Abstract
The CDC73 tumor suppressor gene is mutationally inactivated in hereditary and sporadic parathyroid tumors. Its product, the Cdc73 protein, is a component of the RNA polymerase II and chromatin-associated human Paf1 complex (Paf1C). Here, we show that Cdc73 physically associates with the cleavage and polyadenylation specificity factor (CPSF) and cleavage stimulation factor (CstF) complexes that are required for the maturation of mRNA 3′ ends in the cell nucleus. Immunodepletion experiments indicate that the Cdc73–CPSF–CstF complex is necessary for 3′ mRNA processing in vitro. Microarray analysis of CDC73 siRNA-treated cells revealed INTS6, a gene encoding a subunit of the Integrator complex, as an in vivo Cdc73 target. Cdc73 depletion by siRNA resulted in decreased INTS6 mRNA abundance, and decreased association of CPSF and CstF subunits with the INTS6 locus. Our results suggest that Cdc73 facilitates association of 3′ mRNA processing factors with actively-transcribed chromatin and support the importance of links between tumor suppression and mRNA maturation.
Parafibromin (Cdc73) is encoded by the CDC73 (HRPT2) tumor suppressor gene, mutated in hyperparathyroidism-jaw tumor syndrome (HPT-JT) and sporadic parathyroid tumors. HPT-JT is an autosomal-dominant multiple neoplastic syndrome characterized by parathyroid tumors, ossifying fibromas of the mandible and maxilla, and renal cysts and tumors (1, 2). Mutations consist of truncation, missense, and frameshift alterations within the HRPT2 ORF and are predicted to result in deficient or impaired protein function (1–3).
Recently, we and others (4–6) have shown that, like its yeast counterpart, Cdc73 is a component of the human Paf1 complex (Paf1C). The human Paf1C includes 4 subunits with homology to members of the yeast Paf1C (Cdc73, Paf1, Ctr9, and Leo1) and an additional subunit, Ski8 (6). The human Rtf1 homolog does not appear to be part of the Paf1C, unlike its yeast counterpart. As in yeast, the human Paf1C has a central role in orchestrating cotranscriptional histone modifications. Both the yeast and mammalian Paf1 complexes are required for histone H2B monoubiquitination, which, in turn, is critical for histone lysine 4 (H3-K4) and lysine 79 (H3-K79) methylation (7) In addition, the yeast Paf1C is also required for H3-K36 methylation (8). The human Paf1C has been found to associate with a H3–K4 methyltransferase complex (4).
Genetic studies in yeast suggest that the Paf1C modulates RNA biogenesis. Deletion of Paf1C component genes results in an overall reduction in the poly(A) tail length of mRNA (9). Furthermore, the Paf1C is involved in some way in 3′ end formation of polyadenylated mRNAs (10) and nonpolyadenylated RNAs, such as snoRNAs and snRNAs (11). Most recently, it has been shown that the yeast cleavage and polyadenylation factor Cft1 associates with Paf1C and requires the Paf1C for its interaction with RNA polymerase II (12).
Most eukaryotic mRNA precursors (pre-mRNAs) undergo an extensive maturation process. Processing of the 3′ end occurs cotranscriptionally and can be divided into 2 distinct steps: endonucleolytic cleavage of the nascent mRNA 3′ end followed by synthesis of the poly(A) tail. Multiple protein factors are involved in mammalian mRNA 3′ processing (13–15). The core machinery includes poly(A) polymerase, cleavage and polyadenylation specificity factor (CPSF), cleavage stimulation factor (CstF), and cleavage factors I and II. Both CPSF and CstF are crucial to identify the precise sequence elements on the pre-mRNA where cleavage and subsequent polyadenylation occur (16). Recent studies have shown that a CPSF subunit, CPSF-73, is the pre-mRNA 3′-end-processing endonuclease (17). Importantly, RNA polymerase II, specifically the C-terminal domain of its largest subunit (CTD), is also required for efficient 3′ end formation, likely serving to help link 3′ processing to transcription (18, 19).
Here, we present evidence that Cdc73 is physically associated with CPSF and CstF, connecting the Paf1C directly with RNA 3′ end formation in human cells. Furthermore, we identify Cdc73 target genes and provide evidence that Cdc73 is required for optimal expression and CPSF/CstF recruitment to one of these, the INTS6 gene. Our results suggest that an important role of the tumor suppressor Cdc73 is to help coordinate transcription and RNA processing of specific genes.
