Abstract
Vertebrate cells contain a large number of small nucleolar RNA (snoRNA) species, the vast majority of which bind fibrillarin. Most of the fibrillarin-associated snoRNAs can form 10- to 21-nt duplexes with rRNA and are thought to guide 2′-O-methylation of selected nucleotides in rRNA. These include mammalian UHG (U22 host gene)-encoded U25–U31 snoRNAs. We have characterized two novel human snoRNA species, U62 and U63, which similarly exhibit 15- (with one interruption) and 12-nt complementarities and are therefore predicted to direct 2′-O-methylation of A590 in 18S and A4531 in 28S rRNA, respectively. To establish the function of antisense snoRNAs in vertebrates, we exploited the Xenopus oocyte system. Cloning of the Xenopus U25–U31 snoRNA genes indicated that they are encoded within multiple homologs of mammalian UHG. Depletion of U25 from the Xenopus oocyte abolished 2′-O-methylation of G1448 in 18S rRNA; methylation could be restored by injecting either the Xenopus or human U25 transcript into U25-depleted oocytes. Comparison of Xenopus and human U25 sequences revealed that only boxes C, D, and D′, as well as the 18S rRNA complement, were invariant, suggesting that they may be the only elements required for U25 snoRNA stability and function.
Keywords: [nucleolus, UHGU22 host gene, RNA duplex, fibrillarin]
The nucleolus of eukaryotic cells is the site of ribosome biogenesis; there, rRNA is transcribed, modified, processed, and assembled into the large and small subunits before export to the cytoplasm (reviewed in ref. 1). The newly synthesized RNA polymerase I transcript (≈40 S) consists of the 18S, 5.8S, and 28S rRNAs along with external and internal spacer sequences. In this transcript, a number of uridines are converted to pseudouridines, certain sugar residues are methylated at the 2′ position, and a few nucleotide bases are methylated (reviewed in ref. 2). These modifications are confined solely to the sequences specifying the mature 18S, 5.8S, and 28S rRNAs. Most 2′-O-methylated residues are conserved, with almost no differences among the more than 100 sites found in vertebrates. Yeast rRNA contains 55 2′-O-methylated residues, 18 of which correlate with those in higher eukaryotes. There are three methylated sugar moieties found in Escherichia coli 23S rRNA; two, G2251 and U2552, seem to be universally conserved.
Although it has been proposed that methylated sugar residues protect rRNA from nucleases or provide stability in the ribosome through increased hydrophobic interactions (reviewed in ref. 2), the precise function of 2′-O-methylation has not been established. Because of contradictory results, it is not even clear whether these modifications function primarily during ribosome biogenesis or during translation. In early experiments, methionine starvation of HeLa cells blocked processing of the 32S intermediate (3). Similar results were observed when HeLa cells or rat liver were exposed to the methylation inhibitor ethionine (4, 5). However, other experiments using ethionine in yeast or using an alternative methylation inhibitor, cycloleucine, in HeLa cells indicated that reduced methylation did not alter the pattern of pre-rRNA processing but rather caused less efficient ribosome biogenesis (6, 7). Accordingly, the lack of 2′-O-methylation of G2251 in yeast mitochondrial 21S rRNA (caused by depletion of the Pet56 nuclear protein) prevents large subunit assembly and results in a severe growth defect (8). Severe growth defects are also observed in yeast when mutations in nucleolar protein 1 (NOP1) or NOP77, resulting in undermethylated rRNA, are introduced (9, 10). However, it appears that individual 2′-O-methylated residues are not required for cell viability (ref. 11; M. Fournier, personal communication). Recent data indicate that in vitro-transcribed, prokaryotic rRNA, lacking most modifications, functions in the peptidyl transferase reaction, so it could be that the methylated residues not required for critical steps during translation.
