Abstract
Although many melanomas harbor either activating mutations in BRAF or NRAS, there remains a substantial group of tumors without either mutation about which little is known. Here, we used a genomic strategy to define a novel group of melanoma cell lines with co-overexpression of CDK4 and KIT. Although this sub-group lacked any known KIT mutations, they had high phospho-KIT receptor expression - indicating receptor activity. fuantitative PCR confirmed the existence of a similar KIT/CDK4 sub-group in human melanoma samples. Pharmacological studies showed the KIT/CDK4 overexpressing sub-group to be resistant to BRAF inhibitors but sensitive to imatinib in both in vitro and in vivo melanoma models. Mechanistically, imatinib treatment led to increased apoptosis and G1-phase cell cycle arrest associated with the inhibition of phospho-ERK and increased expression of p27KIP. Other melanoma cell lines, which retained some KIT expression but lacked phospho-KIT, were not sensitive to imatinib, suggesting that KIT expression alone is not predictive of response. We suggest that co-overexpression of KIT/CDK4 is a potential mechanism of oncogenic transformation in some BRAF/NRAS wild-type melanomas. This group of melanomas may be a sub-population for which imatinib or other KIT inhibitors may constitute optimal therapy.
Introduction
As melanoma treatment moves into the era of targeted therapy, there is a growing need to unravel the underlying genetic complexity and cellular signaling heterogeneity of this tumor. It is hoped that an understanding of how genetic and signaling profiles dictate pharmacological response will allow for the selection of optimal patient populations for clinical trials. Following the discovery that many melanomas harbor activating mutations in either BRAF or NRAS, there has been much interest in targeting the BRAF/MEK/ERK mitogen activated protein kinase (MAPK) pathway (1, 2). Although it has been shown that the presence of the BRAF mutation is predictive of response to MEK inhibitors (3), this is unlikely to be the only determinant (4, 5). In a recent study, we demonstrated that there was little correlation between the inhibition of phospho-ERK levels and of melanoma cell line growth following MEK inhibitor treatment (6). This observation suggested that there were likely to be other changes in the pathways responsible for cell proliferation that may be more predictive of response to these small molecule inhibitors when identified. In the current study, we have turned our attention to melanoma lines that respond poorly to BRAF inhibitors.
Entry into the cell cycle is regulated at the G1 restriction checkpoint, a process that becomes deregulated in cancer cells. Progression through the G1 into S-phase of the cell cycle is driven by cyclin dependent kinases (CDK) 4 and 6 which interact with cyclin D1 as well as by CDK2 which interacts with cyclins A/E (7). Uncontrolled growth of melanoma cells results from constitutive MAP kinase activity leading to increased cyclin D1 and reduced p27KIP1 expression (8).
There appears to be differences in the genetic profiles of melanomas that originate from either skin that is chronically sun-damaged (as defined by the appearance of solar elastosis) or skin that lacks sun-induced damage. Thus, melanomas that arise on skin with chronic sun-induced damage have a low incidence of BRAF mutations and instead showed increased cyclin D1 copy number. Frequent amplifications of cyclin D1 also occur in distinct histological subtypes of melanoma. Thus 44% of acral melanomas, 19% of lentigo malignant melanomas and 6% of superficial spreading melanomas are known to have increased cyclin D1 copy number (9). There also is evidence for amplification (10) and mutation of CDK4 (11) in small sub-groups of melanomas (12). Other distinct sub-groups of melanoma have been shown to harbor oncogenic mutations in the receptor tyrosine kinase KIT (13). Again, these aberrations are restricted to certain groups of melanoma, with KIT dysregulation being reported in 36% of acral and 28% of melanomas arising on chronically sun-damaged skin (13). There are no cell lines derived from these rare melanomas, making it difficult to perform the pre-clinical studies essential for guiding clinical trial design.
The current study has identified of a novel sub-set of BRAF inhibitor resistant melanoma cell lines with high expression of both KIT and CDK4. This sub-group of melanomas lack KIT mutations, but have high KIT signaling activity and show sensitivity to imatinib treatment. Similar patterns of CDK4/KIT expression were also found in clinical melanoma specimens. We therefore suggest that pharmacogenomic analysis of melanoma populations may be a suitable strategy for the further sub-classification of melanoma leading to more “personalized” therapy approaches.
Materials and methods
Cell culture
Human melanoma cells and melanocytes were isolated and cultured as described in (14). The lentiviral vector shRNA constructs for KIT were obtained from Dr Levi Garraway (the Broad Institute, Cambridge, MA). The WM1382 cell line was derived from a superficial spreading melanoma. Histopathological data was not available for the WM8 line.
Adherent cell proliferation analysis
Cells were plated into a 96-well plate at a density of 2.5 x104 cells per ml and left to grow overnight. Cells were treated with increasing concentrations SB590885 (GlaxoSmithKline, Collegeville, PA), or imatinib mesylate (Hospital of the University of Pennsylvania) in triplicate, after 72 hrs, the levels of growth inhibition were examined using the MTT assay (6). Data show the mean of at least three independent experiments ± the S.E. mean.
Western blot analysis
Proteins were extracted and blotted for as described in (14). After analysis, Western blots were stripped once and re-probed for β-actin to demonstrate even protein loading. Antibodies to phospho-ERK, total-ERK, phospho-KIT and total KIT were from Cell Signaling Technology (Beverly, MA), the antibody to CDK4 was from Fisher Scientific.
3D spheroid growth
Melanoma spheroids were prepared using the liquid overlay method. Briefly, 200μl of melanoma cells (25,000 cells per ml) were added to a 96-well plate coated with 1.5% agar (Difco, Sparks MD). Plates were left to incubate for 72 hours, by which time cells had organized into 3D spheroids. Spheroids were then harvested using a P1000 pipette. The media was removed and the spheroids were implanted into a gel of bovine collagen I containing EMEM, L-glutamine and 2% FBS. Normal 2% melanoma media was overlayed on top of the solidified collagen. Spheroids were treated with either 3 or 10μM of imatinib before being left to grow for 72 hours. Spheroids were then washed twice in PBS before being treated with calcein-AM, ethidium bromide (Molecular Probes, Eugene, OR) for 1 hour at 37°C, according to the manufacturer’s instructions. After this time, pictures of the invading spheroids were taken using a Nikon-300 inverted fluorescence microscope.
In vivo melanoma xenograft studies
The study protocol was approved by the Wistar Institute Animal Care and Use Committee (IACUC). Each group consisted of 5 severe combined immunodeficient (SCID) CI-1M mice (Charles River Laboratories, Wilmington, MA). Ten mice were inpected subcutaneously with WM1382 cells (2 × 106) in Matrigel ® into the lower back. When animals had developed melanoma nodules of about 5 mm in diameter the study drug administration was initiated (day 1): The SCID mice were randomly assigned to the two experimental groups of 5 animals each: 1) 200 μl vehicle (distilled water), (2) 100 mg/kg imatinib mesylate (in 200 μl distilled water) twice daily by oral gavage over a period of 14 days. Tumors were measured twice a week using digital calipers. Tumor volume was calculated as a product of the three dimensions. Tumor shrinkage was calculated as a fold-change relative to the starting volume. At treatment day 14, one hour after the final drug application, all animals were euthanized.
