Abstract
The role of mitochondrial injury in the pathogenesis of complications of uremia is incompletely defined, although diminished bioenergetic capacity and the accumulation of mitochondrial DNA (mtDNA) mutations have been reported. This study was undertaken to evaluate the prevalence of mtDNA injury in 180 patients who had ESRD and were enrolled into the baseline phase of the HEMO study and to relate these markers to all-cause mortality. The mitochondrial injury markers studied in peripheral blood mononuclear cells were the mtDNA copy number per cell, measured by quantitative PCR, and the presence of the mtDNA4977 mutation. After frequency-matching healthy control subjects for age, mtDNA copy number was lower among older dialysis patients compared with older healthy subjects (P = 0.01). A one-log increase in mtDNA copy number was independently associated with a decreased hazard for mortality (adjusted hazard ratio 0.64; 95% confidence interval 0.46 to 0.89). The mtDNA4977 deletion was present in 48 (31%) patients and was independently associated with a decreased hazard for mortality (adjusted hazard ratio 0.33; 95% confidence interval 0.19 to 0.56). In summary, the mtDNA4977 seems to predict survival in ESRD, but a reduced mitochondrial copy number seems to predict a poor outcome. Although further exploration of these associations is needed, evaluation of mitochondrial DNA copy number and somatic mtDNA mutations may provide simple genomic biomarkers to predict clinical outcomes among patients with ESRD.
The uremic syndrome is characterized by abnormalities in energy metabolism, manifest as changes in basal metabolic rate, relative catabolism, negative nitrogen balance, protein energy malnutrition, insulin resistance, and dyslipidemia.1 Mitochondria are integral to these metabolic processes, which they probably influence through cytokine- and adipokine-mediated mechanisms.2 Early studies with 31-phosphorus nuclear magnetic resonance demonstrated that mitochondrial oxidative capacity was diminished in muscle of patients established on dialysis.3 More recent studies showed a high prevalence of somatic mitochondrial DNA (mtDNA) mutations, specifically the “common deletion” (a 4977-bp deletion between nucleotide positions 8470 and 13,447 [mtDNA4977]) in skeletal muscle of patients with ESRD.4 Human mtDNA is particularly prone to oxidant injury because mitochondria generate reactive oxygen species during ATP production.5 mtDNA injury may be qualitative—deletion or point mutations—or quantitative—abnormalities of mtDNA copy number per cell, mtDNA being polyploid.6 mtDNA mutations are classically studied in skeletal muscle tissue, but recent observations suggested that the use of peripheral blood mononuclear cells (PBMC) may be informative as well.7
This study was a preliminary evaluation of mtDNA injury in PBMC obtained from patients on maintenance hemodialysis (HD) to evaluate the prevalence of the mtDNA4977 mutation and the distribution of mtDNA copy number per cell. We hypothesized that both of these markers were independent predictors of future all-cause mortality (ACM) among HD patients.
RESULTS
Genomic DNA samples were available for 180 patients with ESRD and 36 healthy control subjects. Of the patients with ESRD, 173 were randomly assigned in accordance with the protocol of the parent study, forming four roughly equal groups by the 2 × 2 combinations of dialysis dosage and flux categories. The mean age of the ESRD cohort was 62.2 ± 12.3 yr, and median (interquartile range [IQR]) duration on HD was 2.1 (0.8 to 4.6) years; 46% (83 of 180) were male, 54% (97 of 180) were white, 44% (79 of 180) had diabetes, and 66% (115 of 180) had cardiovascular disease. The mean (±SD) serum albumin was 3.6 ± 0.34 g/dl (36.0 ± 4.0 g/L), and hematocrit was 33.0 ± 4.5%. The mean age of the healthy control subjects was 42.6 ± 16.7 yr, 75% were female, and 36% were older than 50 yr (versus 81% in the ESRD cohort).
mtDNA Copy Number
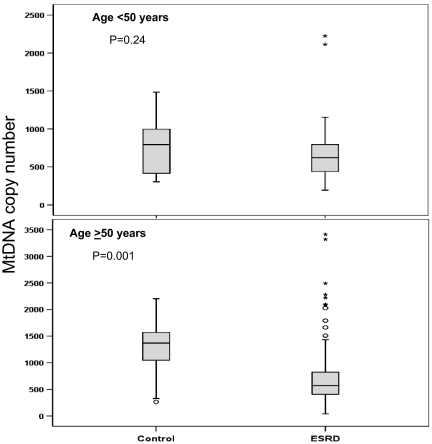
The overall median (IQR) mtDNA copy number per cell was 586 (401 to 817). The corresponding value in healthy control subjects was 914 (445 to 1262), significantly different from patients with ESRD (Mann Whitney test, P = 0.002). Figure 1 shows the distribution of mtDNA copy number per cell by age in the study population versus healthy control subjects. Among older individuals (≥50 yr), the copy number was significantly higher in healthy subjects; no significant difference was seen between younger individuals with and without ESRD. The median copy number increased with age among healthy subjects (by a factor of 1.14 [95% confidence interval (CI) 1.02 to 1.25] for each 10-yr increase in age; P = 0.02). No such increase was seen among patients with ESRD.
Figure 1.