Results
Cdc73 Physically Associates with the CPSF and CstF Complexes.
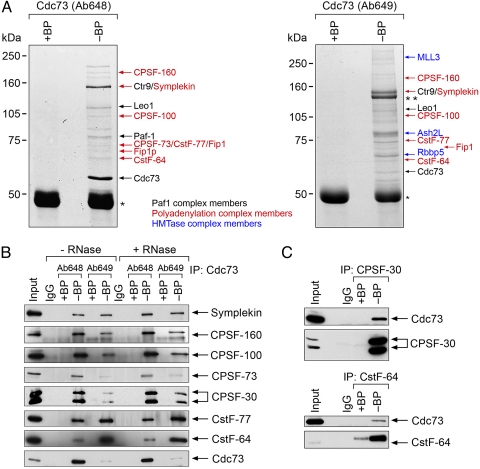
To explore the molecular architecture of human Cdc73 complexes, we used mass spectrometry to identify complex components from anti-Cdc73 immunoprecipitates with an antipeptide polyclonal antibody, Ab648. Among these proteins are the major constituents of the Paf1C (4–6) and the CPSF and CstF 3′ mRNA processing complexes (Fig. 1A and Fig. S1a). Immunoprecipitation with a second anti-Cdc73 antibody, Ab649, which is somewhat less specific as it also immunoprecipitated angiomotin from whole-cell extract (Fig. 1A Right) (4), but not from nuclear extracts (Fig. S1b), identified the same proteins as Ab648 and histone methyltransferase complex (HMTase) components (Fig. 1A Right).
Fig. 1.
Cdc73 associates with CPSF and CstF subunits. (A) Purification of Cdc73 complexes by immunoprecipitation with anti-Cdc73 antibodies, Ab648 (Left) and Ab649 (Right), from 293T cell lysates, with (+BP) or without (−BP) blocking peptide control, shown after gel electrophoresis and Coomassie blue staining. * indicates Ig heavy chain, and ** indicates angiomotin, which cross-reacts with Ab649 (4). Note that the left gel was used to identify the Paf1C components [Reproduced with permission from ref. 4 (Copyright 2005, American Society for Microbiology)]. (B) Immunoprecipitation (IP) with anti-Cdc73 Ab648 or Ab649 from HeLa nuclear extracts and immunoblotting with the indicated antibodies. Lysates were used directly (−RNase) or treated with RNase A (+RNase). Negative controls included immunoprecipitation with normal rabbit serum (IgG) and the respective blocking peptides (+BP). HeLa nuclear extracts were loaded to indicate the position of the endogenous protein (Input). (C) HeLa nuclear extracts were immunoprecipitated with anti-CPSF-30 or anti-CstF-64 antibodies. Immunoprecipitates were resolved and then immunoblotted with either anti-Cdc73 Ab 1 (Upper) or the respective anti-CPSF or anti-CstF antibody (Lower). Normal rabbit IgG and the appropriate blocking peptides (BP) were used as controls.
Because both anti-Cdc73 Ab648 and Ab649 immunoprecipitated multiple components of the CPSF and CstF complexes, we decided to verify these results by performing reciprocal coimmunoprecipitations from HeLa nuclear extracts. Both of the anti-Cdc73 antibodies immunoprecipitated symplekin, known to associate with both CPSF and CstF (20), and multiple CPSF and CstF subunits (Fig. 1B). These interactions remained intact after treatment with RNase (Fig. 1B), suggesting that the Cdc73–CPSF–CstF complex is assembled via protein–protein interactions rather than protein–RNA interactions. Conversely, anti-CPSF and anti-CstF subunit antibodies immunoprecipitated Cdc73 (Fig. 1C). We also found that Cdc73 cofractionated on a glycerol gradient in a high molecular weight complex overlapping components of CPSF and CstF (Fig. S1c). Our results are consistent with the studies by Nordick et al. (12), demonstrating that the yeast Paf1C component, Ctr9, associates with the polyadenylation factor Cft1, the yeast homolog of CPSF160, and indicate that Cdc73 is directly or indirectly associated with the CPSF/CstF complexes. Anti-Paf1- or anti-Leo1-specific antibodies did not immunoprecipitate CPSF or CstF subunits, suggesting either that human Cdc73 may function independently of the Paf1C to interact with CPSF/CstF or that the specific epitopes, to which these anti-peptide Paf1 and Leo1 antibodies are directed, are required for CPSF/CstF interaction that is therefore undetectable with these antibodies.