A growing number of small nucleolar ribonucleoprotein particles (snoRNPs) are emerging as key players in ribosome biogenesis (reviewed in ref. 12). Currently, 57 snoRNAs have been reported, and there are indications that more remain undiscovered. Most snoRNAs associate with the fibrillarin autoantigen (13), whose binding correlates with the presence of conserved sequences termed “box C” (RUGAUGA) and “box D” (CUGA), near their 5′ and 3′ ends, respectively. The major fibrillarin-associated snoRNAs—U3, U8, U14, and U22—have been demonstrated to play critical roles in the processing of pre-rRNA (14–18). Most of the remaining snoRNAs exhibit striking complementarities to highly conserved regions of rRNA; the base-pairing sequences within these snoRNAs are invariably followed by box D or box D′, an internal box D-like sequence (11, 19–21). These antisense snoRNAs have been proposed to function as guides for 2′-O-methylation of rRNA because a 2′-O-methylated residue in mature rRNA is always found in the position that base pairs to the fifth nucleotide upstream of the conserved box D or D′ (11, 21).
In this study, we have confirmed the prediction that complementary snoRNAs direct 2′-O-methylation of rRNA residues in a vertebrate cell, the Xenopus oocyte. We have shown that U25, one of the snoRNAs encoded by the U22 host gene (UHG) in mammals (20), is essential for methylation of G1448 in 18S rRNA. In addition, we have isolated and sequenced two novel human snoRNAs, U62 and U63, as well as genes for multiple Xenopus U25–U31 snoRNA variants. Seven yeast snoRNAs, including U24 and U18, have been shown to guide 2′-O-methylation of selected nucleotides in yeast rRNA (ref. 11; M. Fournier, personal communication).
MATERIALS AND METHODS
Oligonucleotides.
The oligonucleotides used were as follows: a, TGATGAGGACCTTTTCACAGACCTG; b, ACATACCTTTTACAGAACTCC; c, GTCCTCATTGCTTTCAGAA; d, CAGCTTACTATCTCTGAGG; e, TTCAGGCTCTNGCAACAGGCAGC; f, GGTGAAC(C/A)(C/A)ACTGGCTCAACAG; g, ATAGCATGTTAGAGTTCTG; h, CACAAAATCATAAATATAAGCC; i, GTAAAGGTAACCTGTTAAC; j, GACTGGGGCGGTA; k, CCAAGTCTGTTGCTAATGACG; l, GATGCTCAGGAGTTCAAAGCTT; 26, ATCTGGAATCTACCTGCC; XU25-5′, ACAGGTCTGTGAT; XU25-3′, GCCTCAGAGATAGTAAGCTGTC; HU25-3′, CTCCTCAGAGTTATTTATCCTC; XU28-3′, TCCATCAGAACTCCACCA; XaU25, ACCTCTTGCTGGCTTCATAG; 18S-245, GCTGATCCGTTCAGTGTAGC.
Resolving and Sequencing snoRNAs.
Preparation of HeLa cell extracts and immunoprecipitation with antifibrillarin antibody (72B9) were performed as described (22). Precipitated RNAs were [5′-32P]cytidine 3′,5′-bisphosphate labeled (23) and resolved on a 15% denaturing polyacrylamide gel (19:1 acrylamide/bis-acrylamide). Twenty-three bands were excised, and the RNAs were eluted. After ethanol precipitation, the RNAs were further resolved on a 15% denaturing polyacrylamide gel (9:1 acrylamide/bis-acrylamide). Bands were excised and eluted as above, and 3′ end sequences were obtained by direct enzymatic sequencing with RNases T1, U2, PhyM, and Bacillus cereus (24). Dideoxy primer extension sequencing (25) (using primers derived from the obtained 3′ end sequences) provided full snoRNA sequences. The novel sequences were then confirmed by cloning of their cDNAs (26).
Oocyte Injection.
Oocyte injections and dissections were performed essentially as described (18) except that 32 nl of deoxyoligonucleotide at a concentration 2 mg/ml was injected. After 2 days of incubation, the oocytes were dissected, and the RNA was isolated. For “rescue” experiments, germinal vesicles were injected with 25 nl of an in vitro-transcribed RNA (2.5 nmol/ml) 24 h after the oligonucleotide injection. After a further 24-h incubation, the oocytes were dissected, and the RNA was isolated.
