Abstract
Of considerable interest are the evolutionary and developmental origins of complex, adaptive structures and the mechanisms that stabilize these structures. We consider the relationship between the evolutionary process of gene duplication and deletion and the stability of morphogenetic patterns produced by interacting activators and inhibitors. We compare the relative stability of patterns with a single activator and inhibitor (two-dimensional system) against a ‘redundant’ system with two activators or two inhibitors (three-dimensional system). We find that duplication events can both expand and contract the space of patterns. We study developmental robustness in terms of stochastic escape times from this space, also known as a ‘canalization potential’. We embed the output of pattern formation into an explicit evolutionary model of gene duplication, gene loss and variation in the steepness of the canalization potential. We find that under all constant conditions, the system evolves towards a preference for steep potentials associated with low phenotypic variability and longer lifespans. This preference leads to an overall decrease in the density of redundant genotypes as developmental robustness neutralizes the advantages of genetic robustness.
Keywords: genetics, pattern formation, evolution, evodevo, development
1. Introduction
Complex phenotypes evolve through the genetic modification of developmental programmes (Carroll 2001). Developmental programmes consist of temporally varying patterns of gene expression generating networks comprising protein–protein, protein–nucleic acid and nucleic acid–nucleic acid interactions (Wolpert 2002). Research in the evolution of development (evodevo) seeks to determine the genetic causes of phenotypic variability, identify enhancers or filters of genetic variation and understand how mutation, selection and drift modify developmental programmes (Gerhart & Kirschner 1997). One important mechanism for generating variation across generations is gene duplication, deletion and divergence (Ohta 1987). The preservation of newly duplicated genes is thought to depend initially on some form of redundancy—increasing the robustness of phenotypes—with a subsequent establishment of novel protein function (for a review, see Krakauer & Nowak 1999) or the differential degeneration of a multifunction protein with complementary functions in a paralogue preserving the duplicates (van Hoof 2005). Studies of genetic duplication are not typically related to the dynamics of development.
One general class of mechanism for generating variable patterns of gene expression and cell differentiation during the course of development involves heterogeneous diffusion rates of autocatalytic and inhibitory enzymes (Meinhardt 1982). The best known of these are the Turing-type, diffusion-driven instabilities (Turing 1952). The Turing mechanism proposes a minimal model for generating periodic inhomogeneities in morphogen concentrations along a spatial domain. While the relevance of a strict Turing mechanism for understanding development has been questioned, some elements of the diffusive process are clearly essential for establishing spatial patterning (Meinhardt 1992).
Previous theories seeking to explore the consequences of genetic duplication and redundancy on long-term genetic variability, project genotypes onto viability values and emphasize the pleiotropic or regulatory structures leading to the preservation of duplicated genes (Nowak et al. 1997). In this paper, we consider explicitly a developmental dynamics alongside duplication. One advantage of including an explicit developmental dynamics is to better characterize the range of genetic variation under which phenotypes are expected to remain invariant, and provide an intuition for the long-term stability of phenotypes. This relates to a long-standing question in development about how, over the course of days to years, morphological variation is constrained within a range of adaptive function. Some of the earliest work in this area is due to Waddington who wrote about the process of genetic canalization whereby developmental mechanisms buffer against genotypic and phenotypic noise (Waddington 1957). More recently, there has been a growing interest in the many mechanisms of genetic and phenotypic robustness (De Visser et al. 2003).
Redundancy arising through gene duplication is an important mechanism of robustness by allowing for continued function following perturbations through correlated activity of system constituents, although only a subset of duplicates confer this property (Wagner 2000; Zhang 2003; Ay & Krakauer 2006; Ihmels et al. 2007). Genetic duplication is thought in the short term to produce two, identical functional genes capable of substitution in case one gene should be lost. Genes do not however contribute independently to phenotypes but do so through complex interactions. Phenotypes are the outcome of epistatic protein networks, and hence duplication need not simply double the range of parameters over which a system remains viable. To determine the precise stability benefits of gene duplication, it is necessary to model explicitly or measure experimentally the developmental process.