Cell cycle analysis
Cells were plated into 10-cm dishes at 60% confluency and left to grow overnight before being treated with either SB590885 (1 μM), or U0126 (10 μM, Sigma) for 24 hours, or with imatinib (3-10 μM), for 24-48 hours. Cells were analyzed as previously described (6).
Melanoma tumor samples
Melanoma tissue samples were collected according to institutional review board-approved protocols in compliance with HIPPA guidelines at the Memorial Sloan-Kettering Cancer Center (New York), Dana-Farber Cancer Institute (Boston, MA), and University of Vermont (Burlington, vT). For the specimens analyzed in figure 3C and supplemental table 1, seventeen melanoma tumor samples were isolated from fifteen patients as described in (10). The sample set was derived following the previous identification of high CDK4 overexpression in 3 of the samples (10). One of the samples represented subungual primary melanoma (MMF). Specimens MMH and MMI were synchronous anticubital and axillary metastases from the patient with primary melanoma MMF. All the remaining samples were derived from clinically apparent metastases from either superficial spreading or nodular primary melanomas. No mucosal melanomas were analyzed, and no acral melanomas aside from the subungual cases listed above were analyzed. No lentigo maligna melanomas or desmoplastic/neurotropic melanomas were analyzed. All tumor specimens were collected immediately after surgical excision and were rapidly frozen in OCT and were subsequently microdissected to greater than 90% purity following frozen-section verification as shown previously (15).
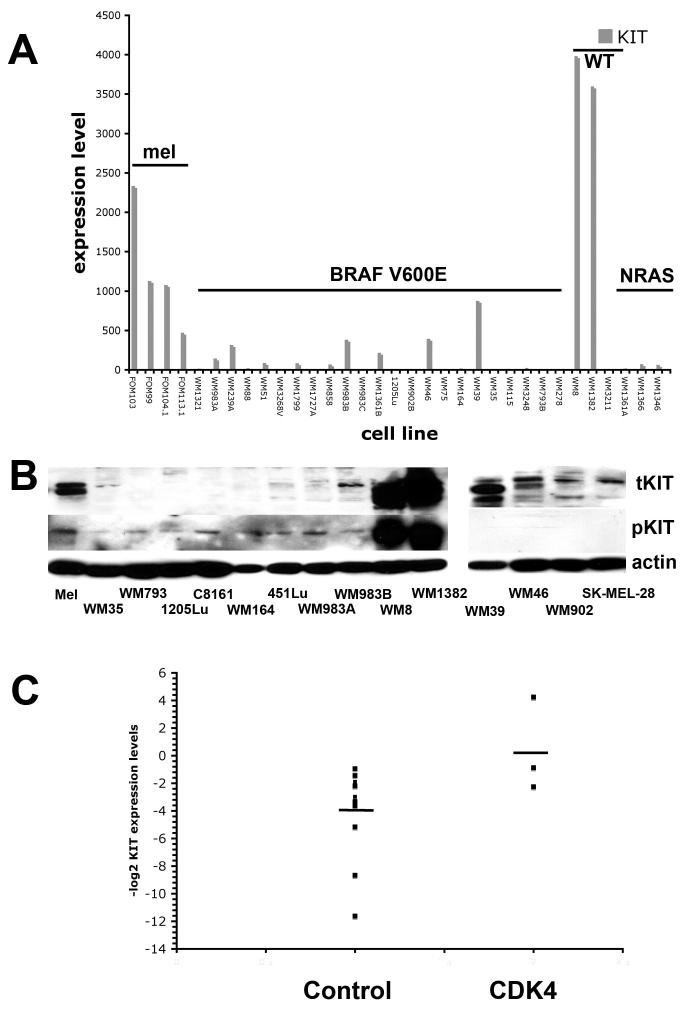
Figure 3. CDK4 overexpressing melanomas also show high KIT expression.
A) CDK4 overexpressing melanoma lines have higher KIT mRNA expression. Microarray analysis, showing KIT mRNA expression in a panel of primary human melanocytes (mel), V600E-mutated melanoma cell lines (BRAF V600E), NRAS/BRAF wild-type melanomas and NRAS mutated melanomas (NRAS). B) CDK4 overexpressing melanoma cell lines have high KIT expression and phospho-KIT activity (Left panel): Protein expression of total KIT (tKIT) and phospho-KIT (pKIT) receptor across a panel of human melanocytes (mel), and human melanoma cell lines (WM35, WM793, 1205Lu, C8161, WM164, 451Lu, WM983A, WM983B, WM8 and WM1382) (Right panel): Expression of KIT in melanoma lines harboring the BRAF V600E mutation. Western blots showing the expression of total KIT and phospho-c-KIT across a panel of melanoma cell lines (WM39, WM46, WM902B, SK-MEL-28) harboring the BRAF mutation. (C) Human melanoma samples with increased CDK4 expression also have higher KIT expression. Data show quantitative RT-PCR results for KIT expression in 14 melanoma samples without CDK4 amplification (control) or 3 with CDK4 amplification confirmed by array CGH (CDK4). Data shown are -log2 values normalized to KIT expression in normal human melanocytes (defined as zero). Bar shows mean value KIT expression for the Control (non-CDK amplified) and CDK4 amplified groups.
Quantitative PCR on melanoma lesions
RNA was isolated from microdissected tumor samples using the RNeasy Plus kit (Qiagen) according to the manufacturer’s recommendations. One microgram of total RNA was used in a reverse transcription reaction using the RETROscript kit (Ambion) according to the manufacturer’s recommendations. Real time quantitative PCR was performed by using 2 1l of 100 fold diluted cDNA template and 0.2 μM gene specific primers against KIT (forward - 5′TCA TTG AGA GTT TTG TCT TGG A 3′ and reverse - 5′ ACT TAC GCC GCT TAT GTA TTT A 3′) in a 25 μl PCR reaction using JumpStart SYBR green kit (Sigma) according to manufacturer’s instructions in an ABI 7700. The reactions were performed in duplicate, dissociation curve analysis was performed to ensure a single product, CT values obtained were normalized to GAPDH levels and quantification was performed using the comparative CT method.
SCF ELISA
Cells were cultured for 24 hours in serum-free tumor media. Supernatants were harvested and measured using a commercially available SCF ELISA kit (R&D Systems). As a positive control we used conditioned media from human skin fibroblast line transduced with a lentiviral vector SCF construct.