The Xenopus U25 rescue transcript and antisense U25 were prepared by transcription of a pGEM3Z.XU25 plasmid with T7 and SP6 polymerase, respectively. The pGEM3Z.XU25 contains Xenopus U25 plus 62 and 25 nt of 5′ and 3′ flanking sequences, respectively, inserted into the SmaI site. For transcription of sense or antisense U25, the plasmid was linearized with HindIII or EcoRI, respectively. The human U25 RNA plus 48 nt at the 5′ and 28 nt at the 3′ end were transcribed by T7 polymerase from the pGEM3Z.HU25 plasmid linearized with HindIII.
Mapping of Ribose Methylation.
Partial alkaline hydrolysis of nuclear RNA was carried out in 150 mM Na2CO3/NaHCO3 (pH 9.2) and 1 mM EDTA for 4 min at 90°C. The partially degraded RNA was recovered by ethanol precipitation. For primer extension analyses, ≈0.1 pmol of 5′ end-labeled 18S rRNA primer was annealed to partially hydrolyzed RNA (either 10 nuclei or 0.1 cytoplasm worth), and the primer extension reaction was carried out as described (25).
RESULTS
Two Novel Human snoRNAs.
To isolate additional human snoRNAs, RNAs precipitated from HeLa cell extracts by antifibrillarin antibody were resolved in two consecutive denaturing polyacrylamide gels that differed in the acrylamide/bis-acrylamide ratio (see Materials and Methods). Ten bands designated in Fig. 1A, representing RNAs in the 70- to 100-nt range, were excised, and ≈30 nt at the 3′ ends were determined by enzymatic sequencing. Two were novel snoRNAs, which we named “U62” and “U63” (Fig. 1B). The sequences of U62 and U63 were completed by dideoxy sequencing and confirmed by sequencing their cDNAs. Both RNAs contained boxes C, D, and D′; box D′ was preceded in each case by a sequence complementary to mature rRNA. A duplex of 15 (with one interruption) or 12 bp could be formed between U62 and 18S or between U63 and 28S rRNA, respectively. In each case, the rRNA residue that base pairs to the fifth nucleotide upstream of box D′ has been demonstrated to carry a 2′-O-methyl group (30, 31). Accordingly, U62 and U63 are predicted to guide 2′-O-methylation of A590 in 18S rRNA and A4531 in 28S rRNA, respectively.
Figure 1.
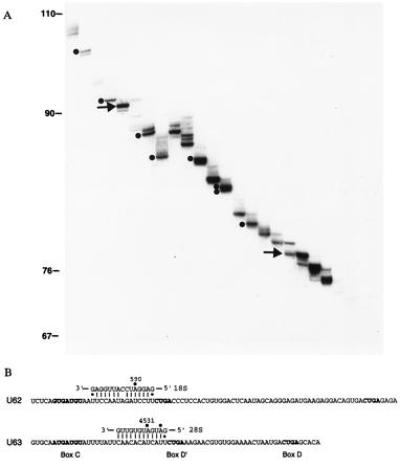
Fibrillarin-associated human snoRNAs. (A) Resolution of RNAs isolated from an antifibrillarin precipitate of HeLa cell extract. The 3′ end-labeled RNAs were first fractionated on a standard 15% denaturing polyacrylamide gel. Twenty-three bands were excised, eluted, and further resolved on a 15% gel with a ratio of acrylamide/bis-acrylamide of 9:1. U62 (Upper) and U63 (Lower) snoRNAs are indicated by arrows. Previously identified snoRNAs are indicated by dots. From largest to smallest they are: U16 (27), U14 (28), U45a (11), U45a variant (11), U20 (29), U28 (20), U26 (20), and U45b (11). (B) Sequences of U62 and U63 snoRNAs and their predicted base-pairing interactions with rRNAs. Conserved boxes C, D, and D′ are shown in boldface type. Filled circles indicate 2′-O-methylated residues in rRNA.