Box plots showing the relative difference in distribution of mtDNA copy number per cell between patients with ESRD and healthy control subjects frequency matched for age. (Top) No significant difference (age <50 yr) between the two groups but in older individuals (age ≥50 yr) (bottom), healthy control subjects showed a higher mtDNA copy number than did patients with ESRD (P = 0.001)
mtDNA copy number did not differ by age, gender, race, number of years spent on HD, Index of Co-Existing Diseases (ICED) score, the presence of diabetes or vascular disease, smoking history, or plasma C-reactive protein (CRP) levels. The copy number was lower among patients who subsequently died during the follow-up period (median [IQR] 524 [352 to 733] versus 642 [486 to 925]; Mann Whitney test, P = 0.003).
mtDNA4977
Genotyping was carried out for 157 patients with satisfactory DNA quality (Figure 2). The mtDNA4977 deletion was present in 48 (31%) patients. Among patients for whom genotyping was carried out, patients with the deletion mutation were older (64.6 ± 10.6 versus 61.2 ± 12.9 yr), but the difference was statistically insignificant. They had started dialysis at an older age (62.1 ± 11.7 versus 57.4 ± 14.5 yr; P = 0.05) and had been on HD for a shorter span of years (median 1.6 versus 2.4 yr; P = 0.01 after log transformation). The prevalence of the mutation was higher among patients with diabetes (37 versus 26%; P = 0.08) and those with preexisting vascular disease (35 versus 22%; P = 0.08), but the differences again were not statistically significant. Patients with the mutation also had lower plasma levels of CRP (median [IQR] 9.9 [2.0 to 17.6] versus 5.8 [4.1 to 22.2] μg/ml; Mann Whitney test, P = 0.10), but the difference did not reach statistical significance. Notably, patients with the mutation had a higher copy number than patients without, but this again did not reach statistical significance (median [IQR] 657 [520 to 814] versus 540 [420 to 825]; Mann-Whitney test, P = 0.08).
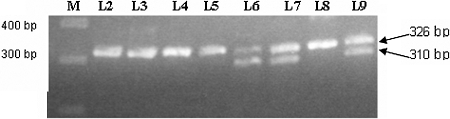
Figure 2.
Agarose gel electrophoresis of the PCR products. Lane 1, a 100-bp molecular weight ladder; lanes 2 through 9, co-amplification products; lanes 6, 7, and 9, two products, the upper band (326 bp) represents wild-type mtDNA and the lower band (301 bp) represents mtDNA4977. In lanes 2 through 5 and 8, the lower band is absent, indicating that there is hardly any mtDNA4977.
Relationship between mtDNA Copy Number per Cell and All-Cause Mortality
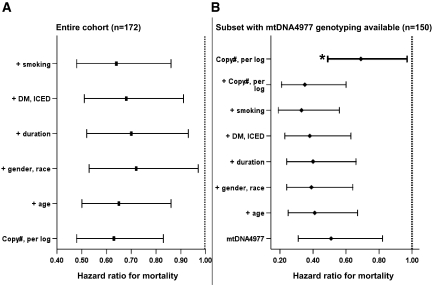
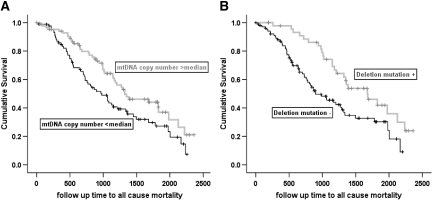
Over the median (IQR) follow up of 980 (510 to 1445) days in 173 patients, there were 102 (59%) deaths. The median (95% CI) survival was 1165 (983 to 1347) days. A one-log increase in mtDNA copy number per cell was associated with a significant decrease in the hazard for mortality (hazard ratio [HR] 0.63; 95% CI 0.48 to 0.83) in univariate analysis. Adjustment for covariates did not affect the relationship (adjusted HR 0.64; 95% CI 0.46 to 0.89; Figure 3A). In a separate model that included (log) CRP and the dosage and flux grouping of the parent study and that was subjected to backward selection, there was a strengthening of the relationship between (log) mtDNA copy number per cell and ACM; variables retained in the model were age, number of years spent on dialysis, smoking, and diabetes (Table 1). Figure 4A shows the Kaplan-Meier survival curves for patients with mtDNA copy number per cell dichotomized about the median.
Figure 3.
Bars in the left panel (A) represent HR for mtDNA copy number (per log), unadjusted and adjusted for all other variables in the model. Bars in the right panel (B) represent HR of mtDNA4977, unadjusted and adjusted for other variables in the model, except for bar marked with *, which represents HR for mtDNA copy number (per log) adjusted for all other variables in the model. The multivariate analysis sequentially adjusted for age, gender, race, duration spent on dialysis, diabetes, comorbidity, and smoking. Comorbidity was expressed as the ICED score.
Table 1.