The Cdc73–CPSF–CstF Complex Is Necessary for 3′ mRNA Processing in Vitro.
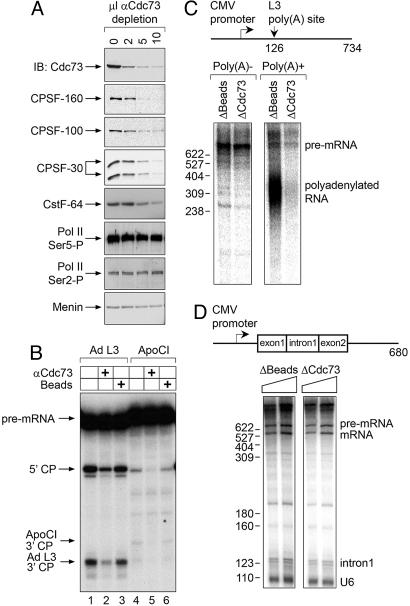
Given that CPSF and CstF are required for mRNA 3′ cleavage activity (17, 21), we wanted to determine whether the Cdc73-associated complex cleaves pre-mRNA substrates. Immunodepletion of Cdc73 from HeLa nuclear extracts resulted in efficient codepletion of CPSF-160, CPSF-100, CPSF-30, and CstF-64, whereas RNA polymerase II isoforms and menin were not depleted (Fig. 2A). Cdc73-depleted, protein A-Sepharose bead-depleted, or untreated nuclear extracts were then used for in vitro cleavage reactions. Compared with control extracts, Cdc73-depleted extracts exhibited decreased cleavage of pre-mRNA 3′ processing substrates derived from the adenovirus type 2 L3 gene (AdL3) and the apoliprotein C-I gene (ApoCI) (Fig. 2B). These results indicate that the CPSF/CstF 3′ mRNA processing activity is associated with Cdc73 but do not imply that Cdc73 is itself essential for in vitro 3′ cleavage. Indeed, extensive purification of the 3′ processing machinery suggests that Cdc73, or Paf1C, is not an essential core 3′ processing factor. Rather, we suspect that the association reflects a mechanism for facilitating linkage of the polyadenylation machinery to other complexes involved in gene expression, i.e., in transcription (14, 22).
Fig. 2.
The Cdc73 complex is associated with 3′ mRNA processing activity. (A) Nuclear extracts (NE) were immunodepleted with the indicated amounts of anti-Cdc73 Ab648 as shown. Depleted NEs were resolved by gel electrophoresis and immunoblotted with the indicated antibodies. (B) Input NE (lanes 1 and 4), NE after immunodepletion with 10 μL of anti-Cdc73 antibody (lanes 2 and 5), or bead-immunodepleted NE (lanes 3 and 6) were used in an in vitro cleavage assay using labeled adenoviral L3 (Ad L3; lanes 1–3) or human ApoCI (lanes 4–6) substrates. Arrows indicate the precursor mRNA (pre-mRNA), the 5′ cleavage products (5′ CP), and the 3′ cleavage products (ApoCI 3′ CP, and Ad L3 3′ CP). (C) A schematic of the pG3CMVL3 DNA template and the position of the polyadenylation site is shown. Cotranscriptional cleavage and polyadenylation were performed on this DNA template with either bead-depleted or Cdc73-depleted HeLa nuclear extract. RNA products were then separated into nonpolyadenylated (poly(A)−) and polyadenylated (poly(A)+) fractions by oligo(dT) selection, and analyzed on a denaturing gel. (D) At the top is a schematic of the pCMVAdML DNA template. The expected sizes of the pre-mRNA and the processed/spliced mRNA are 680 and 560 bp, respectively. Eight microliters or 10 μL of bead-depleted (Left) or Cdc73-depleted (Right) nuclear extract was used to transcribe and splice the DNA template. RNA products were analyzed on denaturing gel.