Expression profiling data
Melanoma samples were prepared for analysis on the Affymetrix U1BBA array platform (16). Cell lines represented in Figure 1A and 3A are (in orderd) FOM103, FOM99, FOM104.1, FOM113.1, WM1321, WM983A, WM239A, WM88, WM51, WM3268A, WM1799, WM1727A, WM858, WM983B, WM983C, WM1361B, 1205Lu, WM902B, WM46, WM75, WM164, WM164, WM39, WM35, WM115, WM3248, WM793, WM278, WM8, WM1382, WM3211, WM1361A, WM1366, WM1346. The data generated from these arrays has been published previously (16, 17) and have been deposited in the NCBIs Gene Expression Omnibus (GEO,http://www.ncbi.nlm.nih.gov/geo/). Data are accessible using GEO Series accession GSE4845. Analysis was performed using GeneSpring® software, where the data from three V600E mutated melanoma cell lines (WM35, WM793 and 1205Lu) was compared to that from the WM8 and WM1382 melanoma cell lines.
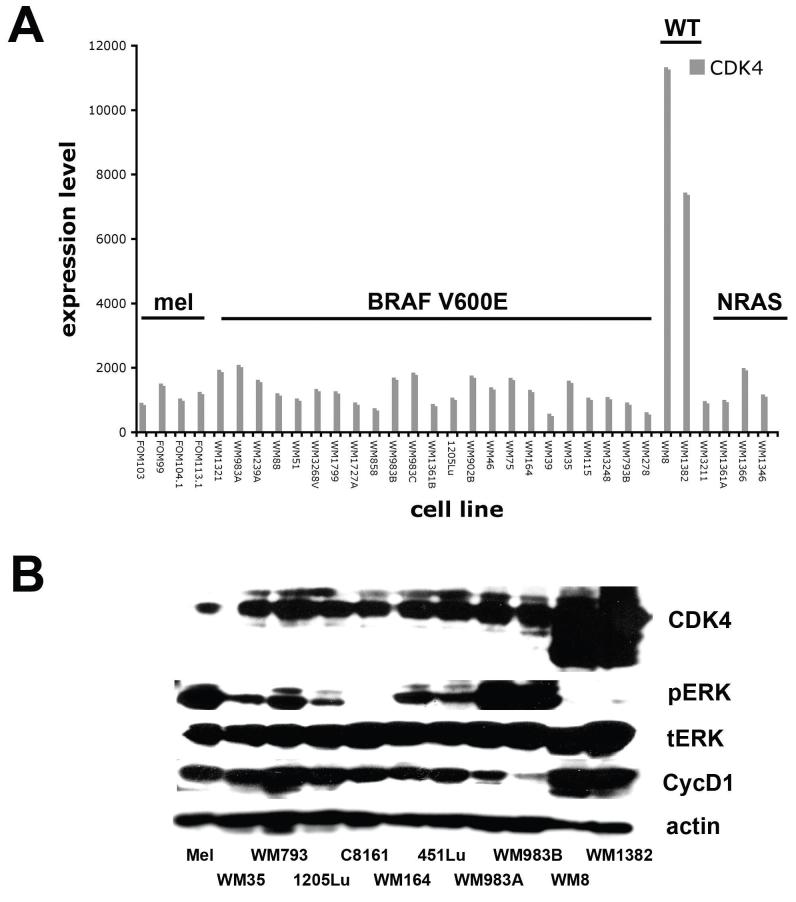
Figure 1. Identification of a sub-group of melanoma lines that express high CDK4 levels.
A) Microarray analysis, showing increased CDK4 mRNA in a panel of BRAF/NRAS wt melanomas. Expression profile shows a panel of primary human melanocytes (mel), V600E-mutated melanoma cell lines (BRAF V600E), NRAS/BRAF wild-type melanomas and NRAS mutated melanomas (NRAS). B) Increased CDK4 expression in the BRAF/NRAS wt cell lines identified from A). Protein expression of CDK4, phospho-ERK (pERK), total ERK (tERK), cyclin D1 (CycD1) across a panel of human melanocytes (mel), and human melanoma cell lines. Note, the WM8 and WM1382 cell lines exhibit high CDK4/cyclin D1 expression and low levels of phospho-ERK.
Mutational testing
Exons 11 and 15 of BRAF and exon 3 of NRAS were screened for mutations by PCR-based sequencing as previously described (18, 19). Mutation screening of KIT for exons 11 and 13, 17 was performed as described in (13). Mutation screening of exon 9 was performed with primers and conditions described in (20). Primers were designed using the Primer3 software (www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi) for exons 14 - F 5′tgaccacccttgggtattt and R 5′gccttgattgcaaaccctta and 18 F 5′catttcagcaacagcagcat and R 5′caaggaagcaggagaccaat. PCR was carried out for 35 cycles (30 sec at 95 °C, 30 sec at 56 °C and 60 sec at 72 °C) on a PCRexpress Thermo Hybaid thermal cycler.
Statistical analysis.
Unless otherwise stated, all experiments show the mean ± S.E. mean of at least three independent experiments. Statistical significance was measured using the Student’s T-Test, where P<0.05 was pudged to be significant.
Results
Identification of melanoma cell lines with high CDK4 expression.
Initial screening of our microrray data identified a panel of melanoma cell lines that were wild-type for BRAF/NRAS with increased expression of CDK4 (Figure 1A). From these data, we identified two cell lines, WM8 and WM1382, with increased levels of CDK4. These cell lines harbored neither a BRAF nor an NRAS mutation (table 1).
Table 1. BRAF, NRAS and KIT mutational status of the melanoma cell line panel.
| Cell line | BRAF | NRAS | KIT Ex 11 | KIT Ex 13 | KIT Ex 14 | KIT Ex 17 | KIT Ex 18 |
|---|---|---|---|---|---|---|---|
| WM35 | V600E | WT | WT | WT | WT | WT | WT |
| WM793 | V600E | WT | WT | WT | WT | WT | WT |
| 1205Lu | V600E | WT | WT | WT | WT | WT | WT |
| WM164 | V600E | WT | WT | WT | WT | WT | WT |
| 451Lu | V600E | WT | WT | WT | WT | WT | WT |
| WM983A | V600E | WT | WT | WT | WT | WT | WT |
| WM983B | V600E | WT | WT | WT | WT | WT | WT |
| WM8 | WT | WT | WT | WT | WT | WT | WT |
| WM1382 | WT | WT | WT | WT | WT | WT | WT |
| WM39 | V600E | WT | WT | WT | WT | WT | WT |
| WM46 | V600E | WT | WT | WT | WT | WT | WT |
| WM902B | V600E | WT | WT | WT | WT | WT | WT |
| SK-MEL-28 | V600E | WT | WT | WT | WT | WT | WT |
WT = wild-type.
Consistent with the lack of BRAF/NRAS mutations, both the WM8 and WM1382 cell lines expressed only low levels of phospho-ERK in Western blotting experiments (Figure 1B). However, further exposure of the blots to film did reveal the presence of some basal phospho-ERK in these lines (not shown). Increased expression of CDK4 was confirmed in the WM8 and WM1382 cell lines at the protein level (Figure 1B), and also was correlated with an increase in cyclin D1 expression.