Cox regression models showing the relationship between mtDNA copy number and mtDNA4977 to ACM
| Parameter | HR | 95% CI | P |
|---|---|---|---|
| Entire cohort (n = 173) | |||
| mtDNA copy number (per log) | 0.60 | 0.44 to 0.82 | 0.001 |
| age (per year) | 1.04 | 1.02 to 1.07 | <0.001 |
| duration on HD (per log-years) | 1.46 | 1.21 to 1.76 | <0.001 |
| smoking | 1.90 | 1.23 to 2.90 | 0.003 |
| diabetes | 1.48 | 0.96 to 2.28 | 0.077 |
| Subset with mtDNA4977 genotyping available (n = 150) | |||
| mtDNA4977 | 0.35 | 0.20 to 0.59 | <0.001 |
| mtDNA copy number (per log) | 0.67 | 0.47 to 0.94 | 0.020 |
| age (per year) | 1.06 | 1.03 to 1.09 | <0.001 |
| duration on HD (per log-years) | 1.62 | 1.27 to 2.08 | <0.001 |
| smoking | 2.23 | 1.41 to 3.54 | <0.001 |
| diabetes | 1.86 | 1.16 to 2.98 | 0.010 |
Figure 4.
Kaplan-Meier survival curves for patients dichotomized as having mtDNA copy number above and below the median (A) and for patients with and without the mtDNA4977 deletion mutation (B).
Relationship between mtDNA4977 Deletion Mutation and All-Cause Mortality
Of 157 patients included in this analysis, 150 underwent randomization in the parent study and had follow-up information. There were a total of 85 (57%) deaths during follow-up. In univariate analysis, the presence of the mutation was associated with a significant decrease in the hazard for mortality (HR 0.51; 95% CI 0.31 to 0.82). Adjustment for covariates (without the inclusion of plasma levels of CRP in the model) strengthened the association considerably (adjusted HR 0.33; 95% CI 0.19 to 0.56). Addition of mtDNA copy number to the model showed that both markers were independent predictors of mortality in this subgroup, without appreciable alteration in their respective estimates (mtDNA4977 HR 0.35 [95% CI 0.21 to 0.60]; mtDNA copy number, per log, HR 0.69 [95% CI 0.49 to 0.97]; Figure 3B). The final model after backward selection of variables including plasma CRP levels and the dialysis and flux grouping of the parent study is shown in Table 1. Figure 4B shows the Kaplan-Meier survival curves for patients with and without the mtDNA4977 deletion mutation.
Relative Predictive Ability of mtDNA Copy Number and mtDNA4977 for Mortality
Table 2 shows the relative predictive ability of mtDNA copy number per cell and the presence of the mtDNA4977 deletion mutation for ACM compared with the other variables in the respective models. The full model provided a c value ≥0.7. When examined alone, the contribution of mtDNA copy number per cell was as strong as smoking history, although less strong than age or duration spent on HD. When both variables were examined together in the second model, the presence of the mutation alone was stronger than diabetes and smoking, as strong as duration spent on HD but less strong than age. Both markers together seemed to add a discriminative ability comparable to age.
Table 2.
Relative contribution of each variable to the final Cox model shown in Table 1
| Parameter | −2LL | AUC |
|---|---|---|
| Entire cohort (n = 173) | ||
| full model | 852 | 0.70 |
| effect of withholding variable on modela | ||
| diabetes | 856 | 0.70 |
| smoking | 862 | 0.68 |
| mtDNA copy number (per log) | 863 | 0.68 |
| duration on HD (per log-years) | 868 | 0.66 |
| age (per year) | 871 | 0.65 |
| Subset with mtDNA4977 genotyping available (n = 150) | ||
| full model | 662 | 0.74 |
| effect of withholding variable on modela | ||
| diabetes | 669 | 0.75 |
| mtDNA copy number (per log) | 668 | 0.74 |
| smoking | 674 | 0.72 |
| mtDNA4977 | 679 | 0.71 |
| duration on HD (per log-years) | 678 | 0.71 |
| age (per year) | 691 | 0.69 |
| both mtDNA4977 and mtDNA copy number (per log) | 688 | 0.70 |
A higher −2LL and lower area under the curve (AUC) suggest greater contribution of the variable to the discriminative ability of the model because this is the effect of leaving the variable out of the model.
Relationship between mtDNA Injury and Other Markers
Plasma 8-OHDG levels were measured as a marker of oxidative stress but showed no relationship to either the mtDNA copy number or the presence of the deletion mutation. Plasma lactate was measured as a crude indicator of redox status and therefore mitochondrial function. The mean plasma lactate level was 1.3 ± 0.8 mmol/L. There was an NS negative correlation between plasma lactate and mtDNA copy number (r = −0.12, P = 0.13) that improved after adjustment for age, diabetes, and (log) CRP (r = −0.17, P = 0.04). Plasma lactate levels did not differ by presence of the deletion mutation.
Sensitivity Analysis
For patients with missing data, the technique of multiple imputation was used to impute values that were missing at baseline for serum cholesterol (18%) and plasma CRP (7%). Cox regression models were re-run with ACM as the outcome for analyses with mtDNA copy number, mtDNA4977, and both, and estimates obtained after inclusion of imputed values did not differ from the original results.
