular events that stratify the disease into clinical outcomes.
The human genome project found that humans have an average of between 20,000 and 25,000 protein-coding genes (68). In addition, genetic variability between individuals is approximately 1% (69), suggesting that interactions between genes, proteins, and the environment contribute to differences in human phenotype, maintenance of health, and susceptibility to disease. Although progress in defining the human genome map has been startling, understanding the underlying mechanisms leading to complex human disease has not been realized. In molecular network regulation, batteries of approximately 22,500 genes and their resulting proteins can be regulated by approximately 500 miRNAs. As opposed to small interfering RNA, which targets specific mRNA molecules, miRNAs may target multiple genes, their proteins, and can target gene networks. Few studies to date have explored their relevance to the pathogenesis of lung disease (Table 1). Our next steps are to define how downstream protein networks are affected by these miRNAs and examine how these targets may be uniquely regulated in different individuals in health and disease.
TABLE 1.
miRNAs IMPLICATED IN LUNG DISEASE
| Disease/Biological Process | MicroRNA |
| Lung development | DICER (34) miR-17-92 (35, 36) |
| Immune response | miR16,21,26,30b,30c,103,125,132,146A,150,155,181,204,let-7 (38–46) |
| Hypoxia (in vitro) | miR-23,24,26,27,103,107,181,210,213 (21) |
| Asthma | miR-148A148B,152 (HLAG SNP) (22) |
| Lung cancer | let-7 (51–53) |
| DICER (54) | |
| miR-128-B (55) | |
| miR-221,222,17–19 (56–58) | |
| 17-3p,21,145,155,let-7a2 (64) | |
| 137,182,372,221,let-7a (65) |
While the potential for miRNAs as therapeutic targets exist, there are few examples that have been translated beyond in vitro and murine models of disease. Potential limitations include proper modes of delivery, tissue specificity, and off-target effects. LNA modified antisense oligonucleotides that target specific miRNAs are currently being utilized in vitro in cell systems of glioblastoma multiforme (miR-21), lung cancer (miR-17–92) and hepatitis C (miR-122) (58, 70, 71). To date, few studies targeting miRNAs have been conducted. A recent investigation demonstrated tissue-specific silencing of miR-122 in primates following intravenous injection of an LNA modified antisense oligonucleotides (72). Despite these studies, in vivo models for therapeutic delivery of miRNAs are in the early stages and require further study.
In summary, new tools provided by systems biology can translate the power of the genome map to functional understanding of human health and disease. miRNAs are critical proximal components of this network regulation and will likely emerge as key regulators of lung health and disease. To accomplish the goal of solving complex human disease, we need to understand individual gene–gene, gene–protein, and protein–protein network interactions in human health and disease. The application of miRNA to regulatory elements in lung protein network regulation using systems biology tools can transform the care and diagnosis of patients with lung disease from reactive and population-based to proactive, preventive, and individualized.
Acknowledgments
The authors thank Timothy Eubank, Ph.D., for his assistance in developing the figures for this manuscript.
Supported by National Institutes of Health Grant #HL077717 (S.P.N.) and Chest/LUNGevity Foundation Grant (S.P.N.).
Originally Published in Press as DOI: 10.1164/rccm.200807-1042PP on September 11, 2008
Conflict of Interest Statement: S.P.N-S. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. M.G.H. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. G.J.N. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. T.D.S. received $2,000 consultant fee from Intergenetics. R.G. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. D.G. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. C.B.M. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993;75:843–854. [DOI] [PubMed] [Google Scholar]
- 2.Chu CY, Rana TM. Small RNAs: regulators and guardians of the genome. J Cell Physiol 2007;213:412–419. [DOI] [PubMed] [Google Scholar]
- 3.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 2007;27:91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mertens-Talcott SU, Chintharlapalli S, Li X, Safe S. The oncogenic microRNA-27a targets genes that regulate specificity protein transcription factors and the G2-M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res 2007;67:11001–11011. [DOI] [PubMed] [Google Scholar]
- 5.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA 2007;104:15805–15810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005;120:15–20. [DOI] [PubMed] [Google Scholar]
- 7.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA 2004;101:2999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med 2007;13:613–618. [DOI] [PubMed] [Google Scholar]
- 9.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 2004;432:226–230. [DOI] [PubMed] [Google Scholar]