Because mRNA processing occurs while the nascent RNA chain is being synthesized by RNA polymerase II (14, 23), we tested whether the Cdc73-associated complex is also necessary for transcription-coupled cleavage and polyadenylation in vitro. To this end, we incubated bead-depleted and Cdc73-depleted nuclear extracts with a DNA template that is transcribed from a CMV promoter and contains the AdL3 poly(A) site. RNA products were then separated into nonpolyadenylated and polyadenylated fractions by oligo(dT) selection. Five percent of the nonpolyadenylated and all of the polyadenylated RNA fractions were analyzed on a denaturing gel. Strikingly, whereas equivalent amounts of nonpolyadenylated AdL3 pre-mRNA were synthesized by all tested nuclear extracts (Fig. 2C Left), indicating that transcription was unaffected by Cdc73 depletion, synthesis of polyadenylated RNA was greatly reduced in the Cdc73-depleted extracts (Fig. 2C Right). In contrast, immunodepletion of the Cdc73-associated complex had no effect on transcription-coupled splicing (Fig. 2D). We conclude that the Cdc73-associated complex is necessary for efficient 3′ end cleavage and polyadenylation, but not splicing, when these processes are coupled with transcription in vitro. Although we did not observe an effect of Cdc73 depletion on transcription of the naked DNA template used in these experiments, it will be of interest to repeat the experiments with chromatin templates.
Cdc73 Depletion Results in Decreased Ints6 Expression and Increased Relative Abundance of 3′-Extended INTS6 Transcripts.
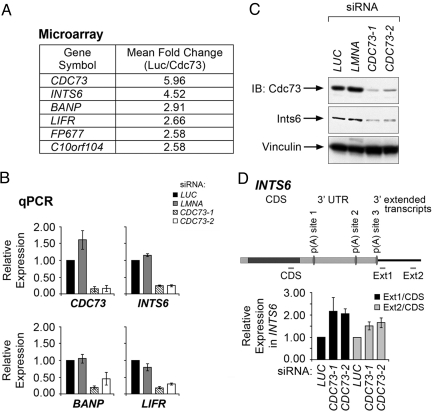
Given that the Cdc73 complex is associated with functional 3′ mRNA processing activity, we reasoned that Cdc73 might modulate levels of cellular transcripts via 3′ processing. To test this possibility, we examined global gene expression patterns in control and CDC73 siRNA-treated HeLa cells by using oligonucleotide array hybridization. Cells treated with 2 different CDC73 siRNAs revealed genes whose expression is down-regulated (Table S1) or up-regulated (Table S2) upon depletion of Cdc73. To verify the expression alterations, we analyzed the abundance of CDC73, INTS6, BANP, and LIFR, which were the most down-regulated transcripts encoding characterized proteins that were represented by at least 3 probe sets on the arrays (Fig. 3A), using quantitative real-time RT-PCR. Cells transfected with CDC73 siRNAs expressed significantly lower levels of CDC73, INTS6, BANP, and LIFR mRNA compared with luciferase (LUC) and lamin (LMNA) siRNA-treated cells (Fig. 3B).
Fig. 3.
Cdc73 regulates the expression of Ints6. (A) Mean fold change for the genes whose expression was most significantly decreased upon knockdown of Cdc73, as assayed by 3 independent probe sets on Affymetrix U133A 2.0 arrays. (B) Expression of CDC73, INTS6, BANP, and LIFR was measured by quantitative real-time RT-PCR in HeLa cells transfected with siRNAs directed to either luciferase (LUC), lamin (LMNA), or CDC73 (CDC73-1, CDC73-2). Transcript levels were normalized to the LUC control. Error bars indicate SD for 3 independent experiments. (C) HeLa cells were transfected with the indicated siRNAs, and cell lysates were subjected to immunoblot analysis (IB) with the indicated antibodies. (D) (Upper) A scheme representing the INTS6 transcript and predicted poly(A) sites located 3,780 bp [p(A) site 1], 6,335 bp [p(A) site 2], and 7,431 bp [p(A) site 3] from the transcriptional start site. Expression of INTS6 transcript was measured by using quantitative real-time RT-PCR with the depicted CDS, Ext1 and Ext2 primers in HeLa cells transfected with the indicated siRNAs. (Lower) Level of extended transcript was calculated by normalizing Ext1 or Ext2 transcript levels to the CDS transcript level. Error bars indicate SEM for 3 independent experiments.
We next focused on Ints6, a component of the Integrator complex that mediates 3′ end processing of snRNAs, because the INTS6 transcript was the most down-regulated mRNA upon depletion of CDC73. Immunoblot analysis showed that depletion of Cdc73 resulted in decreased levels of the Ints6 protein (Fig. 3C and Fig. S2a). Changes in transcript level of additional integrator subunits were not detected in our microarray studies, and immunoblot analysis for several integrator subunits did not reveal changes in their protein expression upon Cdc73 depletion (data not shown). Importantly, cotransfecting an siRNA-insensitive CDC73 mutant together with CDC73 siRNA restored Ints6 protein levels, whereas cotransfection with vector control did not rescue Ints6 expression (Fig. S2b).