DISCUSSION
This study, of a cohort of prevalent maintenance HD patients, showed that markers of mtDNA injury were powerful predictors of ACM. The markers evaluated were the copy number of mtDNA per cell and the presence of the mtDNA4977 deletion mutation. A higher copy number was strongly protective from risk for death, and the presence of the deletion mutation (rather than absence) was strongly protective from risk for death. These relationships were not affected appreciably by adjustment for covariates and indeed with regard to the mutation actually strengthened by adjustment. The mtDNA copy number did not have a significant association with other variables, including age, duration spent on HD, or diabetes. It showed a weak albeit significant inverse relationship with plasma lactate levels after adjustment for age, diabetes, and inflammatory status. The deletion mutation was more frequent among older individuals and those who were older at the onset of HD and was associated with a shorter duration spent on HD. Among patients with the mutation, a higher proportion had diabetes or preexisting vascular disease, lower plasma CRP levels, and, interestingly, a higher mtDNA copy number, but none of these differences reached statistical significance. Compared with healthy control subjects frequency matched for age, the mtDNA copy number among patients with ESRD was significantly lower among older individuals and seemed to have markedly wider variability. The age-associated increase in copy number that was apparent among healthy control subjects was not seen among patients with ESRD.
mtDNA is polyploid (unlike nuclear DNA, which is diploid), indicating that there are multiple copies of mtDNA within a cell. mtDNA replication is independent of nuclear DNA replication but involves coordinated expression of genes in the nucleus and mitochondria. The number of copies and the rate of replication vary by cell type. mtDNA replication is usually associated with but not necessary to the process of mitochondrial biogenesis, which is responsible for maintaining mitochondrial mass and volume during states of normal and altered homeostasis.8 Classically, mtDNA depletion syndromes are associated with myopathies6; recent literature has alluded to a link between decreased mitochondrial biogenesis and diabetes9–11 and the cardiovascular risk associated with the metabolic syndrome.12 mtDNA abundance has been associated with a beneficial muscle response to endurance exercise as well as senescence and states of oxidative stress.13–15 It is indeed conceivable that an increase in mtDNA copy number in the latter situations could occur as a feedback response that compensates for defective mitochondria or mutated mtDNA.
Several acquired mtDNA mutations (point mutations and deletion mutations) have been associated with conditions such as atrial fibrillation, atherosclerosis, senescence, type 2 diabetes, and neurodegenerative diseases such as Alzheimer's and Parkinson's diseases.16–19 mtDNA mutations are heteroplasmic (i.e., both normal and mutant mtDNA are present in the same cell), the phenotype reflecting the proportion of mutant mtDNA molecules and the extent to which the cell type relies on mitochondrial function.5,6 The most frequently encountered mutation is a 4977-bp deletion between nucleotide positions 8470 and 13,447, referred to as the common deletion (mtDNA4977). mtDNA4977 has been established as the most common and abundant large-scale deletion of mtDNA in various human tissues, accounting for 30 to 50% of all deletions, and has been commonly used as an indicator of somatic mtDNA injury. The 4977-bp deleted region encodes for seven of 13 polypeptides that are essential subunits for the respiratory chain enzyme complexes of the oxidative phosphorylation pathway.20 Lim et al. found that the mtDNA4977 deletion mutations were highly prevalent in skeletal muscle of patients with ESRD and correlated positively with the 8-hydroxy 2′ deoxyguanosine (8-OHDG) content (a marker of nucleic acid oxidation) of total skeletal muscle DNA.4 Liu et al.21 also showed that the incidence and proportion of mtDNA with the 4977 deletion in hair follicles both were significantly higher among patients with ESRD on HD compared with age-matched healthy control subjects; however, there has been no systematic investigation of the relationship between markers of mtDNA injury and clinical outcomes among patients with ESRD, and we believe this report is the first to address this area.
Our results, although preliminary, suggest that a higher mtDNA copy number among HD patients is associated with better survival. The observation that the presence of the mtDNA4977 deletion mutation is associated with better survival is counterintuitive. It is possible that a higher mtDNA copy number is needed as a compensatory response to ongoing mitochondrial injury in uremia and therefore is protective.8 Indeed, we did observe that copy number tended to be higher among patients positive for the mtDNA4977 mutation, although statistical significance was not reached. We also observed that plasma lactate levels tended to be higher among patients with lower mtDNA copy number. The absence of an increase in copy number with age among patients with ESRD as was seen with healthy control subjects may also be an indicator of a failed compensatory response. Although the literature is sparse and conflicting, animal studies have shown an increase in mtDNA copy number with age in several organs.22–24
Regarding the mtDNA4977 mutation, one might speculate that it may be a marker of either survival or aging. Although there is a large body of literature associating the mutation with aging and age-related pathology, there is little or no evidence to impute a causal role.18,25,26 The mtDNA4977 mutation may therefore identify individuals who are older and possibly survivors. Alternatively, the mutation may identify less lethal injury to mtDNA, and patients who genotyped negative for this mutation may have had more lethal undiscovered mutations. A mechanistic explanation may be that cells with the mutation produced fewer reactive oxygen species, considering its location in the region encoding the respiratory chain enzyme complexes of the oxidative phosphorylation pathway. Alternatively, accumulation of mutated DNA may reflect deficient or suppressed autophagic mechanisms in uremia, to allow for increased biogenesis.25,27,28 It must be emphasized, however, that the observational nature of this study precludes any definitive inference as to the functional impact of the deletion mutation. The impact of mitochondrial mass and mtDNA mutations, as well HD treatment itself, on key parameters and regulators of mitochondrial function such as mitochondrial calcium uniporter and mitochondrial Na+-Ca2+ exchanger needs to be explored in future studies.