- 10.Fiore R, Siegel G, Schratt G. MicroRNA function in neuronal development, plasticity and disease. Biochim Biophys Acta 2008;1779:471–478. [DOI] [PubMed] [Google Scholar]
- 11.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell 2003;115:787–798. [DOI] [PubMed] [Google Scholar]
- 12.Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics 2007;8:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci USA 2006;103:2746–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang J, Guo JT, Jiang D, Guo H, Taylor JM, Block TM. Liver-specific microRNA miR-122 enhances the replication of hepatitis C virus in nonhepatic cells. J Virol 2008;82:8215–8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du T, Zamore PD. MicroPrimer: the biogenesis and function of microRNA. Development 2005;132:4645–4652. [DOI] [PubMed] [Google Scholar]
- 16.Kim VN, Nam JW. Genomics of microRNA. Trends Genet 2006;22:165–173. [DOI] [PubMed] [Google Scholar]
- 17.Vatolin S, Navaratne K, Weil RJ. A novel method to detect functional microRNA targets. J Mol Biol 2006;358:983–996. [DOI] [PubMed] [Google Scholar]
- 18.Jannot G, Boisvert ME, Banville IH, Simard MJ. Two molecular features contribute to the Argonaute specificity for the microRNA and RNAi pathways in C. elegans. RNA 2008;14:829–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leung AK, Calabrese JM, Sharp PA. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci USA 2006;103:18125–18130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci USA 2007;104:9667–9672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science 2007;318:1931–1934. [DOI] [PubMed] [Google Scholar]
- 22.Saito Y, Jones PA. Epigenetic activation of tumor suppressor microRNAs in human cancer cells. Cell Cycle 2006;5:2220–2222. [DOI] [PubMed] [Google Scholar]
- 23.Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Gosto-Perez FJ, Davuluri, Liu CG, Croce CM, Negrini M, et al. A microRNA signature of hypoxia. Mol Cell Biol 2007;27:1859–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan Z, Randall G, Fan J, Camoretti-Mercado B, Brockman-Schneider R, Pan L, Solway J, Gern JE, Lemanske RF, Nicolae D, et al. Allele-specific targeting of microRNAs to HLA-G and risk of asthma. Am J Hum Genet 2007;81:829–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 2001;294:858–862. [DOI] [PubMed] [Google Scholar]
- 26.Varallyay E, Burgyan J, Havelda Z. Detection of microRNAs by Northern blot analyses using LNA probes. Methods 2007;43:140–145. [DOI] [PubMed] [Google Scholar]
- 27.Wang H, Ach RA, Curry B. Direct and sensitive miRNA profiling from low-input total RNA. RNA 2007;13:151–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, Dumitru CD, Shimizu M, Zupo S, Dono M, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci USA 2004;101:9740–9744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yin JQ, Zhao RC, Morris KV. Profiling microRNA expression with microarrays. Trends Biotechnol 2008;26:70–76. [DOI] [PubMed] [Google Scholar]
- 30.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature 2005;435:834–838. [DOI] [PubMed] [Google Scholar]
- 31.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 2005;33:e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmittgen TD, Jiang J, Liu Q, Yang L. A high-throughput method to monitor the expression of microRNA precursors. Nucleic Acids Res 2004;32:e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang F, Hajkova P, Barton SC, Lao K, Surani MA. MicroRNA expression profiling of single whole embryonic stem cells. Nucleic Acids Res 2006;34:e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, Ciafre SA, Farace MG. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem 2007;282:23716–23724. [DOI] [PubMed] [Google Scholar]
- 35.Kuhn DE, Martin MM, Feldman DS, Terry AV Jr, Nuovo GJ, Elton TS. Experimental validation of miRNA targets. Methods 2008;44:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harris KS, Zhang Z, McManus MT, Harfe BD, Sun X. Dicer function is essential for lung epithelium morphogenesis. Proc Natl Acad Sci USA 2006;103:2208–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic over-expression of the microRNA miR-17–92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol 2007;310:442–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 2008;132:875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams AE, Perry MM, Moschos SA, Lindsay MA. MicroRNA expression in the aging mouse lung. BMC Genomics 2007;8:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA 2006;103:12481–12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tili E, Michaille JJ, Calin GA. Expression and function of micro-RNAs in immune cells during normal or disease state. Int J Med Sci 2008;5:73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA 2007;104:1604–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moschos SA, Williams AE, Perry MM, Birrell MA, Belvisi MG, Lindsay MA. Expression profiling in vivo demonstrates rapid changes in lung microRNA levels following lipopolysaccharide-induced inflammation but not in the anti-inflammatory action of glucocorticoids. BMC Genomics 2007;8:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sonkoly E, Stahle M, Pivarcsi A. MicroRNAs and immunity: novel players in the regulation of normal immune function and inflammation. Semin Cancer Biol 2008;18:131–140. [DOI] [PubMed] [Google Scholar]