We consider the following problem: given a diffusively driven pattern-forming system dependent on some number of variables, what changes are introduced following the duplication of one or more variables? We shall refer to the original system as ancestral, consisting of singleton ancestral genes, and the modified system as derived with duplicated genes. The set of duplicate genes are known as paralogues. The parametric space of solutions under duplication, all of which manifest periodic Turing patterns, is known as the Turing space (Murray 1982). In the results that follow, we seek to characterize the volume of Turing space following morphogen duplication, and determine the evolutionary consequences of mutation for residence time in the Turing space. We consider evolutionary dynamics via a quasi-species formalism which includes stationary developmental probabilities derived from a homeostatic developmental model operating at a faster time scale. We do not consider the equally important implications of variation in the spatial scale and system geometry on stability (Crampin et al. 1999).
2. The model: turing instability with noise
The developmental model we adopt for analysis is the reaction diffusion equation system first proposed by Turing (1952). In this system, we consider the stability of fixed points and the persistence of pattern formation. Start with a minimal Turing two-dimensional field system with mutational noise terms that exhibit pattern formation,
| (2.1) |
| (2.2) |
where u and v are the concentration of activator and inhibitor proteins; d is the diffusion coefficient; and f(·) and g(·) are the general nonlinear functions. These equations are subject to the boundary conditions
| (2.3) |
where ∂B is the closed boundary domain B and n is the unit outward normal vector to ∂B.
Four conditions are necessary for stability and patterning: two conditions for the stability of the fixed points and two for spatial instability, i.e. instability of some k wavevector in the Fourier representation. These conditions are (see appendix A for details):
- stability of the stationary state
(2.4) (2.5) - instability of the homogeneous state for some k>0
(2.6) (2.7)
Here fi and gi (i=u, v) are the first derivatives of the functions f and g evaluated at the fixed point. The size of the spatial domain wherein the reactions take place is assumed to be large enough to support the wavelength of the unstable mode. These are very familiar inequalities in the patterning literature (Nicolis 1995). It is important to be aware that while patterning is guaranteed by the above inequalities, the shape (frequency and amplitude) of the patterns can be different within this space, and will be related to the diffusion parameter and the saturation processes determined by the choice of kinetics.
In order to analyse the robustness of the two-field system including mutational fluctuations, we introduce noise assuming that it acts upon the dynamical terms of the inequalities (2.4)–(2.7). The assumptions behind this type of noise are less restrictive than additive noise—it captures not only external fluctuations but also internal fluctuations engendering structural dynamical changes, via f(·) and g(·), in the vicinity of the stable state.
From the preceding inequalities, we observe that there will be a domain, Ω, where fu, fv, gu, and gv can fluctuate preserving the inequalities given by (2.4)–(2.7). For example, if we fix fu and fv, then the derivatives gv and gu can fluctuate in the plane (gv, gu), within the interior of the region defined by the three points p1, p2, and p3, given by
| (2.8) |
| (2.9) |
| (2.10) |
where
| (2.11) |
The two-dimensional region of stability is given by a triangle of increasing width with increasing values of fu, the rate of autocatalytic activity and decreasing fv, the inhibitor production.
3. System with a replica field
3.1 Stability in the homogeneous state
To capture the impact of genetic duplication of a morphogen, we replicate one component of the system. The generalized stability matrix for the expanded system of equations becomes
| (3.1) |
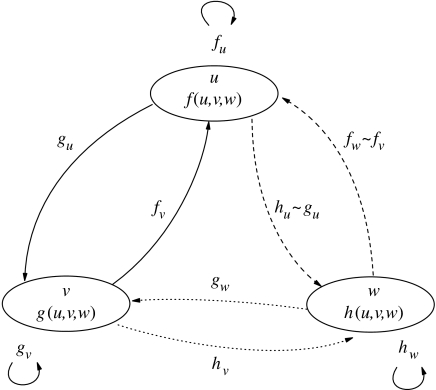
This introduces the new interactions: u–w and v–w. These are represented in figure 1, where the activator and inhibitor are nodes in the graph and kinetic interactions are described by edges. The solid edges represent the ancestral interactions, whereas the dotted lines arise through duplication and are derived.