Although this report is preliminary and the study is exploratory, there are several aspects that help strengthen our observations. The source of DNA was homogeneous, obtained from harvested PBMC, and eliminated platelet contamination. Genotyping and mutation discovery may be accomplished with DNA from whole-blood samples but not quantification; our results were therefore specific to mononuclear cells. The study included a sizable cohort of stable HD patients with long-term follow-up and detailed and accurate ascertainment of both baseline variables and outcomes. Previous patient-level studies of mtDNA injury markers have been relatively smaller and descriptive.4,21 Finally, chronic kidney disease and ESRD are states in which there is dissociation between chronological age and physiologic age, and several uremic complications mimic an accelerated aging process. ESRD is associated with increased levels of oxidative stress similar to senescence and may provide a relevant model to study the significance of mitochondrial injury. We were unable, however, to demonstrate a relationship to serum levels of 8-OHDG; it is possible that levels of this marker in genomic DNA may show a closer relationship to mtDNA copy number or mutations.
The limitations of this study include the secondary nature of the analysis from a preexisting interventional study. Although the gold standard of target tissue to study mitochondria is skeletal muscle, such sampling is invasive. The use of lymphocytes may also be acceptable because they are long-lived cells with low turnover and may allow mutations to accumulate.7 To offset any bias that may have occurred as a result of missing data, we used the statistical technique of multiple imputations to impute missing values and estimates obtained after inclusion of imputed values did not differ from the original results.
In summary, our observations raise intriguing questions as to the role of mitochondrial injury in the genesis of uremic complications and clinical outcomes. Clearly, our observations on somatic mtDNA mutations and mtDNA copy number need to be replicated in a larger patient population, including those with less advanced chronic kidney disease.
CONCISE METHODS
Subjects
The study cohort consisted of 180 patients who had ESRD and were on maintenance HD and were recruited to the baseline phase of the Hemodialysis (HEMO) Study from two Boston centers. This ancillary study was approved by the Human Investigation Review Committee, and all participants provided written informed consent.
The details of the National Institutes of Health–sponsored HEMO Study have been published elsewhere.29,30 Briefly, this study, initiated in 1995, was sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases and was a multicenter, prospective, randomized clinical trial designed to evaluate the effect of dialyzer urea and β2-microglobulin clearances on morbidity and mortality. Eligible patients were between the ages of 18 and 80 yr, were receiving long-term HD three times per week, and had residual renal urea clearance <1.5 ml/min per 35 L of urea distribution volume. Patients who were in acute or chronic care hospitals; had active malignancy or decompensated cardiac, hepatic, or pulmonary disease; had serum albumin <2.5 g/dl; were pregnant, or had a scheduled or recently (<6 mo) failed transplant were excluded. Eligible patients were randomly assigned in a 1:1 ratio with a two-by-two factorial design to either a standard-dosage (single-pool Kt/V of 1.25) or a high-dosage (single-pool Kt/V of 1.65) goal and to dialysis with either a low-flux (minimum values for ultrafiltration coefficient ≤14 ml/h per mmHg and first-use β2-microglobulin clearance <10 ml/min) or a high-flux dialyzer (minimum values for ultrafiltration coefficient >14 ml/h per mmHg and first-use β2-microglobulin clearance >20 ml/min). The planned follow-up ranged from 1 to 6.5 yr depending on the time of randomization.
Data Procurement
Clinical Data.
Demographic, medical, and socioeconomic information was obtained in the baseline phase of the study. HD prescription and monitoring of routine laboratory parameters followed the protocol of the HEMO study. Comorbidities were catalogued using the ICED.31 The highest scores of Index of Disease Severity and Index of Physical Impairment were combined to create the ICED score, from 0 to 3 (0 indicating the absence of disease, and increasing values indicating increasing severity of the disease), and diabetes-related scores were excluded from the final severity scores.
Outcome.
The primary outcome was time to death from any cause (ACM).
Blood Samples
Baseline blood samples were obtained before dialysis within 1 mo of enrollment. Heparinized blood samples (30 ml) were immediately placed on ice and transported to the laboratory.
Healthy Control Subjects.
Blood samples were also obtained from healthy adult subjects to provide a reference for mtDNA quantification compared with patients with ESRD.
PBMC Isolation and DNA Extraction.
Isolated PBMC aliquots were used for DNA extraction and genotyping for this study. PBMC were harvested from whole blood as described previously using Ficoll-Hypaque density gradient separation technique.32 Cells (2.5 × 106 PBMC/ml) were resuspended in RPMI 1640 cell culture medium supplemented with l-glutamine, NaHCO3, HEPES, penicillin, and streptomycin and stored at −80°C until DNA extraction.
Genomic DNA was extracted using a spin-column method (QIAamp DNA Mini Kit; Qiagen, Valencia, CA). In brief, 2.5 × 106 PBMC were treated with 20 μl of proteinase K (Qiagen), followed by the addition of 200 μl of SDS to lyse the cells. The homogeneous solution was incubated at 56°C for 10 min, and 200 μl of 100% ethanol was added to precipitate DNA. The mixture was then applied to the QIAamp spin column, and after two washes with 500 μl of wash buffer, genomic DNA was eluted by the addition of 200 μl of elution buffer. Final DNA concentrations were 50 to 200 ng/ml determined by minigel electrophoresis.