We next sought to determine whether the reduced INTS6 transcript level could be correlated with changes in the 3′ ends of INTS6 transcripts. INTS6 transcripts are predicted to use at least 3 3′ processing sites (see Fig. 3D). We therefore used quantitative real-time RT-PCR to quantify different portions of the INTS6 transcript. Strikingly, after Cdc73 depletion by siRNA, relative levels of INTS6 transcripts with longer 3′ UTRs (as measured by the ratio of signals obtained with primer pairs Ext1 and Ext2 compared with signals obtained with a primer pair within the coding region; CDS, Fig. 3D) was increased. These results suggest that Cdc73 affects INTS6 transcript length and abundance. One scenario is that this effect reflects at least in part a function of Cdc73, and the Paf1C, in facilitating recruitment of the 3′ end processing machinery to active genes.
Cdc73 Depletion in Vivo Results in Decreased Association of CPSF and CstF Subunits with the INTS6 Locus.
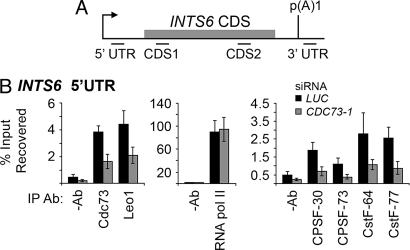
The above results are consistent with the idea that Cdc73 facilitates binding of CPSF and CstF to actively-transcribed chromatin. To test this possibility, we performed ChIPs with antibodies to Cdc73-associated proteins and other chromatin components, with extracts from HeLa cells treated with CDC73 siRNA or luciferase control siRNA. The resulting immunoprecipitates were tested for enrichment of INTS6 locus DNA by quantitative real-time PCR (Fig. 4A). Association of Cdc73 and Leo1 proteins with a DNA fragment spanning the promoter-proximal 5′ UTR of INTS6 was reduced upon Cdc73 depletion (Fig. 4B). Importantly, Cdc73 depletion decreased the association of CPSF-30, CPSF-73, CstF-64, and CstF-77 but not of RNA polymerase II with this INTS6 region (Fig. 4B). Levels of histone H3 lysine 4 methylation were also unaffected at this locus by CDC73 depletion (Fig. S3). Note that the presence of these factors near the promoter supports the view that polyadenylation factors are recruited to active promoters (22). Extending these results, CPSF and CstF cross-linking to the coding region and the 3′UTR of INTS6 was also diminished upon Cdc73 depletion (Fig. S4a). Cdc73 knockdown also resulted in significant reduction in histone H3 lysine 36 trimethylation across the INTS6 locus (Fig. S3). The Paf1C and 3′ processing antibodies did not cross-link to an unexpressed gene, hemoglobin gamma A (HBG1; Fig. S5). These results suggest that Cdc73 regulates INTS6 mRNA levels by facilitating recruitment of CPSF and CstF components to the INTS6 locus and/or or that Cdc73 plays a direct role in INTS6 transcription.
Fig. 4.
Cdc73 facilitates recruitment of CPSF and CstF subunits to the INTS6 locus. (A) Schematic of the INTS6 locus indicating primer sets used to quantify ChIPs. The arrow indicates the transcriptional start site and the gray box indicates the beginning of the coding sequence. (B) ChIPs were performed using the indicated antibodies or without antibody (−Ab) to determine the background level, in HeLa cells treated with LUC or CDC73-1 siRNAs. Precipitated chromatin was used for quantitative real-time PCR amplification with the primer INTS6 5′ UTR primer set. Error bars indicate SEM for 4 independent experiments.
Discussion
Previous studies in yeast have supported a role for the Paf1C in mRNA maturation, although the mechanism by which Paf1C mediates 3′mRNA processing was mostly unknown. Our current results suggest that human Cdc73 regulates transcript processing by binding to CPSF and CstF and facilitating their recruitment to transcribed loci. Our data suggest that Cdc73 exerts primarily a recruitment role. Whether Cdc73 also plays a regulatory role, or exerts some other function on the CPSF/CstF complex, is yet to be determined. Although various alterations in mRNA processing factors have been linked to cancer (24–26), our findings indicate that mRNA maturation may be related to the activity of a tumor suppressor protein.