CDK4 overexpressing melanoma lines are resistant to the BRAF inhibitor SB590885
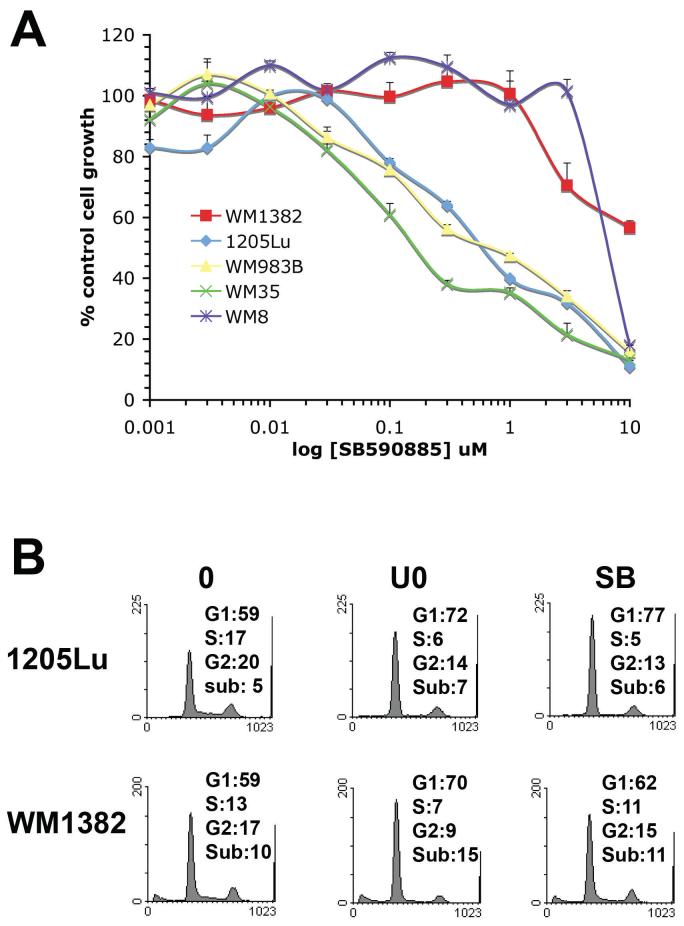
To determine whether CDKE overexpression led to BRAF inhibitor resistance, a panel of melanoma cell lines were treated with increasing concentrations of the BRAF inhibitor SB590885 (21) (Figure 2A). Compared to three melanoma cell lines with BRAF V600E mutations (1205Lu, WM35, WM983B), the two CDK4 overexpressing lines were highly resistant to the BRAF inhibitor SB590885 (21) (Figure 2A). Similarly, SB590885 had little effect upon the cell cycle profile of the WM1382 cells but induced a profound G1-arrest in the BRAF mutated 1205Lu cells (Figure 2B). Interestingly, the U0126 did induce some cell cycle arrest in the WM1382 cells, suggesting that these cells may have low basal phospho-ERK activity that could be responsible for cell cycle entry (Figure 2B).
Figure 2. The CDK4 overexpressing melanoma cell lines are resistant to the anti-proliferative effects of the BRAF inhibitor SB590885.
A) The CDK4 overexpressing melanoma lines are resistant to SB590885 in an MTT assay. Cells were treated with increasing concentrations of SB590885 (1nM - 10 μM) for 72 hrs before being treated with MTT. B) SB590885 preferentially reduces S-phase entry in BRAF V600E mutated melanoma cell lines. The BRAF V600E mutated cell line (1205Lu) and the CDK4 overexpressing cell line (WM1382) were treated with U0126 (10 μM) or SB590885 (1 μM) for 24 hrs, before being fixed, stained with propidium iodide and analyjed using flow cytometry.
CDK4 overexpressing melanomas retain constitutive KIT expression
As melanoma lines that overexpress CDK4 are resistant to BRAF inhibitors we next attempted to identify novel therapeutic targets in the BRAF inhibitor resistant sub-group of melanomas. To address this issue we performed a microarray analysis comparison between three BRAF-mutated melanoma cell lines (NM35, WM793 and 1205Lu) and two CDK4 overexpressing lines (WM8 and WM1382). The analysis was designed to identify any genes with a 3-fold higher or greater expression in the CDK4 overexpressing population. The gene with the highest fold up-regulation in the CDK4 population was the receptor tyrosine kinase KIT (859-fold higher in the CDK4 overexpressing cell linesd.
Stratification of KIT expression according to mutational status (melanocyte, BRAF V600E mutated melanoma, NRAS/BRAF wild-type melanoma or NRAS mutated melanoma) revealed the upregulation of KIT mRNA expression in the BRAF/NRAS wild-type melanoma panel (Figure 3A). The cell lines that we identified (the WM8 and WM1382) were the same two lines with high CDK4 expression. It was also noted that there were several BRAF V600E mutated melanoma lines that also maintained some KIT expression (Figure 3A and not shown).
Western blotting revealed that KIT was highly expressed in the CDK4 overexpressing melanoma cell lines at the protein level and the high levels of phospho-KIT seen indicated that the receptor was constitutively active (Figure 3B). Expression of KIT also was maintained in a sub-group of melanoma cell lines (WM39, WM46, WM 902B and SK-MEL-28) that harbored the BRAF V600E mutation (Figure 3B). However in this instance the receptor was not constitutively active, as shown by the lack of phospho-KIT expression. To investigate the potential mechanism of the KIT receptor activation we performed an ELISA experiment looking for secretion of stem cell factor (SCF) (supplemental figure 1) and noted a lack of SCF secretion from any of the melanoma cell lines tested, including the WM8 and WM1382.
Certain sub-groups of mucosal and acral lentiginous melanomas harbor activating mutations in KIT (13). Mutational analysis testing of KIT at the known mutation hotspots of Exons 11, 13, 14, 17 and 1a revealed that the WM8 and WM1382 cell lines were wild-type (table 1). A recent study from our group identified a high amplification of CDK4 in a series of uncultured melanoma samples (10). Further study of 17 of these samples showed an increase in KIT expression in the 3 samples with CDK4 amplification compared to the non-amplified group (Figure 3C and supplemental table 1). Similar to the CDK4/KIT amplified melanoma cell lines, these 3 samples were also negative for both BRAF and NRAS mutations (supplemental table 1).