Organization of the Xenopus U25–U31 and U22 Genes.
snoRNA function can readily be assessed in the Xenopus oocyte (16, 18). Therefore we characterized Xenopus homologs of human and mouse U25–U31, all of which are candidates for directing 2′-O-methylation of rRNA. Previous work revealed that the mammalian genes for these snoRNAs, as well as for U22, are arranged in tandem within different introns of UHG (20). To clone and sequence the Xenopus U25–U31 snoRNAs genes, we designed a series of primers complementary primarily to highly conserved regions within the mammalian snoRNAs and amplified Xenopus genomic DNA by standard and inverse PCR methods. We obtained several partially overlapping clones that could be arranged into five fragments called “A,” “B,” “C,” “D,” and “E” (Fig. 2A). Sequence comparison with the mammalian UHGs revealed U25–U31 and U22 snoRNA sequences within these fragments, suggesting that they are derived from Xenopus UHG genes. The snoRNA sequences themselves are the only conserved regions between the five Xenopus fragments and the mammalian UHGs.
Figure 2.
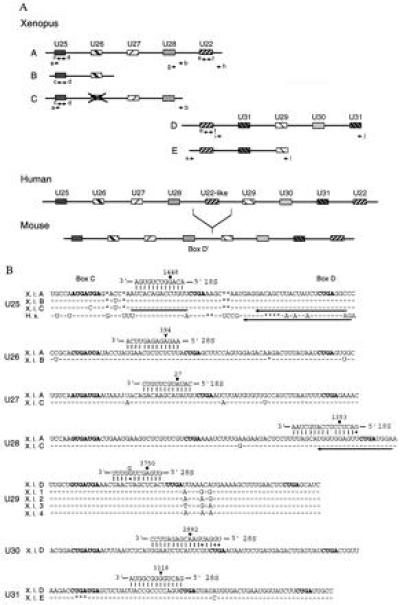
(A) Arrangement of the Xenopus U25–U31 and U22 snoRNA genes (schematic, not drawn to scale). The pairs of arrows indicate primers used for either standard or inverted PCR amplification. Organization of the human and mouse U25–U31 and U22 snoRNA genes (20) are shown for comparison. Boxes filled with identical patterns indicate genes for the same snoRNA. The X over the box in Xenopus fragment C indicates that the U26-like sequence is only 76% identical to human U26 snoRNA and contains several nucleotide changes and deletions within box C and the 18S rRNA complementarity, respectively. (B) Xenopus U25–U31 snoRNA sequences and their predicted base pairing interactions with rRNAs. The sequences of all Xenopus U25–U28, U30, and U31 snoRNA variants, as well as the U29D variant, were deduced from genomic sequences by comparison with their mammalian counterparts, whereas the sequences of the other U29 variants were determined by analyzing their cDNAs. Human U25 is shown for comparison. Identical and missing nucleotides are represented by dashes and asterisks, respectively. Conserved boxes C, D, and D′ are shown in bold. A bar and arrows indicate complementary oligonucleotides used in oocyte injections and for primer extension experiments, respectively. Filled circles or arrowheads indicate rRNA residues that are reported or predicted to be 2′-O-methylated, respectively.
The genomic arrangement of the U25–U31 and U22 snoRNA genes within the presumed Xenopus UHG homologs is similar to that of their mammalian counterparts (Fig. 2A), except that an additional U31 sequence is found between U22 and U29. Of interest, the position of Xenopus U22 corresponds to that of the human U22-like sequence, which is completely absent in mouse UHG. It is not clear at present whether a second Xenopus U22 gene exists 3′ to U31, as in the mammalian UHGs. Moreover, it has not been ruled out that one of the A, B, or C fragments is contiguous with D or C. Attempts to amplify DNA from several pairs of primers (in which one primer was complementary to either A, B, or C and another to either D or C), however, yielded negative results. Taken together, these data indicate that Xenopus UHG is a multicopy gene. This supposition is further supported by the fact that, in addition to U29D, four other U29 clones were generated by reverse transcription-PCR amplification (Fig. 2B).
The characterized variants of Xenopus U25–U31 snoRNAs differ at most in three positions, usually in the evolutionarily least conserved regions. The exception is U31, where variant E carries a UGA deletion within box C (Fig. 2B). It is not known whether U31E RNA accumulates in vivo; the U31E sequence could simply represent a nonfunctional pseudogene. Xenopus U25–U31 snoRNAs all exhibit complementarities to rRNAs; in the cases of U25, U27, U28, U29, and U30, the rRNA residue paired to the fifth position upstream of box D or D′ has been reported to carry 2′-O-methyl (30, 31). The lack of reported 2′-O-methyl groups within the 28S rRNA regions predicted to base pair with U26 and U31 may simply reflect incomplete localization of 2′-O-methylated nucleotides in vertebrate rRNA (2).