Mitochondrial copy number was estimated by determining relative amounts of nuclear DNA and mtDNA by quantitative real-time PCR (Stratagene Mx4000Multiplex QPCR System; Stratagene, La Jolla, CA).33 The ratio of mtDNA to nuclear DNA reflects the tissue concentration of mitochondria per cell. A 120-bp-long mtDNA fragment within the ND1 gene and a 120-bp region of the lipoprotein lipase gene (LPL) were amplified. The ND1 forward primer used was (5′ to 3′) CCCTAAAACCCGCCACATCT, and reverse primer was GAGCGATGGTGAGAGCTAAGGT. The LPL (accession no. NM_000237) forward primer used was CGAGTCGTCTTTCTCCTGATGAT and reverse primer was TTCTGGATTCCAATGCTTCGA. The quantification assay was performed in a total reaction volume of 25 μl containing 12.5 μl of 2× SYBR Green, 1.25 μl of each primer, 1 μl of sample DNA, and 9 μl of water. Amplification and detection were performed in a Stratagene Mx4000 Multiplex Quantitative PCR System. PCR was initiated with 15 min at 95°C, followed by 40 cycles of 15 s at 95°C, 30 s at 49°C, and 30 s at 72°C, followed by 1 min at 95°C and 41 cycles starting at 49°C for 30 s, escalating by 1°C per cycle. Each sample was assayed in triplicate, and fluorescence spectra were continuously monitored by the Mx4000 system and sequence detection software. Data analysis was based on measurement of the cycle threshold (CT), and the difference in CT values was used as the measure of relative abundance: CT(ND1) − CT(LPL) or ΔCT, a quantitative measure of the mitochondrial genome. Results were expressed as the copy number of mtDNA/cell, provided by 2 × 2−ΔCT and, being a ratio, was unitless.
Genotyping.
The common deletion (mtDNA4977) was genotyped in a subset of 157 patients for whom DNA was of sufficient quality, using a nested PCR protocol.16 The amplification target was the segment between 8224 and 13501 bp. The primer pairs for mtDNA4977 and wild-type mtDNA were used in the same tube of reaction (co-amplification). In the presence of the deletion mutation, two products were obtained: A 301-bp product from mtDNA4977 and a 326-bp product from wild-type mtDNA. Figure 2 shows the agarose gel electrophoresis of the PCR products.
Plasma levels of CRP were measured using a commercially available high-sensitivity immunoassay (Hemagen Diagnostics, Columbia, MD).
Levels of 8-OHDG and lactate were measured in plasma using commercially available assays. Plasma 8-OHDG was quantified using a competitive enzyme immunoassay (Cell Biolabs, San Diego, CA). Briefly, plasma samples or 8-OHDG standards are first added to an 8-OHDG/BSA conjugate preabsorbed enzyme immunoassay plate. After a brief incubation, an anti–8-OHDG mAb is added, followed by an horseradish peroxidase–conjugated secondary antibody. The 8-OHDG content in the plasma samples was then determined by comparison with the 8-OHDG standard curve. The assay has a detection sensitivity range of 100 pg/ml to 20 ng/ml. Lactate levels were measured with a colorimetric assay (BioVision Research Products, Mountain View, CA).
For each individual patient, samples from each time point were analyzed in the same assay. All samples for a given assay were tested simultaneously, in duplicate and in appropriate dilutions.
Statistical Analysis
Analysis was performed using SAS 9.1 (SAS Institute, Cary, NC). Data were expressed as means and SD for continuous variables that were normally distributed and medians and ranges for non-normally distributed data (mtDNA copy number per cell, CRP levels, duration of HD, and body mass index). Categorical data were expressed as proportions. The unadjusted relationships of either mtDNA copy number per cell or the mtDNA4977 deletion mutation to patient characteristics including relationship to plasma CRP levels were examined using nonparametric statistics or correlation, with transformed data as applicable.
Cox proportional hazards regression was used to evaluate the effect of mtDNA copy number per cell on ACM outcome. Data were censored at the time of transplantation, but, in keeping with the intention-to-treat principle of the parent study, data were not censored when patients left the study because of transfer to a nonparticipating center or alternative method of dialysis. The proportional hazards assumption was tested using Schoenfeld residuals, a time-varying coefficient model, and by examination of log (−log survival) curves for parallelism and was met for all covariates except diabetes. The covariates explored were age, gender, race (white versus black), diabetes, duration on HD, the ICED score as a measure of comorbidity, body mass index, smoking history, and the dialysis dosage and flux grouping of randomization. The effect of adding plasma CRP level as a covariate was subsequently studied in a separate model. The final models were arrived at by backward selection of variables to limit the number of variables and avoid overfitting in the models.
Cox models were also run for the subset of patients for whom genotyping data for the deletion mutation mtDNA4977 were available to examine its relationship to ACM outcome. An additional model was constructed whereby the effect of mtDNA4977 on ACM outcome was adjusted for mtDNA copy number per cell. Both unadjusted estimates and estimates adjusted for covariates selected as described previously were generated and a separate model was constructed with the inclusion of plasma CRP level.
