Abstract
Inferring the relationships among Bilateria has been an active and controversial research area since Haeckel. The lack of a sufficient number of phylogenetically reliable characters was the main limitation of traditional phylogenies based on morphology. With the advent of molecular data, this problem has been replaced by another one, statistical inconsistency, which stems from an erroneous interpretation of convergences induced by multiple changes. The analysis of alignments rich in both genes and species, combined with a probabilistic method (maximum likelihood or Bayesian) using sophisticated models of sequence evolution, should alleviate these two major limitations. We applied this approach to a dataset of 94 genes and 79 species using CAT, a previously developed model accounting for site-specific amino acid replacement patterns. The resulting tree is in good agreement with current knowledge: the monophyly of most major groups (e.g. Chordata, Arthropoda, Lophotrochozoa, Ecdysozoa, Protostomia) was recovered with high support. Two results are surprising and are discussed in an evo–devo framework: the sister-group relationship of Platyhelminthes and Annelida to the exclusion of Mollusca, contradicting the Neotrochozoa hypothesis, and, with a lower statistical support, the paraphyly of Deuterostomia. These results, in particular the status of deuterostomes, need further confirmation, both through increased taxonomic sampling, and future improvements of probabilistic models.
Keywords: phylogenomics, deuterostomes, systematic error, taxon sampling
1. Introduction
(a) The limits of morphology
The inference of the animal phylogeny from morphological data has always been a difficult issue. Although a rapid consensus was obtained on the definition of phyla (with a few exceptions; e.g. vestimentiferans, pogonophores and platyhelminths), the relationships among them have long remained unsolved (Brusca & Brusca 1990; Nielsen 2001). The dominant view, albeit far from being universally accepted, was traditionally biased in favour of the Scala Naturae concept of Aristotle, which postulates an evolution from simple to more complex organisms (Adoutte et al. 1999). Briefly, acoelomates (platyhelminths and nemertines) were considered as emerging first, followed by pseudocoelomates (nematodes) and then coelomates, representing the ‘crown-group’ of Bilateria. A similar gradist view was proposed for deuterostomes, with the successive emergence of Chaetognatha, Echinodermata, Hemichordata, Urochordata and Cephalochordata culminating in Vertebrata (e.g. Conway Morris 1993).
Irrespective of its underlying ‘ideological’ preconceptions, however, this traditional bilaterian phylogeny was based on very few morphological and developmental characters (position of the nerve cord, cleavage patterns, modes of gastrulation, etc.) whose phylogenetic reliability may sometimes be disputable (owing to either the description or the coding and analysis; see Jenner 2001). This overall lack of homologous characters is essentially related to the wide disparity observed between body plans. For some phyla, such as echinoderms, the body plan is nearly exclusively characterized by idiosyncrasies, leaving few characters to compare with other bilaterian phyla. Traditional animal phylogenies based on morphological data were thus hampered by an insufficient amount of reliable primary signal.
(b) The difficult beginning of molecular phylogeny
Great hopes were placed in the use of molecular data for phylogeny reconstruction (Zuckerkandl & Pauling 1965). Unfortunately, the first phylogenies based on ribosomal RNA (rRNA) turned out to be quite controversial (Field et al. 1988). They contained some difficulty to accept results, such as the polyphyly of animals. We will not review in detail this turbulent early history, but rather note that these trees were based on scarce taxon sampling and inferred using overly simple methods (e.g. Jukes and Cantor distance). Consequently, tree-building artefacts were not infrequent. The problem was mainly addressed through improved taxon sampling; over a period of approximately 10 years, rRNAs were sequenced from several hundreds of species. In part owing to the improvement not only due to denser taxonomic sampling, but also due to a systematic selection of the slowest evolving representatives of the majority of animal phyla, a consensus rapidly emerged, reducing the diversity of Bilateria into three main clades: Deuterostomia, Lophotrochozoa (Halanych et al. 1995) and Ecdysozoa (Aguinaldo et al. 1997). The statistical support for most of the nodes was nevertheless non-significant (Philippe et al. 1994; Abouheif et al. 1998), thus preventing any firm conclusions.