Depletion of U25 snoRNA Inhibits 2′-O-methylation of G1448 in 18S rRNA.
To examine the role of Xenopus snoRNAs in rRNA 2′-O-methylation, we chose U25. Both mammalian and Xenopus U25 contain a 12-nt sequence upstream of box D′ that is complementary to 18S rRNA; 2′-O-methylated G1490 in human and the corresponding residue G1448 in Xenopus 18S rRNA base pair to the fifth position counting from box D′ of U25 (ref. 20; Fig. 2B).
We targeted U25 for degradation by injecting an antisense deoxyoligonucleotide into Xenopus oocytes. A single injection into the cytoplasm of an oligonucleotide complementary to positions 18–30 in U25 snoRNA (Fig. 3A, lane 3), but not of a control oligonucleotide (lane 2), caused efficient degradation of U25 as assayed by primer extension. The level of another snoRNA, U28, remained unchanged.
Figure 3.
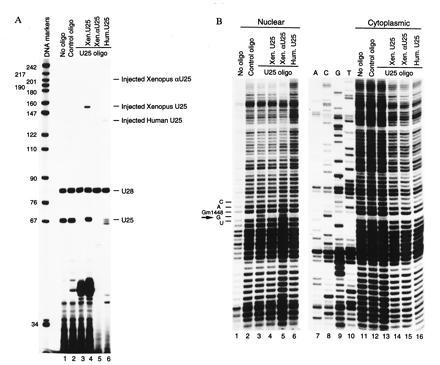
Depletion of U25 snoRNA from Xenopus oocytes inhibits 2′-O-methylation of G1448 in 18S rRNA. Oocytes were cytoplasmatically injected with either U25-5′ or a nonspecific oligonucleotide. Sixteen hours later, a Xenopus or human U25 transcript or a Xenopus antisense U25 transcript was injected into germinal vesicles as indicated at the top. After 24 h, the oocytes were dissected, and RNA was isolated from both the nuclear and cytoplasmic compartments. (A) Primer extension analysis of U25 and U28 snoRNAs in the nuclei of uninjected oocytes (lane 1) and oocytes injected either with the nonspecific (lane 2) or U25-5′ (lanes 3–6) oligonucleotide. Oocytes in lanes 4, 5, and 6 were also injected with Xenopus U25, human U25, and Xenopus antisense U25 transcripts, respectively. A mixture of XU25-3′ and XU28-3′ primers was used in lanes 1–4. A combination of XαU25 and XU28-3′ or HU25-3′ and XU28-3′ primers was used in lane 5 or 6, respectively. (B) Mapping ribose methylation at G1448 in 18S rRNA. Nuclear (lanes 1–6) or cytoplasmic (lanes 11–16) RNA was subjected to partial alkaline hydrolysis, and 18S sequences were analyzed by primer extension using 18S-245 primer. Lanes 1 and 11, uninjected oocytes; lanes 2 and 12, oocytes injected with nonspecific oligonucleotide 26; lanes 3–6 and 13–16, oocytes injected with U25-5′ oligonucleotide. Oocytes in lanes 4 and 14, 5 and 15, or 6 and 16 were also injected with Xenopus U25, human U25, or Xenopus antisense U25 transcripts, respectively. Lanes 7–10 show dideoxy sequencing of 18S rRNA performed on the cytoplasmic RNA. An arrow indicates a gap in the ladders of primer extension products caused by 2′-O-methylation of G1448. The primer extension products were resolved on 8% denaturing polyacrylamide gels.