Inhibition of KIT following using Imatinib has selective anti-tumor effects upon the CDK4/KIT overexpressing melanoma cell lines
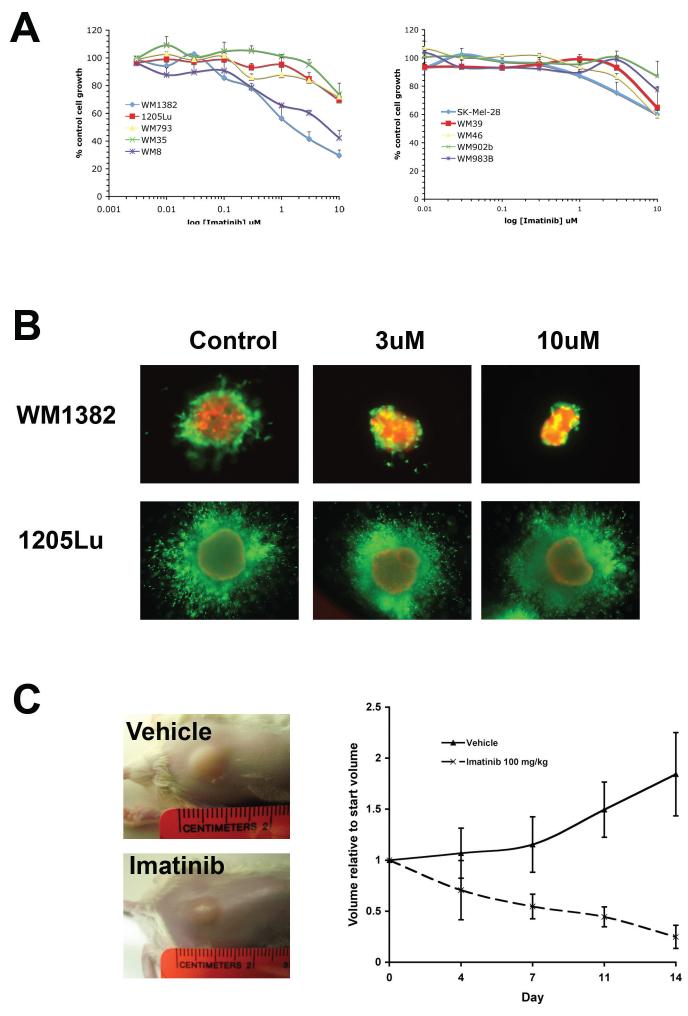
The high expression of phospho-KIT in the CDK4 overexpressing melanoma lines suggests that KIT activity is essential for their proliferation and survival. Imatinib is a receptor tyrosine kinase inhibitor which selectively blocks the activity of Bcr-Abl, the platelet derived growth factor receptor (PDGFR) and KIT (22). In a series of MTT assays it was found that the two CDK4/KIT overexpressing melanoma lines were markedly growth inhibited following 72 hr drug treatment with imatinib (Figure 4A). The panel of melanoma cell lines that harbored the BRAF V600E mutation in the absence of any significant KIT expression (WM793, 1205Lu, WM983B and WM35) were resistant to the effects of imatinib and showed very little growth inhibition (Figure 4A). Likewise, the melanoma cell lines with a BRAF V600E mutation that also maintained KIT expression (SK-Mel-28, WM39, WM46, WM902B) were also resistant to imatinib (Figure 4A).
Figure 4. CDK4/KIT overexpressing melanomas are sensitive to imatinib treatment.
(A) CDK4/KIT overexpressing cells were sensitive to the growth inhibitory effects of imatinib in an MTT assay. Cells were treated with increasing concentrations of imatinib (10 nM - 10 μM) for 72 hrs before being treated with MTT. Absorbances were read at 570 nm and expressed as a percentage of control absorbance. Data show the mean of three independent experiments +/- S.E. mean. Only the WM8 and WM1382 were sensitive to imatinib treatment (B) Imatinib reduces the viability and survival of melanoma cells grown as 3D collagen-implanted spheroids. Pre-formed spheroids that either overexpressed CDK4 (WM1382) or harbored the BRAF V600E mutation (1205Lu) were embedded into collagen and overlayed with medium. Cells were then treated with Imatininb (3 and 10 μM) for 72 hrs, before being treated with calcein-AM and propidium iodide. Green, viable cells; red, dead cells. Lack of green staining also indicates loss of viability. (C) Imatinib treatment induces regression of established CDK4/KIT melanoma xenografts. CDK4/KIT overexpressing (WM1382) cells were grown as tumor xenografts in SCID mice. After tumor establishment, mice were dosed twice daily with either vehicle (distilled water) or imatinib mesylate (100 mg/kg in distilled water) by oral gavage for 14 days. Tumor volumes were measured twice per week. Left panel: Photographs of representative vehicle and imatinib treated tumors taken after day 14 of treatment. Right panel: Growth curves were normalized to the start volumes. Imatinib treatment led to significant regression of the established xenografts.
Our previous studies have indicated that melanoma cell lines that are sensitive to targeted therapy agents in 2D monolayer culture often become highly drug resistant when grown as 3D collagen-implanted spheroids (5). Here we show that the CDK4 overexpressing WM1382 melanoma cell line is highly sensitive to imatinib under 3D culture conditions (Figure 4B). Treatment of the WM1382 spheroids with imatinib (3 and 10 μM) for 72 hrs was associated with reduced cell invasion into the surrounding collagen as well as marked decreases in cell viability.
Next, we grew the WM1382 cells as tumor xenografts in SCID mice. After tumor establishment (5 × 5 mm), mice were dosed twice daily with either vehicle (distilled water) or imatinib mesylate (100 mg/kg in distilled water) by oral gavage. After 14 days it was found that imatinib treatment had suppressed tumor growth and led to a significant level of regression (Figure 4C), demonstrating the utility of imatinib treatment in the CDK4/KIT overexpressing melanomas.
Imatinib blocks the proliferation of CDK4 overexpressing melanoma cells through the inhibition of KIT mediated MAP kinase signaling
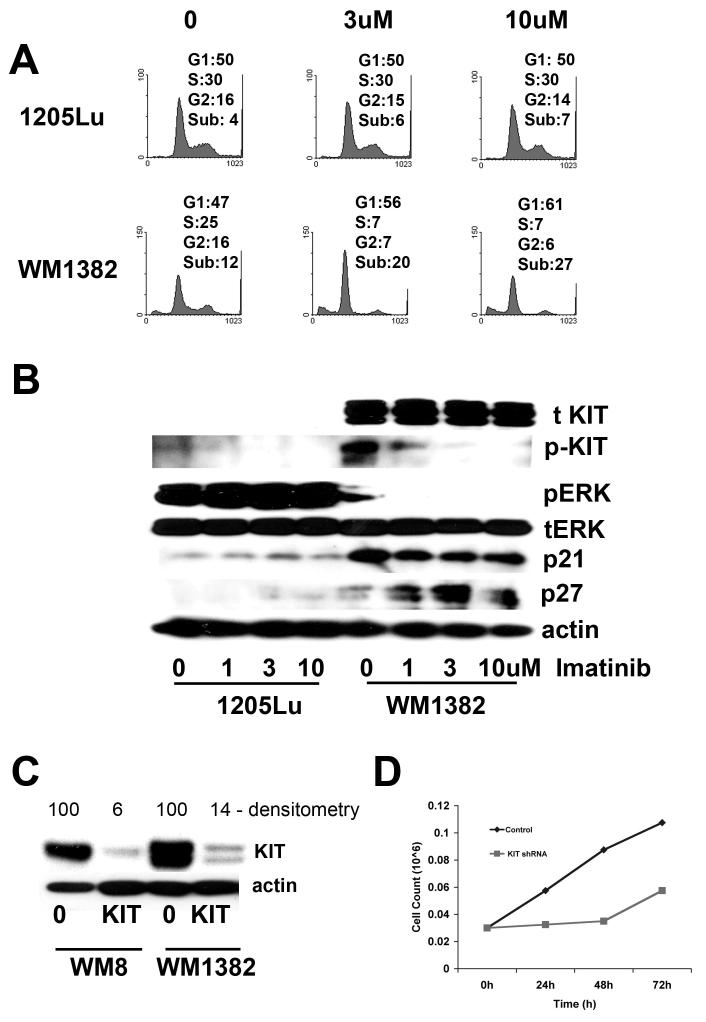
Next, we turned our attention to the mechanism of action of imatinib in our CDK4/KIT overexpressing melanoma population. Treatment of the BRAF V600E mutated 1205Lu cells with imatinib had very little effect upon the cell cycle profile and did not induce any apoptosis (Figure 5A). Increasing concentrations of imatinib (3 and 10 μM) induced a G1-phase cell cycle arrest and some apoptosis in the CDK4 overexpressing WM1382 cells, with 27% apoptosis being induced following 10 μM imatinib treatment (Figure 5A).