This brief historical overview provides a clear illustration of phylogenetic inference problems. The resolution of the morphological and rRNA trees is limited because too few substitutions occurred during the evolution of this set of conserved characters, yielding too few synapomorphies. At the same time, unequal rates of evolution across characters imply that some characters accumulate numerous multiple substitutions (convergences and reversions). These multiple substitutions can be misinterpreted by tree reconstruction methods and lead to incorrect results. In particular, the well-known long-branch attraction (LBA) artefact (Felsenstein 1978) implies an erroneous branching of fast-evolving taxa, often resulting in an apparent earlier emergence (Philippe & Laurent 1998). For instance, this is the reason for the initial inability to recover the monophyly of the animals, with some Bilateria evolving too fast and being attracted towards the out-group. Similarly, the recognition of the LBA problem played a major role in establishing the Ecdysozoa hypothesis (Aguinaldo et al. 1997); that is, when the fast-evolving Caenorhabditis is considered, nematodes emerge at the base of Bilateria, but when the slowly evolving Trichinella is included, nematodes cluster with arthropods. Compositional heterogeneity can also generate artefacts, especially for trees based on mitochondrial sequences (Foster & Hickey 1999).
Twenty years ago, there were great expectations for the use of genomic data. The underlying assumption was that the joint analysis of numerous genes potentially provides numerous synapomorphies, thus eliminating the problem of stochastic errors. Yet, although it is true that stochastic errors will naturally vanish in a phylogenomic context, systematic errors, which are due to the inconsistency of tree-building methods, will not disappear. Indeed, they should become even more apparent (Philippe et al. 2005a).
We recognized the following two main avenues to circumvent systematic errors (Philippe & Laurent 1998): (i) the use of rare, and putatively slowly evolving, complex characters, such as gene order (Boore 2006), which should be homoplasy-free and (ii) the use of numerous genes combined with inference methods that deal efficiently with multiple substitutions, which should avoid artefacts due to homoplasy. We will briefly review the application of the second approach to the question of the monophyly of Ecdysozoa as a way of demonstrating the importance of using numerous species and models that handle the heterogeneity of the evolutionary process across positions.
(c) Illustration of the misleading effect of multiple substitutions in the case of Ecdysozoa
The first phylogenies based on numerous genes (up to 500) significantly rejected the monophyly of Ecdysozoa (e.g. Blair et al. 2002; Dopazo et al. 2004; Wolf et al. 2004). To exclude the possibility that this was due to an LBA artefact, the use of putatively rarely changing amino acids was proposed (Rogozin et al. 2007), an approach that also supported Coelomata (i.e. arthropods sister group of vertebrates). At first, phylogenomics seems to strongly reject the new animal phylogeny, which was mainly based on rRNA.
However, these phylogenomic analyses were characterized by very sparse taxon sampling, and used only simple tree reconstruction methods, rendering them potentially sensitive to systematic errors. We will show that, as in the first rRNA phylogenies, the monophyly of Coelomata was an artefact due to the attraction of the fast-evolving Caenorhabditis by the distant out-group (e.g. fungi). As detailed below, three different and independent approaches that reduce the misleading effect of multiple substitutions lead to change in the topology from Coelomata to Ecdysozoa.