The relative predictive ability of mtDNA copy number per cell and the presence of the mtDNA4977 deletion mutation for ACM compared with other known risk variables were tested using the likelihood ratio χ2 statistic as the change in −2log likelihood of each model upon addition of each marker. The concordance statistic (area under the curve, or “c index”) was used as a measure of discrimination by each marker for mortality outcome. The discriminative ability indicates how well a model can distinguish between patients with different survival expectations. The c statistic for survival data estimates the probability that for a randomly chosen pair of patients, the one having the higher predicted survival is the one who survives longer. A predictive model with a c of 0.5 has no predictive value, whereas a model with a c of 1.0 discriminates perfectly between patients differing in survival.
Because data were complete for 93 and 82% of the study population for the baseline covariates plasma CRP and serum cholesterol, respectively, the technique of multiple imputation was used to impute missing values for these variables. Cox regression models were re-run, and estimates were compared with those from the models without imputed data in the form of a sensitivity analysis.
All tests were two-tailed, and P < 0.05 was considered significant. All CI were calculated at the 95% level.
DISCLOSURES
None.
Acknowledgments
This study was supported by a grant from the National Institutes of Health (DK 2819-O1A1). Additional support was provided by a grant from Satellite Healthcare.
This study was presented at the annual meeting of the American Society of Nephrology; November 2 through 5, 2007; San Francisco, CA.
Published online ahead of print. Publication date available at www.jasn.org.
References
- 1.Kalantar-Zadeh K: Recent advances in understanding the malnutrition-inflammation-cachexia syndrome in chronic kidney disease patients: What is next? Semin Dial 18: 365–369, 2005 [DOI] [PubMed] [Google Scholar]
- 2.Heilbronn L, Smith SR, Ravussin E: Failure of fat cell proliferation, mitochondrial function and fat oxidation results in ectopic fat storage, insulin resistance and type II diabetes mellitus. Int J Obes Relat Metab Disord Suppl 4: S12–S21, 2004 [DOI] [PubMed] [Google Scholar]
- 3.Thompson CH, Kemp GJ, Taylor DJ, Ledingham JG, Radda GK, Rajagopalan B: Effect of chronic uraemia on skeletal muscle metabolism in man. Nephrol Dial Transplant 8: 218–222, 1993 [PubMed] [Google Scholar]
- 4.Lim PS, Ma YS, Cheng YM, Chai H, Lee CF, Chen TL, Wei YH: Mitochondrial DNA mutations and oxidative damage in skeletal muscle of patients with chronic uremia. J Biomed Sci 9: 549–560, 2002 [DOI] [PubMed] [Google Scholar]
- 5.Wei Y: Mitochondrial DNA alterations as ageing-associated molecular events. Mutat Res 275: 145–155, 1992 [DOI] [PubMed] [Google Scholar]
- 6.Rotig A, Munnich A: Genetic features of mitochondrial respiratory chain disorders. J Am Soc Nephrol 14: 2995–3007, 2003 [DOI] [PubMed] [Google Scholar]
- 7.Mohamed SA, Wesch D, Blumenthal A, Bruse P, Windler K, Ernst M, Kabelitz D, Oehmichen M, Meissner C: Detection of the 4977 bp deletion of mitochondrial DNA in different human blood cells. Exp Gerontol 39: 181–188, 2004 [DOI] [PubMed] [Google Scholar]
- 8.Chabi B, Adhihetty PJ, Ljubicic V, Hood DA: How is mitochondrial biogenesis affected in mitochondrial disease? Med Sci Sports Exerc 37: 2102–2110, 2005 [DOI] [PubMed] [Google Scholar]
- 9.Heilbronn LK, Gan SK, Turner N, Campbell LV, Chisholm DJ: Markers of mitochondrial biogenesis and metabolism are lower in overweight and obese insulin-resistant subjects. J Clin Endocrinol Metab 92: 1467–1473, 2007 [DOI] [PubMed] [Google Scholar]
- 10.Rolo AP, Palmeira CM: Diabetes and mitochondrial function: Role of hyperglycemia and oxidative stress. Toxicol Appl Pharmacol 212: 167–178, 2006 [DOI] [PubMed] [Google Scholar]
- 11.Ukropcova B, Sereda O, de Jonge L, Bogacka I, Nguyen T, Xie H, Bray GA, Smith SR: Family history of diabetes links impaired substrate switching and reduced mitochondrial content in skeletal muscle. Diabetes 56: 720–727, 2007 [DOI] [PubMed] [Google Scholar]
- 12.Nisoli E, Clementi E, Carruba MO, Moncada S: Defective mitochondrial biogenesis: A hallmark of the high cardiovascular risk in the metabolic syndrome? Circ Res 100: 795–806, 2007 [DOI] [PubMed] [Google Scholar]
- 13.Hood DA, Saleem A: Exercise-induced mitochondrial biogenesis in skeletal muscle. Nutr Metab Cardiovasc Dis 17: 332–337, 2007 [DOI] [PubMed] [Google Scholar]