Figure 5. KIT inhibition blocks MAP kinase activity and induces cell cycle arrest and apoptosis in CDK4/KIT overexpressing melanomas.
(A) Imatinib preferentially reduces S-phase entry and induces apoptosis in a KIT/CDK4 overexpressing melanoma cell line but not BRAF V600E mutated melanoma cell line. The BRAF V600E mutated cell line (1205Lu) and the CDK4 overexpressing cell line (WM1382) were treated with Imatinib (3 and 10 μM) for 24 hrs, before being fixed, stained with propidium iodide and analyzed using flow cytometry. (B) Imatininb treatment preferentially reduces phospho-KIT, and phospho-ERK expression in CDK4 overexpressing cells (WM1382) but not in BRAF V600E mutated cells (1205Lu). Cells were treated with imatinib (1-10 μM) for 24 hrs, protein was then harvested and resolved by Western blotting. Figure shows expression of total KIT (tKIT), phospho-KIT (pKIT), phospho-ERK (pERK), total ERK (tERK), p21 and p27. Equal protein loading was confirmed by stripping the blot once and probing for actin expression. (C) Knockdown of KIT levels using a lentiviral shRNA construct. CDK4 overexpressing cells were infected with a lentivirus encoding for shRNA against KIT and were selected using puromycin. Blot shows knockdown of KIT protein and was scored using a densitometer. (D) Knockdown of KIT reduces the growth of WM1382 cells. CDK4 overexpressing cells were infected with lentiviral shRNA against KIT. A scrambled shRNA sequence was used as the control. Following drug selection, 30,000 cells/ml were plated out and were counted at 24, 48 and 72 hrs.
Treatment of the WM1382 cells with imatinib led to progressive reduction in the level of KIT receptor activity, as shown by reduced phospho-KIT expression (Figure 5B). At the same time, imatinib also inhibited the low basal level of phospho-ERK activity in the WM1382 cells, indicating that imatinib blocked the MAP kinase pathway the CDK4/KIT expressing cells. Consistent with the ability of imatinib to block the MAP kinase signaling, it was found that drug treatment also increased the expression of the CDK inhibitor p27KIP1 (Figure 5B). To demonstrate the ability of the KIT receptor to stimulate phospho-ERK signaling, we treated primary human melanocytes with the KIT ligand for increasing periods of time (0 - 30 minutes) and showed a robust increase in phospho-ERK signaling (supplemental figure 2).
Imatinib inhibits the activity of multiple receptor tyrosine kinases. To determine whether the specific effects of imatinib were mediated through KIT inhibition, we generated lentiviral shRNAs against KIT that produced effective knockdown (>85%) of protein levels (Figure 5C). Infection of the WM8 cell line with the KIT shRNA led to very high levels of apoptosis (data not shown), and significantly reduced the growth of the WM1382 cell lines (Figure 5D), confirming the role of KIT in the survival and growth of the CDK4 overexpressing melanoma lines.
Discussion
The past two years have seen progress in defining new sub-categories of melanoma based upon patterns of gene amplification and mutation (3, 12, 13). How these different mutational profiles and molecular sub-groupings can be translated into novel strategies for treatment has been little explored. Our initial screen of a panel of early passage melanomas identified two cell lines with very high expression of CDK4 that lacked BRAF and NRAS mutations. Pharmacological studies showed that the CDK4 overexpressing melanoma lines were resistant to BRAF inhibition. Typically, BRAF inhibitors block cell cycle entry through the increase of cyclin D1 expression and through the suppression of the CDK inhibitor p27KIP1 (5, 8). As the CDK4 overexpressing melanomas have much lower phospho-ERK levels and higher baseline expression of both CDK4 and cyclin D1, high activity in the MAP kinase pathway appeared to be less critical for growth in these lines.
We next used existing microarray data to identify KIT expression as a possible pharmacological target within the CDK4 overexpressing sub-group. The receptor tyrosine kinase KIT is a critical regulator of growth, differentiation, migration and proliferation in the hematopoeitic, germ cell and melanocytic systems (23, 24). Functional KIT/SCF signaling is essential for melanocyte development and plays an important role in pigmentation. Thus, dysfunctional KIT signaling is associated with pigmentary defects, resulting from impaired melanocyte survival and migration (25).
Activating mutations in KIT are well described in cancer and are implicated in the development of gastrointestinal stromal tumors (GIST), some forms of leukemia (AML) and testicular seminomas (26). The role of KIT in melanoma is more complex, with a number of studies demonstrating the loss of receptor expression during tumor progression (27). In some melanoma cell lines, the forced overexpression of KIT leads to induction of apoptosis (28). The recent years have seen renewed interest in the role of KIT in melanoma following the work by Bastian and colleagues identifying activating KIT mutations in defined histological sub-sets of melanomas (13).
A number of small molecule receptor tyrosine kinase (RTK) inhibitors have been developed that target KIT activity, the best studied of which being imatinib mesylate (Gleevec ®), an RTK inhibitor with activity against Bcr-Abl, PDGFR and KIT (22, 29). Although imatinib is now routinely used in the treatment of patients with chronic myeloid leukemia and GIST, its activity in melanoma has been very disappointing (30). Recent phase II clinical trials of imatinib in patients with metastatic melanoma, unselected in regard to KIT mutation or amplification, revealed no obpective responses, poor survival rates and significant toxicity (30). Expression of KIT alone across our cell line panel was not indicative of an imatinib response. Of the cell lines tested, only the BRAF/NRAS wild-type cell lines with CDK4/KIT overexpression showed any anti-proliferative response following imatinib treatment. Responsive cell lines were found to have high phospho-KIT expression, whereas the non-responding lines lacked any phospho-KIT. We therefore suggest that presence of KIT alone is not necessarily predictive of response to imatinib therapy.