(d) Removal of the fast-evolving positions
An obvious way to reduce systematic errors is to remove the fastest evolving characters from the alignment (Olsen 1987). In principle, the phylogeny has to be known to compute the evolutionary rate, rendering simplistic circular approaches potentially hazardous (Rodriguez-Ezpeleta et al. 2007). The slow–fast (SF) method (Brinkmann & Philippe 1999) partially circumvents this issue by computing rates within predefined monophyletic groups. Only the relationships among these groups can be studied and an equilibrated species sample should be available for each predefined group. When the SF method is applied to a large alignment of 146 genes with four representatives each from Fungi, Arthropoda, Nematoda and Deuterostomia (Delsuc et al. 2005), the removal of fast-evolving sites leads to an almost total disappearance of the support in favour of Coelomata. Interestingly, this does not correspond to a loss of phylogenetic signal, since support in favour of Ecdysozoa regularly increases (up to a bootstrap value of 91%). The simplest interpretation of this experiment is that the misleading effect of multiple substitutions creates an LBA artefact that disappears when fast-evolving positions are discarded. Note that this way of selecting slowly evolving characters (SF method) differs from the one of Rogozin et al. (2007) by the use of rich taxon sampling that allows positions to be selected that more likely reflect the ancestral state of the predefined monophyletic group, therefore reducing the risk of convergence along the long terminal branch.
(e) Improvement of taxon sampling
Another obvious way of reducing the misleading effect of multiple substitutions is to incorporate more species, breaking long branches (Hendy & Penny 1989) and thus allowing one to detect convergences and reversions more easily. In the case of Bilateria, simply adding a close out-group (Cnidaria) to an alignment containing only a distant out-group (Ascomycetes) is sufficient to change strong support for Coelomata to strong support for Ecdysozoa. This is true for the analysis of both complete primary sequences (Delsuc et al. 2005) and rare amino acid changes (Irimia et al. 2007). Undetected convergences between the fast-evolving nematodes and the distant out-group therefore create a strong but erroneous signal that biases tree-building methods. Accordingly, all phylogenomic studies that have used dense taxon sampling did not find any support in favour of Coelomata (e.g. Philippe et al. 2005b; Marletaz et al. 2006; Matus et al. 2006).
(f) Improvement of tree-building method
Probabilistic methods are now widely recognized as the most accurate methods for phylogenetic reconstruction (Felsenstein 2004). However, to avoid the problem of systematic errors, they require good models of sequence evolution. We recently developed a new model, named CAT, which partitions sites into categories, so as to take into account site-specific amino acid preferences (Lartillot & Philippe 2004). When applied to the difficult case of the bilaterian tree rooted by a distant out-group (Fungi), the CAT model provides strong support for Ecdysozoa, whereas the WAG model (Whelan & Goldman 2001) strongly favours Coelomata (Lartillot et al. 2007). Posterior predictive analyses demonstrate that the CAT model predicts homoplasies more accurately than the WAG model. In other words, the CAT model detects multiple substitutions more efficiently and is therefore less sensitive to systematic errors.
The greater robustness of CAT against the Coelomata artefact is related to the fact that it better accounts for site-specific restrictions of the amino acid alphabet. Amino acid replacements in most proteins tend to be biochemically very conservative, with typical variable positions in a protein accepting substitutions among only two or three amino acids. This has important consequences for phylogenetic reconstruction using amino acid sequences, since it implies that convergences and reversions (homoplasies) are much more frequent than would be expected if all amino acids were considered equally acceptable at any given position. In practice, the typical number of amino acids observed per position is indeed overestimated by classical site-homogeneous models, based on empirical matrices such as WAG, which in turn result in a poor anticipation of the risk of homoplasy, and thereby in a greater prevalence of artefacts. In contrast, site-specific models such as CAT better anticipate these problems, and will be less prone to systematic errors (Lartillot et al. 2007).
Of course, there are many other potential causes of error, all of which can in principle be traced back to model misspecification problems: in all cases, it is a matter of correctly modelling various features of the substitution process that may potentially lead to an increase of the level of homoplasy (e.g. compositional biases). Improving the models of sequence evolution is thus an essential requirement for phylogenetics and is currently a very active area of research. In principle, it should be preferred to the two other approaches detailed above because (i) it avoids the risk of stochastic errors implied by the use of rare, slowly evolving positions and (ii) it applies even when the taxon sampling is by necessity sparse (e.g. coelacanths in a vertebrate framework).