Two days after the oligonucleotide injection, the methylation status of G1448 was assayed by performing primer extension on partially alkali-hydrolyzed RNA isolated from either the nucleus or the cytoplasm of injected oocytes (Fig. 3B); 2′-O-methyl groups confer resistance to alkaline hydrolysis, so each modified nucleotide is preceded by a “gap” in the ladder of extension products. In both uninjected oocytes and control oligonucleotide-injected oocytes, G1448 was preceded by such gaps in both nuclear (lanes 1 and 2) and cytoplasmic (lanes 11 and 12) RNA, confirming 2′-O-methylation of this nucleotide in the Xenopus oocyte. Depletion of U25 snoRNA caused the appearance of a band in this position in the nuclear RNA (lane 3) but not in the cytoplasmic (lane 13) RNA. This demonstrates a lack of 2′-O-methylation of G1448 in newly synthesized 18S rRNA, which is the predominant species in the nucleus, whereas the bulk of the cytoplasmic rRNA was made before U25 depletion.
The methylation of G1448 can be restored by injection into U25-depleted oocytes of either Xenopus (Fig. 3B, lane 4) or human (lane 6) in vitro-transcribed U25 snoRNA but not by in vitro-transcribed Xenopus antisense U25 (lane 5). Note that both Xenopus and human U25 transcripts, which contain flanking sequences at both ends, were processed after injection to U25-sized RNAs (Fig. 3A, lanes 4 and 6, and data not shown). In contrast, the antisense U25 transcript was totally degraded (Fig. 3A, lane 5).
DISCUSSION
The number of distinct fibrillarin-associated snoRNA species in vertebrates is likely to approach 100. Most of these can form long (10–21 nt), uninterrupted duplexes with rRNAs. This feature, in collaboration with the adjacent box D or D′ in the snoRNA, is thought to direct 2′-O-methylation of specific residues in rRNA (11, 21, 32). Two novel human snoRNA species characterized in this paper, U62 and U63, also can form such duplexes; a 15- (with one interruption) or 12-nt base-pairing interaction, each followed by box D′, can be predicted for U62 and 18S or U63 and 28S rRNA, respectively. Because 2′-O-methylated A590 of 18S and A4531 of 28S rRNA are located within their respective helices at canonical positions for 2′-O-methylation (i.e., 5 nt upstream from boxes D′), U62 and U63 are proposed to guide methylation of these nucleotides. A590 2′-O-methylation is highly conserved; it has been reported in both vertebrate and yeast 18S (2).
An unusual feature of the U62/18S rRNA helix is that it contains one mismatch. Although the vast majority of the proposed snoRNA/rRNA helices are uninterrupted, single mismatches are predicted to occur in yeast U14/18S rRNA (17) and Xenopus U28/28S rRNA (Fig. 2B) interactions. Likewise, a bulge is likely to interrupt the U29/28S rRNA duplex (Fig. 2B). The effect of these imperfections on the functioning of snoRNAs remains to be determined.
Partial cloning and sequencing revealed that the Xenopus U25–U31 and U22 snoRNA genes are organized in tandem in an order similar to that of their mammalian counterparts. This suggested that they are contained within introns of Xenopus homologs of the mammalian UHG gene although currently we cannot be certain whether exons reside between the Xenopus UHG snoRNAs. However, unlike their mammalian counterparts, the Xenopus U25–U31 and U22 snoRNA genes are found in multiple copies (except U30). This is partially due to the multiplication of the presumed Xenopus UHG homolog itself. On the other hand, duplication of the U31 gene within a particular Xenopus UHG locus provides a further example of the multiplication and mobility of intron-encoded snoRNA genes between different introns of their host genes (12).
Depletion of U25 snoRNA from Xenopus oocytes prevented methylation of G1448 in 18S rRNA. This defect can be rescued by injection into U25-depleted oocytes of either the Xenopus or human U25 transcript. Human U25 is 75% identical to its Xenopus counterpart, with nucleotide changes scattered throughout the molecule except in boxes C, D, D′, and the region complementary to 18S rRNA, which are unaltered. This argues that these conserved sequences may be sufficient for both the stability and function of U25. In other systems (33, 34), boxes C and D are required for the stability of snoRNAs. Thus, the 18S rRNA complement and box D′ may be the only elements within U25 directly involved in methylation of G1448. The essentiality of the rRNA complement and box D′ for determining the position of the residue to be 2′-O-methylated has been demonstrated for yeast U24 snoRNA (11).