Figure 1.
Schematic illustration of the interactions among morphogens in a three-component system. Solid lines, interactions in the ancestral system; dashed lines, derived interactions with a duplicated inhibitor; dotted lines, interactions between the two inhibitors. Autocatalytic interactions, both positive and negative, are represented with curved arrows. The instance of a duplicated activator is not shown. Variables (partial derivatives) are defined in §§2 and 3.
The conditions for stability of the fixed point (not the patterned state) are
| (3.2) |
| (3.3) |
| (3.4) |
From these inequalities, we observe that the following characteristics are added by the duplicated gene. Condition (3.2) acts as a constraint on hw. Under conditions leading to the inequality (2.4) not being satisfied, the parameter hw can by assuming negative values render condition (3.2) fulfilled. Kinetically this implies a greater constraint on w near the fixed point. In general, only non-autocatalytic reactions involving the replica component in the three-field system will be more robust in the homogeneous state. We also observe that the first two terms of equations (3.2) and (3.3) are identical to inequalities (2.4) and (2.5). The additional terms arising from the new interaction pertain to u–w and v–w. The final inequality describes the mixed interaction.
3.2 Spatial instability in the extended system
Previously we have analysed the stability conditions for the matrix S, at the homogeneous steady state. When spatial instability is taken into account, the stability matrix includes perturbation terms,
| (3.5) |
where k is the wavevector associated with the Fourier decomposition of the fields u, v, and w, and D is the diagonal matrix characterized by its diagonal values: D11=1, D22=d2 and D33=d3.
To ensure instability and hence patterning for some k>0, we require that the real part of the dominant eigenvalue calculated for the expanded stability matrix is positive. This condition is met when a number of inequalities derived from the characteristic polynomial are satisfied. The inequalities make use of the following functions:
| (3.6) |
or
| (3.7) |
with
and
We are interested in the critical instability threshold and find this by looking for one real part positive eigenvalue near zero. The relevant quantity in this case will be the point at which F1(k) becomes positive. In order to achieve spatial instability, the function F1 should present a maximum greater than zero for some value of k.
We require that at least one of the two following inequalities is true:
| (3.8) |
| (3.9) |
and the following two inequalities are true:
| (3.10) |
with km such that
| (3.11) |
These are general results that apply to any three-dimensional system. In §§3.3.1 and 3.3.2, we shall substitute into these expressions results pertaining to particular morphogens.
3.3 Specific replicated systems
Here we consider the impact of duplication of either the inhibitor or the activator. We consider these independently, as they contribute to the kinetics differently and can thereby cause both an expansion and a contraction of the space of stable solutions and stable patterning.
3.3.1 Inhibitor
We consider an inhibitor replicated without error and introduce the new variable w, with potential interactions: w–w, u–w. The ancestral interactions remain unchanged u–v. We set the derivatives near the stationary state to
| (3.12) |
| (3.13) |
| (3.14) |
The inequalities for the stability of the fixed point will be given by (primes indicate the duplicated components)
| (3.15) |
| (3.16) |
| (3.17) |
For the fixed point, since the inhibitor must have a positive rate of decay, the duplicated inhibitor will also have a negative derivative . This increases the stability values of the first inequality in relation to the ancestral state and allows for greater fluctuation in gv and fu. For the second and third inequalities, we can demonstrate this effect more clearly by collecting some of the terms of the inequalities into the variables A and B. The inequalities have the form with A>0, and with B1∼B2 and Bi>0 (i=1, …, 4), under no change of sign of the ancestral variables. Once again the duplication of the inhibitor has the effect of expanding the range of values that the ancestral variables can experience without loss of the fixed point, as this increases likelihood that the two-dimensional inequality is fulfilled.