In addition to its potential to modulate mRNA 3′ end formation, the Paf1C is associated with a variety of activities that modify chromatin. In yeast, the histone methylation and polyadenylation complexes appear to be linked, because the Saccharomyces cerevisiae protein Swd2 is a component of both the CPF complex and the Set1 complex (27). The Cdc73 defects that predispose to tumor formation could result from alterations in chromatin structure, alterations in 3′ mRNA processing, or a combination of these events. In this regard, it is intriguing that mutation of the MEN1 tumor suppressor gene also results in parathyroid tumors, and that the protein product of MEN1, menin, is similarly complexed with the MLL histone methyltransferases (28, 29).
The Paf1C is localized to transcriptionally active genes, suggesting a possible global role in transcription (30, 31). However, it does not appear to be required for expression of most genes (6, 7, 10), and the human Paf1C specifically regulates expression of only a subset of genes, including Hox genes (7) and genes encoding certain growth factors (32). 3′ mRNA processing factors have been reported to associate with transcribed loci in yeast and mammalian cells. It still remains unclear whether these factors bind to the entire coding region, including the promoter, throughout transcription (33, 34) or become enriched preferentially at the 3′ ends of genes (35). Although our results provide support for the former possibility, genomewide maps for 3′ mRNA processing factors using microarrays (ChIP-chip) or sequencing (ChIP-seq) will help to clarify this divergence.
Why are only specific target genes affected by loss of Cdc73? One possibility is that both yeast and human Paf1C target genes have a specific sequence or structure at their poly(A) site that is particularly sensitive to loss of cotranscriptional poly(A) factor recruitment. It will be important to determine whether Cdc73-sensitive transcripts in fact share some particular features of their poly(A) site or 3′UTR. More broadly, depletion of Cdc73 followed by mRNA analysis on whole-genome tiling arrays could identify additional Paf1C-target genes, including those, like INTS6, that generate 3′-extended transcripts in the absence of Cdc73. However, an interesting possibility is that expression of some, perhaps many, genes is not significantly affected when cotranscriptional recruitment of 3′ processing factors is compromised.
It remains to be determined whether the 3′ mRNA processing functions associated with Cdc73, shown here, are indeed essential to prevent tumor formation. The development of suitable model systems in which to study parathyroid tumorigenesis and the role of Cdc73, such as tumor-derived cell lines or animal models, will help to resolve this question. Further elucidation of Cdc73 function is likely to contribute to a better understanding of the links between tumor suppression, histone modification, and pre-mRNA processing.
Materials and Methods
Antibodies.
Anti-Cdc73, anti-CPSF, anti-CstF, and anti-Ints6 antibodies were generated by Bethyl Laboratories.
Immunopurification and Mass Spectrometry.
Purification of Cdc73 interacting proteins was performed as described (4). Trypsin digestion and mass spectrometry were carried out at the Taplin Biological Mass Spectrometry Facility (Harvard Medical School).
Immunoprecipitation.
Nuclear extracts (Accurate Chemicals) were incubated for either 3 h or overnight with 3–5 μL of the indicated antibodies or with negative controls that included antibodies blocked with peptide (1:1 mass ratio) or rabbit IgG, and 50 μL of protein A-Sepharose beads (GE HealthCare). RNase A (280 μg) (Sigma) was added to the indicated Cdc73 immunoprecipitations shown in Fig. 1B.
In Vitro 3′ Cleavage Assays and in Vitro Transcription-Coupled Processing Assays.
32P-labeled apolipoprotein C-I (ApoCI) and adenovirus type 2 L3 gene (AdL3) pre-mRNAs and HeLa nuclear extracts were prepared as described (36, 37). Cleavage assays using untreated, protein A-Sepharose bead-depleted, and anti-Cdc73 immunodepleted nuclear extracts were performed as described (36). Transcription-coupled polyadenylation was carried out, and RNA products were separated into nonpolyadenylated and polyadenylated fractions by oligo(dT) selection, thereafter 5% of nonpolyadenylated and all of the polyadenylated fractions were analyzed on 6% denaturing gel.
RNA Interference.
Ten-centimeter plates of HeLa or U2OS cells were transfected with the indicated siRNA duplexes by using Oligofectamine (Invitrogen) according to the manufacturer's instructions and as described (38). Cells were harvested 72 h after transfection.
RNA Isolation, Quantitation, and Expression Analysis.