Although depletion of U25 from the Xenopus oocyte abolishes 2′-O-methylation of G1448 in 18S rRNA, it has little effect on the relative amounts of rRNA-processing products or intermediates that are generated (data not shown). Thus, the 2′-O-methyl group at G1448 is not absolutely required for cleavage of the pre-rRNA. Similarly, genetic depletion of either U24 or U18 snoRNAs, which also direct 2′-O-methylation of selected rRNA nucleotides, does not affect yeast cell growth (ref. 11; M. Fournier, personal communication). Likewise, U18 is not absolutely required for rRNA processing in the Xenopus oocyte (35). In contrast, the lack of the highly conserved 2′-O-methyl group at G2251 in the peptidyl transferase center of yeast mitochondrial 21S rRNA prevents assembly of the large ribosomal subunit (8).
The mechanism of the 2′-O-methylation reaction remains entirely unexplored. Do U25 and other snoRNAs form a scaffold for a proteinaceous methyltransferase, or do they themselves play a role in catalysis? The Xenopus oocyte provides a powerful complement to the yeast system for answering these questions.
Acknowledgments
We thank B. Peculis for Xenopus genomic DNA, the Steitz lab for many stimulating discussions, and D. Crothers, A. Weiner, E. Scharl, and J. Mermoud for critical reading of the manuscript. This work was supported by Grant GM26154 from the National Institutes of Health.
Footnotes
Abbreviations: snoRNA, small nucleolar RNA; UHG, U22 host gene.
Data deposition: The sequences reported in this paper (U62, U63, U25-A, U25-B, U25-C, U26-A, U26-B, U27-A, U27-C, U28-A, U28-C, U29-D, U29-1, U29-2, U29-3, U29-4, U30-D, U31-D, and U31-E snoRNAs) have been deposited in the GenBank data base (accession nos. U72851–U72869U72851U72852U72853U72854U72855U72856U72857U72858U72859U72860U72861U72862U72863U72864U72865U72866U72867U72868U72869).
References
- 1.Hadjiolov A A. In: Cell Biology Monographs. Alfert M, Beermann W, Goldstein L, Porter K R, Sitte P, editors. Vol. 12. Vienna: Springer; 1985. pp. 1–268. [Google Scholar]
- 2.Maden B E H. Prog Nucleic Acids Res Mol Biol. 1990;39:241–303. doi: 10.1016/s0079-6603(08)60629-7. [DOI] [PubMed] [Google Scholar]
- 3.Vaughan M H, Soeiro R, Jr, Warner J R, Darnell J E., Jr Proc Natl Acad Sci USA. 1967;58:1527–1534. doi: 10.1073/pnas.58.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swann P F, Peacock A C, Bunting S. Biochem J. 1975;150:335–344. doi: 10.1042/bj1500335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolf S F, Schlessinger D. Biochemistry. 1977;16:2783–2791. doi: 10.1021/bi00631a031. [DOI] [PubMed] [Google Scholar]
- 6.Tollervey D, Lehtonen H, Jansen R, Kern H, Hurt E C. Cell. 1993;72:443–457. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- 7.Caboche M, Bachellerie J-P. Eur J Biochem. 1977;74:19–29. doi: 10.1111/j.1432-1033.1977.tb11362.x. [DOI] [PubMed] [Google Scholar]
- 8.Sirum-Connoly K, Mason T L. Science. 1993;262:1886–1889. doi: 10.1126/science.8266080. [DOI] [PubMed] [Google Scholar]
- 9.Tollervey D, Lehtonen H, Carmo-Fonseca M, Hurt E C. EMBO J. 1991;10:573–583. doi: 10.1002/j.1460-2075.1991.tb07984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berges T, Petfalski E, Tollervey D, Hurt E C. EMBO J. 1994;13:3136–3148. doi: 10.1002/j.1460-2075.1994.tb06612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiss-Laszlo Z, Henry Y, Bachellerie J-P, Caizergues-Ferrer M, Kiss T. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 12.Maxwell E S, Fournier M J. Annu Rev Biochem. 1995;35:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 13.Ochs R L, Lischwe M A, Spohn W H, Busch H. Biol Cell. 1985;54:123–134. doi: 10.1111/j.1768-322x.1985.tb00387.x. [DOI] [PubMed] [Google Scholar]