To determine whether we observe patterns, we need to look at the inequalities defined by the functions F1, with i=0, 2, 4, 6. The region of spatial instability (patterns formation) associated with duplication of the inhibitor is reduced as a result of the negative sign of gv acting on F1,4. If the diffusion constants are large, and the duplicated components preserve their ancestral diffusion coefficients (i.e. d=d2=d3), then from the requirement that F1,4>0 we find that for sufficiently large values of d we require that .
3.3.2 Activator
In this section, we perform the same analysis as above, but instead consider duplication of the activator. The duplicate element gives the following first derivatives at the stationary points:
| (3.18) |
| (3.19) |
| (3.20) |
This gives rise to the following inequalities for the stationary solution:
| (3.21) |
| (3.22) |
| (3.23) |
The activator duplication creates an additional constraint, , which when not satisfied eliminates the stability of the homogeneous state. The second inequality, including the aggregated variables A and B, as in the previous case describing the inhibitor, has the form with A1>0 and , as we assume approximately perfect duplication. This implies that increases the domain of stability for the fixed point. The structure of the third inequality is given by , with B1∼B2 and B1, B2, B3>0 under no change of signs of the variables. These are the constraints acting on (fw, hu) which increase the range of permissible fluctuations preserving the fixed points of the system. For the patterning solution, and for large diffusion constants, the spatial instability increases as a result of the positive sign of fu acting on F1,4.
3.3.3 Summary of duplication results
For the two specific cases described above, analysing the effects of a redundant inhibitor and a redundant activator, we see that the functional consequence in terms of stability of either the fixed point or the patterning solution is far from redundant. Redundant inhibitors decrease the Turing space but increase the range of values promoting stability for the fixed point. Redundant activators increase the Turing space but decrease the range of values promoting stability for the fixed point. A stable fixed point is a necessary precondition for patterning, and so it is not trivial to determine which of these two duplication events will make the greater contribution to robust patterns. This will depend on the nature of the fluctuations, in particular the variables that are most influenced by the noise. We also find that duplication can preserve a stable solution assuming that there is some degeneration of the ancestral morphogen and some complementation by the derived paralogue. Hence, duplication can promote stability by either expanding a stable basin of attraction or by preserving a basin when the kinetics of duplicate morphogens are perturbed in opposing or ‘complementary’ directions.
4. Developmental escape time from Turing space
As we have observed, duplication can lead to an increase in the concentration or variety of morphogens. Formally this adds dimensions in the phase space. This expansion introduces the possibility of a large increase in the corresponding parameter space (as every edge in the reaction graph is associated with a kinetic constant) and to changes in the volume of stable patterning. We have not explained however what drives the values of the elements of the stability matrix into the patterning region. Typically this will be explained in terms of a mutation selection process, whereby natural selection filters out variants with properties placing them outside of an adaptive, patterned phenotype. It would be more accurate however to consider evolution as having established robustness mechanisms that allow adaptive states to be restored during development when they are locally perturbed.
In this section, we map the dynamics of ontogenetic errors acting on morphogen kinetics onto a stochastic process describing developmental canalization. Previously we considered these errors in terms of general linear perturbations of the elements of the stability matrix as a means of characterizing regions of patterning in the phase plane. Here we model the fluctuation in the mean value of the dynamical parameters in S using Gaussian white noise ξ(t). We consider a homeostatic dynamic such that any deviation in the stationary values of the stability matrix tends to be driven back to their set points. The strength of this homeostatic process is modulated by the value of an evolutionary parameter γ. This parameter is a coarse-grained variable capturing a suite of mechanisms all able to dampen intrinsic sources of noise (the evolution of γ is treated in §5).
We introduce an equation that describes each of the fluctuating variables in the duplicated system,
| (4.1) |
where xi stands in for any of the derivatives in the matrix S. Thus, what is fluctuating randomly are the dependencies of the morphogens on each other.
Such a functional form can be interpreted in a straightforward way as describing an escape processes from a quadratic, attractive single-well potential. The steepness of the potential is governed by the value of γ, which captures evolved properties of the enzyme kinetics capable of dampening fluctuations. This can be thought of as the strength of homeostasis. The stable/patterned phenotypes lie within the space shown in figure 2, whereas the non-stable/patterned phenotypes lie outside the space.