Finally, it should be noted that incorrect handling of multiple substitutions does not necessarily lead to a robust incorrect tree (as in the case of Coelomata), but possibly to an unresolved tree. For instance, an analysis based on 50 genes using a sparse taxon sampling (21 species, with most of the animal phyla being represented by a single, often fast-evolving, species) and a simple model of sequence evolution (RtREV+I+Γ), resulted in a poorly resolved tree, in which even the monophyly of Bilateria was not supported (Rokas et al. 2005). Since the approach used does not allow an efficient detection of multiple substitutions, we decided to do a comparable study in which we simultaneously improved the species sampling (from 21 to 57, including many slowly evolving species) and the model of sequence evolution (i.e. using the CAT model). Interestingly, the statistical support was high (e.g. bootstrap values more than 95% for Bilateria, Ecdysozoa and Lophotrochozoa) in the resulting tree (Baurain et al. 2007). This illustrates that incorrect handling of multiple substitutions can create an artefactual signal that although not strong enough to overcome the genuine phylogenetic signal, is sufficient to lead to a poorly resolved tree.
In summary, a combination of many positions, corresponding to multiple genes, and a dense taxonomic sampling are necessary to obtain reliable phylogenies. Ideally, these sequences should then be analysed with probabilistic models that correctly describe the true evolutionary patterns of the sequences under study. In practice, one may perform analyses with alternative models of evolution among those currently available, compare the fit of those models, check for possible model violations and test the robustness of the analyses by site and taxon resampling. This is the method that we apply to the phylogeny of Bilateria.
2. Results
(a) Comparison of phylogenies based on CAT and WAG models
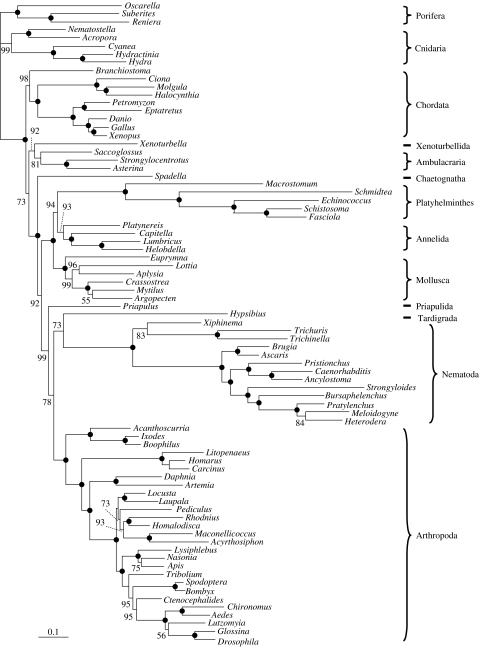
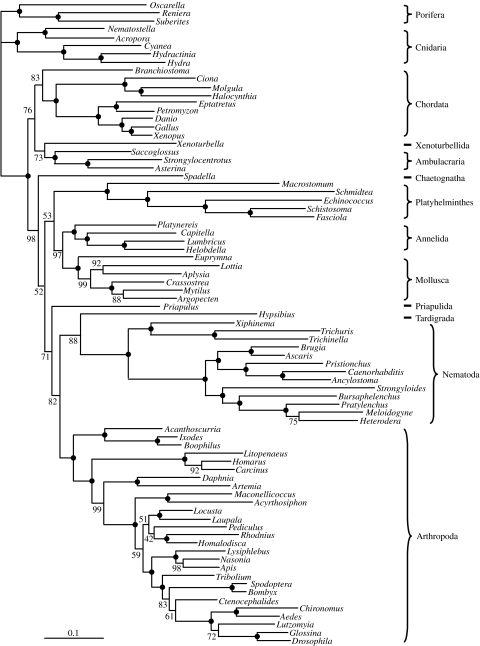
We analysed our large dataset (79 animal species and 19 993 positions) using two alternative models of amino acid replacement: the WAG empirical matrix (Whelan & Goldman 2001), which is currently one of the standard models (Ronquist & Huelsenbeck 2003; Jobb et al. 2004; Hordijk & Gascuel 2005; Stamatakis et al. 2005) and the CAT mixture model (see above). The trees obtained under CAT (figure 1) and WAG (figure 2) models are very similar and in good agreement with current knowledge (Halanych 2004). The following major aspects can be noted:
Ecdysozoa and Lophotrochozoa receive a stronger bootstrap support under CAT (bootstrap proportion (BP) of 99 and 100%) than under WAG (53 and 71%). Under WAG, platyhelminths are slightly attracted by nematodes, as can be seen by the low bootstrap support values along the path between the two groups. The attraction is nevertheless less marked than with a poorer taxon sampling (Philippe et al. 2005b).