- 14.Kass S, Tyc K, Steitz J A, Sollner-Webb B. Cell. 1990;60:897–908. doi: 10.1016/0092-8674(90)90338-f. [DOI] [PubMed] [Google Scholar]
- 15.Hughes J M X, Ares M., Jr EMBO J. 1991;10:4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peculis B A, Steitz J A. Cell. 1993;73:1233–1245. doi: 10.1016/0092-8674(93)90651-6. [DOI] [PubMed] [Google Scholar]
- 17.Liang W-Q, Fournier M J. Genes Dev. 1995;9:2433–2443. doi: 10.1101/gad.9.19.2433. [DOI] [PubMed] [Google Scholar]
- 18.Tycowski K T, Shu M-D, Steitz J A. Science. 1994;266:1558–1561. doi: 10.1126/science.7985025. [DOI] [PubMed] [Google Scholar]
- 19.Bachellerie J-P, Michot B, Nicoloso M, Balakin A, Ni J, Fournier M J. Trends Biochem Sci. 1995;20:261–264. doi: 10.1016/s0968-0004(00)89039-8. [DOI] [PubMed] [Google Scholar]
- 20.Tycowski K T, Shu M-D, Steitz J A. Nature (London) 1996;379:464–466. doi: 10.1038/379464a0. [DOI] [PubMed] [Google Scholar]
- 21.Nicoloso M, Qu L-H, Michot B, Bachellerie J-P. J Mol Biol. 1996;260:178–195. doi: 10.1006/jmbi.1996.0391. [DOI] [PubMed] [Google Scholar]
- 22.Tyc K, Steitz J A. EMBO J. 1989;8:3113–3119. doi: 10.1002/j.1460-2075.1989.tb08463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.England T E, Bruce A G, Uhlenbeck O C. Methods Enzymol. 1980;65:65–74. doi: 10.1016/s0076-6879(80)65011-3. [DOI] [PubMed] [Google Scholar]
- 24.Donis-Keller H, Maxam A M, Gilbert W. Nucleic Acids Res. 1977;4:2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Black D L, Pinto A L. Mol Cell Biol. 1989;9:3350–3359. doi: 10.1128/mcb.9.8.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tarn W-Y, Steitz J A. Cell. 1996;84:801–811. doi: 10.1016/s0092-8674(00)81057-0. [DOI] [PubMed] [Google Scholar]
- 27.Fragapane P, Prislei S, Michienzi A, Caffarelli E, Bozzoni I. EMBO J. 1993;12:2921–2928. doi: 10.1002/j.1460-2075.1993.tb05954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dworniczak B, Mirault M-E. Nucleic Acids Res. 1987;15:5181–5197. doi: 10.1093/nar/15.13.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicoloso M, Caizergues-Ferrer M, Michot B, Azum M C, Bachellerie J-P. Mol Cell Biol. 1994;14:5766–5776. doi: 10.1128/mcb.14.9.5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maden B E H. J Mol Biol. 1986;189:681–699. doi: 10.1016/0022-2836(86)90498-5. [DOI] [PubMed] [Google Scholar]
- 31.Maden B E H. J Mol Biol. 1988;201:289–314. doi: 10.1016/0022-2836(88)90139-8. [DOI] [PubMed] [Google Scholar]
- 32.Tollervey D. Science. 1996;273:1056–1057. doi: 10.1126/science.273.5278.1056. [DOI] [PubMed] [Google Scholar]
- 33.Huang G M, Jarmolowski A, Struck J C R, Fournier M J. Mol Cell Biol. 1992;12:4456–4463. doi: 10.1128/mcb.12.10.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caffarelli E, Fatica A, Prislei S, De Gregorio E, Fragapane P, Bozzoni I. EMBO J. 1996;15:1121–1131. [PMC free article] [PubMed] [Google Scholar]
- 35.Dunbar D A, Ware V C, Baserga S M. RNA. 1996;2:324–333. [PMC free article] [PubMed] [Google Scholar]
- 36.Green R, Noller H F. RNA. 1996;2:1011–1021. [PMC free article] [PubMed] [Google Scholar]