Figure 2.
Canalization potential illustrating the regions of attraction for patterned phenotypes within which the kinetics of the morphogens can fluctuate without losing adaptive pattern. The coordinate x measures the value of the components of the matrix S. The coordinate measures the energetic distance from the optimal configuration. ΔΩ measures the increase in the width of the potential contributed by the duplicate. Γ is the boundary of the potential defined by the ancestral system.
To describe the effect of noise, the dynamically fluctuating functions can be associated with a one-dimensional random walk, related to a coordinate x, under an attractive potential U(x). The equation of motion for the probability p(x, t) of this particle, equation (4.1), is described using the Fokker–Planck equation,
| (4.2) |
where p(x, t) is the probability of finding the particle at position x at time t; is the first derivative of the potential; and D is the diffusion parameter. The position of the Brownian particle in the selective potential determines the value of the dynamical linear term (near the fixed point) of the system plus the replica field.
Here we are assuming D is constant (a constant rate of developmental noise), whereas the diffusion term could depend on the state of the system, i.e. D=D(x).
We consider the neutral region Ω of parameters in the Turing space where the system can diffuse without loss of stability/patterning. Increases in the volume of Turing space correspond as a first approximation to stretching the borders of the canalization potential leaving the topological structure within the basin as it was in the ancestral (low-dimensional) state. We can expand the potential's border, Γ, as
| (4.3) |
For any of the chosen variables susceptible to fluctuations, we can calculate the escape time, τ, or time taken to randomly walk out of the potential into a region without pattern, as the Kramer time,
| (4.4) |
where ΔU2D (ΔU3D) is the potential barrier for the two (three)-element system. We find that an increment ΔΩ in the volume of the parameter space leads to an exponential increase in the escape time (time for the system to lose stability and k-instability properties) from the canalization potential. Thus, gene duplication events expanding the Turing space (such as duplication of an activator) can have an exponential impact on the robustness of phenotypes to fluctuations in say the binding or catalytic properties of the morphogens. It should be stressed that we are defining robustness in binary terms—pattern or no pattern, and it could be the case that variation in the pattern is important. If this were the case, then the adaptive value of the pattern could be lost before the system escaped from the canalization potential.
5. Evolutionary dynamics of turing space
In §§3 and 4, we have analysed the impact of morphogen duplications on the volume of stable, patterning steady states. We subsequently determined how long, assuming stochasticity in morphogen behaviour, it takes for patterns to be lost when these phenotypes are canalized. Here we couple the fast time scales of development and canalization to the slower time scale of evolutionary change.
We consider explicitly the dynamics of duplication and deletion of morphogen genes and continuous mutational variation in the value of γ within an adaptive dynamics framework. We ask what happens to the value of γ when we allow it to evolve on a genetic background of non-redundant and redundant genotypes.
For simplicity, we consider two genome configurations according to the number of copies of morphogens that they encode—two or three—an ancestral state, X2D and a derived, duplicate state, X3D. Both states are associated with a value of the canalization parameter γ.
The haploid evolutionary dynamics of the morphogens are described by a coupled system,
| (5.1) |
| (5.2) |
where r modulates the relative rate of growth; μ1 determines the rate of duplication, and μ2 deletions; and ϕ=F(Ω) is a parameter capturing the volume of the Turing space. The final bracketed terms are the flux terms that contribute density-dependent regulation to the dynamics. The parameter ϕ is essentially a measure of the reciprocal of the volume of the space of patterning derived from the single-well potential in §4. The parameter γ tunes the steepness of the canalization potential. From §§3 and 4, we consider those cases of duplication where the duplicated morphogen expands the Turing space, such that ϕ<1. This model has three separated time scales: the fastest time scale is given by the Turing system; the next fastest is the duplication and deletion dynamics; and the slowest time scale describes the evolution of γ.
This system has three steady states, only two of which are of interest (have non-negative densities), one of which is the trivial steady state at zero. We denote the positive steady as and and include the variable γ in the argument holding all other parameters constant.