- 14.Menshikova EV, Ritov VB, Fairfull L, Ferrell RE, Kelley DE, Goodpaster BH: Effects of exercise on mitochondrial content and function in aging human skeletal muscle. J Gerontol A Biol Sci Med Sci 61: 534–540, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rasbach KA, Schnellmann RG: Signaling of mitochondrial biogenesis following oxidant injury. J Biol Chem 282: 2355–2362, 2007 [DOI] [PubMed] [Google Scholar]
- 16.Lai LP, Tsai CC, Su MJ, Lin JL, Chen YS, Tseng YZ, Huang SK: Atrial fibrillation is associated with accumulation of aging-related common type mitochondrial DNA deletion mutation in human atrial tissue. Chest 123: 539–544, 2003 [DOI] [PubMed] [Google Scholar]
- 17.Botto N, Berti S, Manfredi S, Al-Jabri A, Federici C, Clerico A, Ciofini E, Biagini A, Andreassi MG: Detection of mtDNA with 4977 bp deletion in blood cells and atherosclerotic lesions of patients with coronary artery disease. Mutat Res 570: 81–88, 2005 [DOI] [PubMed] [Google Scholar]
- 18.Pieczenik SR, Neustadt J: Mitochondrial dysfunction and molecular pathways of disease. Exp Mol Pathol 83: 84–92, 2007 [DOI] [PubMed] [Google Scholar]
- 19.Wallace DC, Schoffner JM, Trounce I, Brown MD, Ballinger SW, Corral-Debrinski M, Horton T, Jun AS, Lott MT: Mitochondrial DNA mutations in human degenerative diseases and aging. Biochim Biophys Acta 1271: 141–151, 1995 [DOI] [PubMed] [Google Scholar]
- 20.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG: Sequence and organization of the human mitochondrial genome. Nature 290: 457–465, 1981 [DOI] [PubMed] [Google Scholar]
- 21.Liu CS, Ko LY, Lim PS, Kao SH, Wei YH: Biomarkers of DNA damage in patients with end-stage renal disease: Mitochondrial DNA mutation in hair follicles. Nephrol Dial Transplant 16: 561–565, 2001 [DOI] [PubMed] [Google Scholar]
- 22.Frahm T, Mohamed SA, Bruse P, Gemünd C, Oehmichen M, Meissner C: Lack of age-related increase of mitochondrial DNA amount in brain, skeletal muscle and human heart. Mech Ageing Dev 126: 1192–1200, 2005 [DOI] [PubMed] [Google Scholar]
- 23.Masuyama M, Iida R, Takatsuka H, Yasuda T, Matsuki T: Quantitative change in mitochondrial DNA content in various mouse tissues during aging. Biochim Biophys Acta 1723: 302–308, 2005 [DOI] [PubMed] [Google Scholar]
- 24.Miller FJ, Rosenfeldt FL, Zhang C, Linnane AW, Nagley P: Precise determination of mitochondrial DNA copy number in human skeletal and cardiac muscle by a PCR-based assay: Lack of change of copy number with age. Nucleic Acids Res 31: e61, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krishnan KJ, Greaves LC, Reeve AK, Turnbull DM: Mitochondrial DNA mutations and aging. Ann N Y Acad Sci 1100: 227–240, 2007 [DOI] [PubMed] [Google Scholar]
- 26.McKenzie D, Bua E, McKiernan S, Cao Z, Aiken JM, Wanagat J: Mitochondrial DNA deletion mutations: A causal role in sarcopenia. Eur J Biochem 269: 2010–2015, 2002 [DOI] [PubMed] [Google Scholar]
- 27.Jin S: Autophagy, mitochondrial quality control, and oncogenesis. Autophagy 2: 80–84, 2006 [DOI] [PubMed] [Google Scholar]
- 28.Kim I, Rodriguez-Enriquez S, Lemasters JJ: Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys 462: 245–253, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eknoyan G, Beck GJ, Cheung AK, Daugirdas JT, Greene T, Kusek JW, Allon M, Bailey J, Delmez JA, Depner TA, Dwyer JT, Levey AS, Levin NW, Milford E, Ornt DB, Rocco MV, Schulman G, Schwab SJ, Teehan BP, Toto R, Hemodialysis (HEMO) Study Group: Effect of dialysis dose and membrane flux in maintenance hemodialysis. N Engl J Med 347: 2010–2019, 2002 [DOI] [PubMed] [Google Scholar]
- 30.Greene T, Beck GJ, Gassman JJ, Gotch FA, Kusek JW, Levey AS, Levin NW, Schulman G, Eknoyan G: Design and statistical issues of the hemodialysis (HEMO) study. Control Clin Trials 21: 502–525, 2000 [DOI] [PubMed] [Google Scholar]
- 31.Miskulin DC, Athenites N, Yan G, Martin AA, Ornt DB, Kusek JW, Meyer KB, Levey AS; Hemodialysis (HEMO) Study Group: Comorbidity assessment using the Index of Coexistent Diseases in a multicenter clinical trial. Kidney Int 60: 1498–1510, 2001 [DOI] [PubMed] [Google Scholar]
- 32.Poutsiaka DD, Clark BD, Vannier E, Dinarello CA: Production of interleukin-1 receptor antagonist and interleukin-1 beta by peripheral blood mononuclear cells is differentially regulated. Blood 78: 1275–1281, 1991 [PubMed] [Google Scholar]
- 33.He L, Chinnery PF, Durham SE, Blakely EL, Wardell TM, Borthwick GM, Taylor RW, Turnbull DM: Detection and quantification of mitochondrial DNA deletions in individual cells by real-time PCR. Nucleic Acids Res 30: e68, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]