- 45.Vigorito E, Perks KL, Breu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, Das PP, Miska EA, Rodriguez A, Bradley A, et al. MicroRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity 2007;27:847–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodriguez AE, Vigorito S, Clare MV, Warren P, Couttet DR, Soond DS, van Grocock RJ, Das PP, Miska EA, Vetrie D, et al. Requirement of bic/microRNA-155 for normal immune function. Science 2007;316:608–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cameron JE, Yin Q, Fewell C, Lacey M, McBride J, Wang X, Lin Z, Schaefer BC, Flemington EK. Epstein-Barr virus latent membrane protein 1 induces cellular MicroRNA miR-146a, a modulator of lymphocyte signaling pathways. J Virol 2008;82:1946–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi JT, Braich R, Manoharan M, Soutschek J, Ohler U, et al. A viral microRNA functions as an orthologue of cellular miR-155. Nature 2007;450:1096–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Di Leva G, Calin GA, Croce CM. MicroRNAs: fundamental facts and involvement in human diseases. Birth Defects Res.C.Embryo. Today 2006;78:180–189. [DOI] [PubMed] [Google Scholar]
- 50.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 2005;65:7065–7070. [DOI] [PubMed] [Google Scholar]
- 51.Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene 2006;25:2537–2545. [DOI] [PubMed] [Google Scholar]
- 52.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell 2005;120:635–647. [DOI] [PubMed] [Google Scholar]
- 53.Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res 2007;67:7713–7722. [DOI] [PubMed] [Google Scholar]
- 54.Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non–small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci USA 2008;105:3903–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res 2004;64:3753–3756. [DOI] [PubMed] [Google Scholar]
- 56.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet 2007;39:673–677. [DOI] [PubMed] [Google Scholar]
- 57.Weiss GJ, Bemis LT, Nakajima E, Sugita M, Birks DK, Robinson WA, Varella-Garcia M, Bunn PA Jr, Haney J, Helfrich BA, et al. EGFR regulation by microRNA in lung cancer: correlation with clinical response and survival to gefitinib and EGFR expression in cell lines. Ann Oncol 2008;19:1053–1059. [DOI] [PubMed] [Google Scholar]
- 58.Matsubara H, Takeuchi T, Nishikawa E, Yanagisawa K, Hayashita Y, Ebi H, Yamada H, Suzuki M, Nagino M, Nimura Y, et al. Apoptosis induction by antisense oligonucleotides against miR-17–5p and miR-20a in lung cancers overexpressing miR-17–92. Oncogene 2007;26:6099–6105. [DOI] [PubMed] [Google Scholar]
- 59.Garofalo M, Quintavalle C, Di LG, Zanca C, Romano G, Taccioli C, Liu CG, Croce CM, Condorelli G. MicroRNA signatures of TRAIL resistance in human non-small cell lung cancer. Oncogene 2008;27:3845–3855. [DOI] [PubMed] [Google Scholar]
- 60.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T. A polycistronic microRNA cluster, miR-17–92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res 2005;65:9628–9632. [DOI] [PubMed] [Google Scholar]
- 61.Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, Li Y. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene 2008;27:4373–4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res 2008;18:350–359. [DOI] [PubMed] [Google Scholar]
- 63.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 2008;27:2128–2136. [DOI] [PubMed] [Google Scholar]
- 64.Potti A, Mukherjee S, Petersen R, Dressman HK, Bild A, Koontz J, Kratzke R, Watson MA, Kelley M, Ginsburg GS, et al. A genomic strategy to refine prognosis in early-stage non-small-cell lung cancer. N Engl J Med 2006;355:570–580. [DOI] [PubMed] [Google Scholar]
- 65.Rahman SM, Shyr Y, Yildiz PB, Gonzalez AL, Li H, Zhang X, Chaurand P, Yanagisawa K, Slovis BS, Miller RF, et al. Proteomic patterns of preinvasive bronchial lesions. Am J Respir Crit Care Med 2005;172:1556–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006;9:189–198. [DOI] [PubMed] [Google Scholar]
- 67.Yu SL, Chen HY, Chang GC, Chen CY, Chen HW, Singh S, Cheng CL, Yu CJ, Lee YC, Chen HS, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell 2008;13:48–57. [DOI] [PubMed] [Google Scholar]
- 68.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature 2004;431:931–945. [DOI] [PubMed] [Google Scholar]
- 69.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell, Evans CA, Holt RA, et al. The sequence of the human genome. Science 2001;291:1304–1351. [DOI] [PubMed] [Google Scholar]
- 70.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res 2005;65:6029–6033. [DOI] [PubMed] [Google Scholar]
- 71.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 2005;309:1577–1581. [DOI] [PubMed] [Google Scholar]
- 72.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, et al. LNA-mediated microRNA silencing in non-human primates. Nature 2008;452:896–899. [DOI] [PubMed] [Google Scholar]