In order to determine whether the steady state value of γ is stable, we ask whether a rare mutant with a value of γ′=γ+ϵ can invade into the steady-state population. To do so, we consider the linearized system consisting of the wild-type system at steady state (A(γ)) and a rare γ′ mutant. We now track the densities of the mutant genotypes, and :
| (5.3) |
| (5.4) |
where . This system has a single equilibrium at zero. For invasion, we consider the sign of the dominant eigenvalue λ1 of the stability matrix (J) at the zero equilibrium of the mutant
| (5.5) |
We find that . This states that any mutant that experiences an increase in the value of γ relative to that of the wild-type will increase in frequency when rare. Intuitively, an increase in the value of γ leads to an increase in the expected lifetime of the genotype. This result has been confirmed for a larger system of equations where we allow for N discrete values of γi and track the densities of the 2N, and variables. This allows to confirm fixation of the mutant and not only invasion.
Whereas the value of γ is strictly increasing under adaptive dynamics, the consequences on the abundance of the two genotypes can be fairly complex. A few results are shown in figure 3. At low values of γ>0 when developmental noise is large, the redundant genomes (three-dimensional) have the advantage in terms of increased expected lifetimes. As the value of γ increases, the relative difference in the death rates decrease, and the slight advantage that the two-dimensional system derives from carrying fewer genes (r>1) is enough to allow it to outcompete the three-dimensional system. When the rate of duplication is greater than the rate of deletion, the redundant, three-dimensional, genome is able to maintain an advantage over a greater range of values of γ. Clearly the upper bound on the value of γ is going to determine whether the redundant or non-redundant genome dominates in the population and any extrinsic increase in noise, effectively decreasing the value of γ, is going to favour redundancy.
Figure 3.
Steady-state densities of non-redundant (two-dimensional) and redundant (three-dimensional) genomes as a function of the canalization parameter γ. Parameter values: (a) r=1.5, ϕ=10−1, μ1=5×10−2, μ2=5×10−3 and (b) r=1.5, ϕ=10−1, μ1=5×10−3, μ2=5×10−2.
6. Discussion
Genotypes have evolved many mechanisms for increasing the robustness of phenotypes, from simple duplications of genes to more elaborate mechanisms of repair and error buffering. In this paper, we have shown that simple duplication cannot guarantee increased robustness, sometimes it reduces robustness. This is a consequence of the dynamics of regulation. Furthermore, when there are several mechanisms of robustness evolving, we can observe that one mechanism dominates, and the additional, redundant mechanisms can be neutralized. We have explored these ideas with an idealized model of development, the Turing model, and an idealized model of evolution, replicator dynamics.
We consider a space of stable, developmental pattern, the Turing space. This space is defined as that volume in parameter space of a dynamical system capable of generating diffusion-driven inhomogeneities. We have explored the consequences on the volume of Turing space of duplicating morphogens in the simple case of one duplicated morphogen. We have also analysed a simple homeostatic model including parameter fluctuations in order to calculate escape times from Turing space over the course of development. We use the escape time from the developmental model to parameterize the death rate in an evolutionary model describing the adaptive dynamics of canalization in the presence of morphogen duplication and deletion. We arrive at the following conclusions.
Duplication of a morphogen can lead to either a contraction of the Turing space—increasing the likelihood that random fluctuations impinging on morphogens lead to a loss of patterning—or an increase in the space. Expansion is associated with duplication of activators and contraction with duplication of inhibitors.
While activators and inhibitors have positive and negative influences on the stability of pattern, they contribute inversely to the stability of the non-spatial, steady state. Activators reduce the domain of stability and inhibitors increase the domain of stability.
An additional benefit of duplicates arises when the ancestral morphogen is mutated and the duplicate can maintain the pattern through compensation. Under these conditions, the degeneration of a morphogen function can be complemented by a suitable modification of its derived paralogue.