It is unclear at this juncture how the high phospho-KIT activity is maintained in the CDK4 overexpressing melanoma cell lines. Neither of the CDK4 overexpressing cell lines were found to harbor activating KIT mutations at known mutational hotspots, nor did any of these lines secrete autocrine SCF. One possible explanation for the constitutive activity of KIT in this system comes from studies on epidermal growth factor receptor (EGF) signaling, which have shown that increased receptor expression through gene amplification/overexpression also can lead to increased signaling activity (31). Indeed, there is evidence that overexpression of EGFR is predictive of response to EGF inhibitors in non-small cell lung carcinoma (32, 33).
Imatinib is known to inhibit the activity of at least three receptor tyrosine kinases, all of which are expressed in melanoma (34). Functional studies revealed that imatinib treatment led to reduced S-phase entry and apoptosis in the CDK4/KIT overexpressing melanoma line WM1382. To determine whether these effects were through the inhibition of KIT rather than Bcr-Abl and/or PDGFR we stably knocked down KIT expression in the WM8 and WM1382 cell lines using a lentiviral shRNA construct. We found that KIT knockdown markedly reduced the growth of the WM1382 and led to a total loss of cell viability in the WM8 line.
Having demonstrated that the CDK4/KIT overexpressing melanoma lines possess a limited amount of phospho-ERK activity and could be partly growth arrested, we next investigated whether KIT signaling regulated MAP kinase signaling in these cell lines. Studies of melanocytes showed that SCF treatment led to a robust increase in phospho-ERK expression, demonstrating that the receptor can activate this pathway. In CDK4 overexpressing melanoma cells, treatment with increasing concentrations of imatinib led to a progressive decrease in phospho-KIT and phospho-ERK. Treatment of the CDK4 overexpressing cells with imatinib was also found to increase p27KIP1 expression, suggesting that imatinib was working primarily in these cells through inhibition of the MAP kinase pathway (3, 8). This finding raises the intriguing possibility that all melanomas rely upon MAP kinase signaling activity, even in the absence of BRAF and NRAS mutations, suggesting that MAP kinase inhibition needs to be part of any optimized future melanoma treatment strategy.
These findings lead us to propose an alternative model for melanoma progression on a BRAF/NRAS wild-type background whereby a limited MAP kinase signaling via KIT can co-operate with increased CDK4/cyclin D1 to drive cell cycle entry. We hypothesize that the combination of overexpression of CDK4 and KIT may play a similar role to the high MAP kinase activity driven through either an activating BRAF V600E or NRAS mutation. Further studies are required to determine whether the combination of KIT/CDK4 is sufficient to fully transform human melanocytes. Although there is evidence of increased melanoma formation in CDK4 R24C knock-in mice, there also is a requirement for other factors (35). Most published studies suggest that inactivation of the p53 pathway and the INK4A/retinoblastoma protein axis also are required to achieve full oncogenic transformation (36, 37). Indeed, previous work from our own group suggests that CDK4 and MDM2 amplifications occur in parallel (10).
The current study has identified a new panel of melanomas that lack BRAF/NRAS mutations and instead have co-amplification of CDK4 and KIT. Unlike previously identified sub-groups of acral and mucosal melanomas, this novel group was discovered in metastases from superficial spreading and nodular melanomas, not in mucosal or acral lentiginous melanomas that have higher rates of KIT amplification. As this is not an exhaustive pathological study, it is currently difficult to pudge the prevalence of this genetic profile across the whole melanoma population. Based on our data, we suggest that melanomas with KIT/CDK4 overexpression may also be suitable for imatinib treatment. It is hoped that the continued molecular sub-classification of melanoma will lead to the identification of focused patient groups with the best likelihood of clinical response to defined agents.
Acknowledgements
This work was supported by grants from the National Institutes of Health (CA 76674, CA 25874, CA 10815, CA 93372, CA 47159, CA 80999, CA 098101, CA117881, GM071695) and by funds from the Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health. KSMS is the recipient of a Career Development Award from the NCI SPORE-CA93372.
References
- 1.Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007;445:851–7. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- 2.Smalley KSM. A pivotal role for ERK in the oncogenic behaviour of malignant melanoma? International Journal of Cancer. 2003;104:527–32. doi: 10.1002/ijc.10978. [DOI] [PubMed] [Google Scholar]
- 3.Solit DB, Garraway LA, Pratilas CA, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358–62. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verhaegen M, Bauer JA, Martin de la Vega C, et al. A novel BH3 mimetic reveals a mitogen-activated protein kinase-dependent mechanism of melanoma cell death controlled by p53 and reactive oxygen species. Cancer Res. 2006;66:11348–59. doi: 10.1158/0008-5472.CAN-06-1748. [DOI] [PubMed] [Google Scholar]
- 5.Smalley KS, Haass NK, Brafford PA, Lioni M, Flaherty KT, Herlyn M. Multiple signaling pathways must be targeted to overcome drug resistance in cell lines derived from melanoma metastases. Mol Cancer Ther. 2006;5:1136–44. doi: 10.1158/1535-7163.MCT-06-0084. [DOI] [PubMed] [Google Scholar]
- 6.Smalley KS, Contractor R, Haass NK, et al. Ki67 expression levels are a better marker of reduced melanoma growth following MEK inhibitor treatment than phospho-ERK levels. Br J Cancer. 2007;96:445–9. doi: 10.1038/sj.bjc.6603596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sherr CJ. G1 phase progression: cycling on cue. Cell. 1994;79:551–5. doi: 10.1016/0092-8674(94)90540-1. [DOI] [PubMed] [Google Scholar]
- 8.Bhatt KV, Spofford LS, Aram G, McMullen M, Pumiglia K, Aplin AE. Adhesion control of cyclin D1 and p27Kip1 levels is deregulated in melanoma cells through BRAF-MEK-ERK signaling. Oncogene. 2005;24:3459–71. doi: 10.1038/sj.onc.1208544. [DOI] [PubMed] [Google Scholar]
- 9.Sauter ER, Yeo UC, von Stemm A, et al. Cyclin D1 is a candidate oncogene in cutaneous melanoma. Cancer Research. 2002;62:3200–6. [PubMed] [Google Scholar]