Total RNA was extracted from HeLa cells by using Trizol (Invitrogen), followed by DNase treatment (DNA-free; Ambion), and purification on RNeasy columns (Qiagen). The extracted RNA was converted to cDNA by using Sprint PowerScript random hexamer preprimed single shots (Clontech) and used for real-time PCR. Total RNA was isolated and hybridized to Human Genome U133A 2.0 Arrays (Affymetrix) according to standard protocols. Data were analyzed with dChip, arrays were normalized by using default parameters, and expression levels were calculated with the PM-MM model.
ChIPs.
ChIP and real-time PCR quantitation of coimmunoprecipitated DNA fragments were performed as described (39). Briefly, siRNA-treated HeLa cells were formaldehyde-cross-linked 72 h after transfection, and the DNA was sheared by sonication. Approximately 100 μg of cross-linked whole-cell extract was used per immunoprecipitation. After overnight immunoprecipitation, each sample was washed with sonication buffer, high-salt buffer, LiCl buffer, and Tris-EDTA. The DNA was eluted, and phenol/chloroform was extracted after proteinase K digestion. DNA fragments of each sample were quantitated in triplicate by using Power SYBR green PCR Master Mix (Applied Biosystems). ChIP assays were performed on 4 independent transfections.
See SI Text for detailed methods and primer sequences.
Supplementary Material
Acknowledgments.
We thank J. Jaehning, S. Buratowski, and V. Runner for sharing unpublished results and discussions; S. Rozenblatt, and M. Keogh for discussions; A. Bass, S. A. Woo, W. Lin, and A. Alexander for critical reading of the manuscript; E. McIntush at Bethyl Laboratories for antibody design and preparation of polyclonal antibodies; R. Tomaino at Taplin Biological Mass Spectrometry Facility for mass spectrometric analysis; R. Lührmann and C. Merz (Max Planck Institute, Gottingen, Germany) for providing plasmids; and S. Nannepaga for technical assistance. This work was supported by gifts from the Caring for Carcinoid Foundation and Drs. R. and B. Sackler (to M.M.) and National Institutes of Health Grant GM28983 (to J.L.M.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812023106/DCSupplemental.
References
- 1.Carpten JD, et al. HRPT2, encoding parafibromin, is mutated in hyperparathyroidism-jaw tumor syndrome. Nat Genet. 2002;32:676–680. doi: 10.1038/ng1048. [DOI] [PubMed] [Google Scholar]
- 2.Shattuck TM, et al. Somatic and germ-line mutations of the HRPT2 gene in sporadic parathyroid carcinoma. N Engl J Med. 2003;349:1722–1729. doi: 10.1056/NEJMoa031237. [DOI] [PubMed] [Google Scholar]
- 3.Howell VM, et al. HRPT2 mutations are associated with malignancy in sporadic parathyroid tumours. J Med Genet. 2003;40:657–663. doi: 10.1136/jmg.40.9.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rozenblatt-Rosen O, et al. The parafibromin tumor suppressor protein is part of a human Paf1 complex. Mol Cell Biol. 2005;25:612–620. doi: 10.1128/MCB.25.2.612-620.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yart A, et al. The HRPT2 tumor suppressor gene product parafibromin associates with human PAF1 and RNA polymerase II. Mol Cell Biol. 2005;25:5052–5060. doi: 10.1128/MCB.25.12.5052-5060.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu B, et al. The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev. 2005;19:1668–1673. doi: 10.1101/gad.1292105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu B, et al. Monoubiquitination of human histone H2B: The factors involved and their roles in HOX gene regulation. Mol Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 8.Chu Y, et al. Regulation of histone modification and cryptic transcription by the Bur1 and Paf1 complexes. EMBO J. 2007;26:4646–4656. doi: 10.1038/sj.emboj.7601887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mueller CL, Porter SE, Hoffman MG, Jaehning JA. The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol Cell. 2004;14:447–456. doi: 10.1016/s1097-2765(04)00257-6. [DOI] [PubMed] [Google Scholar]
- 10.Penheiter KL, et al. A posttranscriptional role for the yeast Paf1-RNA polymerase II complex is revealed by identification of primary targets. Mol Cell. 2005;20:213–223. doi: 10.1016/j.molcel.2005.08.023. [DOI] [PubMed] [Google Scholar]