In the duplicated activator system, the space of unstable wavevectors increase. This is due to the fact that the degree of the polynomial function associated with the linear stability analysis is smaller in the ancestral system. This will have implications for the diversity of patterned states, with a potentially greater number of patterns in the duplicated system.
Assuming that patterned phenotypes are canalized, a linear increase in the volume of Turing space corresponds to an exponential increase in the residence time in adaptive patterning in the presence of developmental noise.
When the degree of canalization is free to evolve, this evolves to its maximum value, corresponding to a large reduction in the variance of phenotypes and a significant increase in the expected lifetime.
The increase in developmental canalization leads to a reduction in the advantages accrued through genetic redundancy, causing non-redundant genotypes with slight growth advantage to outcompete redundant genotypes.
It is often assumed that gene duplication is an important mechanism of robustness by promoting functional redundancy. This assumption rests on a simple model of gene interaction in which two identical genes are indistinguishable from one gene. In reality, genes interact to promote phenotypic patterns and interactions are unlikely to be linear. We have shown through a simple developmental process that duplication can in fact reduce the robustness of a system to mutations, by contracting the space of viable patterning. In order for duplication to substantially increase the space of viable patterning (the Turing space), the derived duplicate should be an activator and preferentially one with an increased rate of decay than the ancestral form. This result is somewhat counter-intuitive as this would seem to correspond to a reduction in protein function.
Force et al. (1999) have argued that an imperfect duplication process is essential for preserving duplicates and is described as a model of complementary, degenerative mutation (CDM). Degenerative mutations in regulatory sub-functions promote stability of duplicates by functionally partitioning the expression of alleles. In this way the partitions can come under independent normalizing selection. This hypothesis has received experimental support (van Hoof 2005). While this is a related hypothesis to ours, it does not provide an explanation for the expanded space of stability in our models. In our results duplicates can be beneficial for two reasons: (i) owing to the reduced sensitivity to developmental noise and (ii) complementation of a function by derived duplicates in response to a degenerate ancestral function. Unlike the CDM model, we do not require that the proteins are multifunctional.
In addition to increasing the robustness of patterning, some duplicates allow patterns to be established more readily. In other words, duplicate systems make the patterned phenotype more accessible to small fluctuations originating in the non-patterned state. Thus, duplicates, under the right conditions, can both increase the efficiency of evolutionary search and increase the robustness of the patterned phenotype. This runs counter to the intuition that increased accessibility should reduce stability, as this might seem to imply easier loss.
Genotypes have evolved many mechanisms for increasing the robustness of phenotypes, from simple duplications of genes to more elaborate mechanisms of repair and error buffering. We observe that several of these mechanisms interact negatively, such that the evolution of effective developmental canalization is able to render simple genetic redundancy, redundant.
In summary, without an empirical or theoretical model of development, it is not possible to determine the influence of genetic duplication on the robustness of phenotypes. As a result of nonlinearity and more sophisticated mechanisms of repair, simple duplication can reduce robustness as well as increase both the accessibility and the robustness of phenotypic patterns. While the Turing model is highly idealized, it provides us with one formal means of integrating developmental and evolutionary dynamics and makes testable predictions about the incidence of duplicated patterning genes. In particular, where we observe patterning genes we expect that paralogues should typically be activators.
Appendix A.
Homogeneous and spatial instability can be analysed using a standard linear stability analysis. Given the set of equations (2.1) and (2.2), the homogeneous stability can be determined by considering
| (A1) |
where (u0, v0) is the stationary state. Then, near (u0, v0) equations (2.1) and (2.2) become
| (A2) |
where S is the stability matrix defined in the text. The solution will be of the form , where λ is the eigenvalue. Substituting this into equation (A 2) the values of λ will be given by
| (A3) |
where I is the identity matrix. Inequalities (2.4) and (2.5) arise from the previous expression and ensure the stability of the homogeneous state, i.e. .