- 10.Muthusamy V, Hobbs C, Nogueira C, et al. Amplification of CDK4 and MDM2 in malignant melanoma. Genes Chromosomes Cancer. 2006;45:447–54. doi: 10.1002/gcc.20310. [DOI] [PubMed] [Google Scholar]
- 11.Wolfel T, Hauer M, Schneider J, et al. A p161NK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science. 1995;269:1281–4. doi: 10.1126/science.7652577. [DOI] [PubMed] [Google Scholar]
- 12.Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–47. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 13.Curtin JA, Busam K, Pinkel D, Bastian BC. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol. 2006;24:4340–6. doi: 10.1200/JCO.2006.06.2984. [DOI] [PubMed] [Google Scholar]
- 14.Smalley KSM, Brafford P, Haass NK, Brandner JM, Brown E, Herlyn M. Up-regulated expression of zonula occludens protein-1 in human melanoma associates with N-cadherin and contributes to invasion and adhesion. American Journal of Pathology. 2005;166:1541–54. doi: 10.1016/S0002-9440(10)62370-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muthusamy V, Duraisamy S, Bradbury CM, et al. Epigenetic silencing of novel tumor suppressors in malignant melanoma. Cancer Res. 2006;66:11187–93. doi: 10.1158/0008-5472.CAN-06-1274. [DOI] [PubMed] [Google Scholar]
- 16.Hoek KS, Schlegel NC, Brafford P, et al. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res. 2006;19:290–302. doi: 10.1111/j.1600-0749.2006.00322.x. [DOI] [PubMed] [Google Scholar]
- 17.Tsavachidou D, Coleman ML, Athanasiadis G, et al. SPRY2 is an inhibitor of the ras/extracellular signal-regulated kinase pathway in melanocytes and melanoma cells with wild-type BRAF but not with the V599E mutant. Cancer Res. 2004;64:5556–9. doi: 10.1158/0008-5472.CAN-04-1669. [DOI] [PubMed] [Google Scholar]
- 18.Edwards RH, Ward MR, Wu H, et al. Absence of BRAF mutations in UV-protected mucosal melanomas. Journal of Medical Genetics. 2004;41:270–2. doi: 10.1136/jmg.2003.016667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spittle C, Ward MR, Nathanson KL, et al. Application of a BRAF pyrosequencing assay for mutation detection and copy number analysis in malignant melanoma. J Mol Diagn. 2007;9:464–71. doi: 10.2353/jmoldx.2007.060191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sihto H, Sarlomo-Rikala M, Tynninen O, et al. KIT and platelet-derived growth factor receptor alpha tyrosine kinase gene mutations and KIT amplifications in human solid tumors. J Clin Oncol. 2005;23:49–57. doi: 10.1200/JCO.2005.02.093. [DOI] [PubMed] [Google Scholar]
- 21.King AJ, Patrick DR, Batorsky RS, et al. Demonstration of a genetic therapeutic index for tumors expressing oncogenic BRAF by the kinase inhibitor SB-590885. Cancer Res. 2006;66:11100–5. doi: 10.1158/0008-5472.CAN-06-2554. [DOI] [PubMed] [Google Scholar]
- 22.Druker BJ, Talpaz M, Resta DJ, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–7. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 23.Lennartsson J, Jelacic T, Linnekin D, Shivakrupa R. Normal and oncogenic forms of the receptor tyrosine kinase kit. Stem Cells. 2005;23:16–43. doi: 10.1634/stemcells.2004-0117. [DOI] [PubMed] [Google Scholar]
- 24.Grichnik JM. Kit and melanocyte migration. J Invest Dermatol. 2006;126:945–7. doi: 10.1038/sj.jid.5700164. [DOI] [PubMed] [Google Scholar]
- 25.Wehrle-Haller B, Weston JA. Soluble and cell-bound forms of steel factor activity play distinct roles in melanocyte precursor dispersal and survival on the lateral neural crest migration pathway. Development. 1995;121:731–42. doi: 10.1242/dev.121.3.731. [DOI] [PubMed] [Google Scholar]
- 26.Kitamura Y, Hirotab S. Kit as a human oncogenic tyrosine kinase. Cell Mol Life Sci. 2004;61:2924–31. doi: 10.1007/s00018-004-4273-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang S, Jean D, Luca M, Tainsky MA, Bar-Eli M. Loss of AP-2 results in downregulation of c-KIT and enhancement of melanoma tumorigenicity and metastasis. Embo J. 1998;17:4358–69. doi: 10.1093/emboj/17.15.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang S, Luca M, Gutman M, et al. Enforced c-KIT expression renders highly metastatic human melanoma cells susceptible to stem cell factor-induced apoptosis and inhibits their tumorigenic and metastatic potential. Oncogene. 1996;13:2339–47. [PubMed] [Google Scholar]
- 29.Heinrich MC, Griffith DJ, Druker BJ, Wait CL, Ott KA, zigler AJ. Inhibition of c-kit receptor tyrosine kinase activity by STI 571, a selective tyrosine kinase inhibitor. Blood. 2000;96:925–32. [PubMed] [Google Scholar]
- 30.Ugurel S, Hildenbrand R, Zimpfer A, et al. Lack of clinical efficacy of imatinib in metastatic melanoma. Br J Cancer. 2005;92:1398–405. doi: 10.1038/sj.bjc.6602529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.zandi R, Larsen AB, Andersen P, Stockhausen MT, Poulsen HS. Mechanisms for oncogenic activation of the epidermal growth factor receptor. Cell Signal. 2007;19:2013–23. doi: 10.1016/j.cellsig.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 32.Liu W, Wu X, Zhang W, et al. Relationship of EGFR Mutations, Expression, Amplification, and Polymorphisms to Epidermal Growth Factor Receptor Inhibitors in the NCI60 Cell Lines. Clin Cancer Res. 2007;13:6788–95. doi: 10.1158/1078-0432.CCR-07-0547. [DOI] [PubMed] [Google Scholar]
- 33.Cappuzzo F, Varella-Garcia M, Shigematsu H, et al. Increased HER2 gene copy number is associated with response to gefitinib therapy in epidermal growth factor receptor-positive non-small-cell lung cancer patients. J Clin Oncol. 2005;23:5007–18. doi: 10.1200/JCO.2005.09.111. [DOI] [PubMed] [Google Scholar]
- 34.Ivan D, Niveiro M, Diwan AH, et al. Analysis of protein tyrosine kinases expression in the melanoma metastases of patients treated with Imatinib Mesylate (STI5M1, Gleevec) J Cutan Pathol. 2006;33:280–5. doi: 10.1111/j.0303-6987.2006.00432.x. [DOI] [PubMed] [Google Scholar]
- 35.Sotillo R, Garcia JF, Ortega S, et al. Invasive melanoma in CDK4-targeted mice. Proc Natl Acad Sci U S A. 2001;98:13312–7. doi: 10.1073/pnas.241338598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chudnovsky Y, Adams AE, Robbins PB, Lin Q, Khavari PA. Use of human tissue to assess the oncogenic activity of melanoma-associated mutations. Nat Genet. 2005;37:745–9. doi: 10.1038/ng1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gupta PB, Kuperwasser C, Brunet JP, et al. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat Genet. 2005;37:1047–54. doi: 10.1038/ng1634. [DOI] [PMC free article] [PubMed] [Google Scholar]