- 11.Sheldon KE, Mauger DM, Arndt KM. A requirement for the Saccharomyces cerevisiae Paf1 complex in snoRNA 3′ end formation. Mol Cell. 2005;20:225–236. doi: 10.1016/j.molcel.2005.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nordick K, Hoffman MG, Betz JL, Jaehning JA. Direct interactions between the Paf1 complex and a cleavage and polyadenylation factor are revealed by dissociation of Paf1 from RNA polymerase II. Eukaryot Cell. 2008;7:1158–1167. doi: 10.1128/EC.00434-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 14.Proudfoot N. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr Opin Cell Biol. 2004;16:272–278. doi: 10.1016/j.ceb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirose Y, Manley JL. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- 17.Mandel CR, et al. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature. 2006;444:953–956. doi: 10.1038/nature05363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirose Y, Manley JL. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- 19.McCracken S, et al. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 20.Takagaki Y, Manley JL. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol Cell Biol. 2000;20:1515–1525. doi: 10.1128/mcb.20.5.1515-1525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takagaki Y, Ryner LC, Manley JL. Four factors are required for 3′-end cleavage of pre-mRNAs. Genes Dev. 1989;3:1711–1724. doi: 10.1101/gad.3.11.1711. [DOI] [PubMed] [Google Scholar]
- 22.Calvo O, Manley JL. Strange bedfellows: Polyadenylation factors at the promoter. Genes Dev. 2003;17:1321–1327. doi: 10.1101/gad.1093603. [DOI] [PubMed] [Google Scholar]
- 23.Zorio DA, Bentley DL. The link between mRNA processing and transcription: Communication works both ways. Exp Cell Res. 2004;296:91–97. doi: 10.1016/j.yexcr.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 24.Cools J, et al. The FIP1L1-PDGFRalpha kinase in hypereosinophilic syndrome and chronic eosinophilic leukemia. Curr Opin Hematol. 2004;11:51–57. doi: 10.1097/00062752-200401000-00008. [DOI] [PubMed] [Google Scholar]
- 25.Kleiman FE, Manley JL. The BARD1-CstF-50 interaction links mRNA 3′ end formation to DNA damage and tumor suppression. Cell. 2001;104:743–753. doi: 10.1016/s0092-8674(01)00270-7. [DOI] [PubMed] [Google Scholar]
- 26.Topalian SL, et al. Identification and functional characterization of neo-poly(A) polymerase, an RNA processing enzyme overexpressed in human tumors. Mol Cell Biol. 2001;21:5614–5623. doi: 10.1128/MCB.21.16.5614-5623.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng H, He X, Moore C. The essential WD repeat protein Swd2 has dual functions in RNA polymerase II transcription termination and lysine 4 methylation of histone H3. Mol Cell Biol. 2004;24:2932–2943. doi: 10.1128/MCB.24.7.2932-2943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hughes CM, et al. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol Cell. 2004;13:587–597. doi: 10.1016/s1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- 29.Yokoyama A, et al. Leukemia protooncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24:5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pokholok DK, Hannett NM, Young RA. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol Cell. 2002;9:799–809. doi: 10.1016/s1097-2765(02)00502-6. [DOI] [PubMed] [Google Scholar]
- 31.Simic R, et al. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 2003;22:1846–1856. doi: 10.1093/emboj/cdg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang P, et al. Parafibromin, a component of the human PAF complex, regulates growth factors and is required for embryonic development and survival in adult mice. Mol Cell Biol. 2008;28:2930–2940. doi: 10.1128/MCB.00654-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calvo O, Manley JL. The transcriptional coactivator PC4/Sub1 has multiple functions in RNA polymerase II transcription. EMBO J. 2005;24:1009–1020. doi: 10.1038/sj.emboj.7600575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venkataraman K, Brown KM, Gilmartin GM. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev. 2005;19:1315–1327. doi: 10.1101/gad.1298605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim M, et al. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 2004;23:354–364. doi: 10.1038/sj.emboj.7600053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaneko S, Rozenblatt-Rosen O, Meyerson M, Manley JL. The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3′ processing and transcription termination. Genes Dev. 2007;21:1779–1789. doi: 10.1101/gad.1565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takagaki Y, Ryner LC, Manley JL. Separation and characterization of a poly(A) polymerase and a cleavage/specificity factor required for pre-mRNA polyadenylation. Cell. 1988;52:731–742. doi: 10.1016/0092-8674(88)90411-4. [DOI] [PubMed] [Google Scholar]
- 38.Scacheri PC, et al. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc Natl Acad Sci USA. 2004;101:1892–1897. doi: 10.1073/pnas.0308698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chakrabarti SK, Mirmira RG. Transcription factors direct the development and function of β cells. Trends Endocrinol. 2003;14:78–84. doi: 10.1016/s1043-2760(02)00039-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.