In the case of spatial instability, near the steady state, we find for equations (2.1) and (2.2)
| (A4) |
The solution of this equation will have the form
| (A5) |
where is the eigenfunction corresponding to the wavenumber k and is solution of the time-independent equation,
| (A6) |
subject to appropriate boundary conditions. As with the case of the homogeneous solution, substituting the function (A 5) into (A 4) determines the stability of a given mode by reference to the sign of the root of the characteristic polynomial
| (A7) |
Equations (2.5) and (2.6) are derived from the last equality and ensure instability of a given mode k, with k>0.
References
- Ay N, Krakauer D.C. Geometric robustness theory and biological networks. Theory Biosci. 2006;125:93–121. doi: 10.1016/j.thbio.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Carroll S.B. Chance and necessity: the evolution of morphological complexity and diversity. Nature. 2001;409:1102–1109. doi: 10.1038/35059227. [DOI] [PubMed] [Google Scholar]
- Crampin E.I, Gaffney E.A, Maini P.K. Reaction and diffusion on growing domains: scenarios for robust pattern formation. Bull. Math. Biol. 1999;61:1093–1120. doi: 10.1006/bulm.1999.0131. [DOI] [PubMed] [Google Scholar]
- De Visser J.A, et al. Perspective: evolution and detection of genetic robustness. Evolution. 2003;57:1959–1972. doi: 10.1111/j.0014-3820.2003.tb00377.x. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett F.B, Amores A, Yan Y, Pstelthwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart J, Kirschner M. Blackwell; Oxford, UK: 1997. Cells, embryos and evolution: toward a cellular and developmental understanding of phenotypic variation and evolutionary adaptation. [Google Scholar]
- Ihmels J, Collins S.R, Schuldiner M, Krogan N.J, Weissman J.S. Backup without redundancy: genetic interactions reveal the cost of duplicated genes. Mol. Syst. Biol. 2007;3:1–11. doi: 10.1038/msb4100127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakauer D.C, Nowak M.A. Evolutionary preservation of redundant duplicated genes. Semin. Cell Dev. Biol. 1999;10:555–559. doi: 10.1006/scdb.1999.0337. [DOI] [PubMed] [Google Scholar]
- Meinhardt H. Academic Press; London, UK: 1982. Models of biological pattern formation. [Google Scholar]
- Meinhardt H. Pattern formation in biology: a comparison of models and experiments. Rep. Prog. Phys. 1992;55:797–849. doi: 10.1088/0034-4885/55/6/003. [DOI] [Google Scholar]
- Murray J.D. Parameter space for turing instability in reaction diffusion mechanisms: a comparison of models. J. Theor. Biol. 1982;98:143–163. doi: 10.1016/0022-5193(82)90063-7. [DOI] [PubMed] [Google Scholar]
- Nicolis G. Cambridge University Press; Cambridge, UK: 1995. Introduction to nonlinear science. [Google Scholar]
- Nowak M.A, Boerlijst M.C, Cooke J, Maynard Smith J. Evolution of genetic redundancy. Nature. 1997;338:167–171. doi: 10.1038/40618. [DOI] [PubMed] [Google Scholar]
- Ohta T. Simulating evolution by gene duplication. Genetics. 1987;115:207–213. doi: 10.1093/genetics/115.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turing A.M. The chemical basis of morphogenesis. Phil. Trans. R. Soc. B. 1952;237:37–72. doi: 10.1098/rstb.1952.0012. [DOI] [Google Scholar]
- van Hoof A. Conserved functions of yeast genes support the duplication, degeneration and complementation model for gene duplication. Genetics. 2005;171:1455–1461. doi: 10.1534/genetics.105.044057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington C.H. MacMillan; New York, NY: 1957. The strategy of the genes. [Google Scholar]
- Wagner A. Robustness against mutations in genetic networks of yeast. Nat. Genet. 2000;24:355–361. doi: 10.1038/74174. [DOI] [PubMed] [Google Scholar]
- Wolpert L. Oxford University Press; New York, NY: 2002. Principles of development. [Google Scholar]
- Zhang J. Evolution by gene duplication: an update. Trends Ecol. Evol. 2003;18:292–298. doi: 10.1016/S0169-5347(03)00033-8. [DOI] [Google Scholar]