Within Lophotrochozoa, many phyla are absent, but the three that are present (platyhelminths, molluscs and annelids) are reasonably well sampled. Interestingly, platyhelminths and annelids are sister groups with the CAT model (94% BP), while the analysis under WAG recovers a more traditional grouping of annelids and molluscs (Neotrochozoa, 97% BP). A sister-group relationship between annelids and platyhelminths had already been observed in a combined large subunit–small subunit (LSU–SSU) analysis (Passamaneck & Halanych 2006), and in an analysis based on mitochondrial gene order (Lavrov & Lang 2005), but was not found in previous analyses based on expressed sequence tags (ESTs) (Philippe et al. 2005b).
The relationships among Ecdysozoa are not well resolved. This is mainly due to the fluctuating position of the tardigrade and the priapulid, whose sequences are incomplete (39.9 and 75.8% of missing data, respectively). The most likely configuration places priapulids at the base of all other Ecdysozoa, and tardigrades sister group of nematodes, but two major alternatives are also proposed by the bootstrap analysis: tardigrades sister group of priapulids, together at the base of nematodes and arthropods, or priapulids at the base of nematodes.
The chaetognath appears at the base of all protostomes (92% CAT BP, 52% WAG BP), which is in accordance with Marletaz et al. (2006).
The monophyly of deuterostomes is weakly supported under the WAG model (76% BP), whereas the CAT model favours paraphyly, also weakly supported, with chordates emerging first (73% BP, only 19% for deuterostome monophyly).
Chordates are monophyletic (98% CAT BP, 83% WAG BP), receiving stronger support than in previous phylogenomic studies (Bourlat et al. 2006; Delsuc et al. 2006). In addition, under both analyses, urochordates are closer to vertebrates than cephalochordates with 100% BP confirming the monophyly of Olfactores (Delsuc et al. 2006).
The phylogenetic position of Xenoturbellida, as sister group of echinoderms + hemichordates (Bourlat et al. 2006), is also recovered (92% CAT BP, 73% WAG BP).
Figure 1.
Phylogeny inferred using the CAT model. The alignment consists of 19 993 unambiguously aligned positions (94 genes and 79 species). The tree was rooted using sponges and cnidarians as an out-group. Nodes supported by 100% bootstrap values are denoted by black circles while lower values are given in plain text. The scale bar indicates the number of changes per site.
Figure 2.
Phylogeny inferred using the WAG model. See figure legend 1 for details.
In summary, the WAG and CAT models agree with each other on 73 nodes, and disagree for a minor change at the base of insects and two major points: the monophyly of deuterostomes, and the relative order of molluscs, annelids and platyhelminths. In the case of lophotrochozoans, the two models strongly disagree, whereas concerning deuterostomes, the difference is not statistically significant.
(b) Model comparison and evaluation
The discrepancy between the two models indicates the presence of artefacts due to systematic errors. A statistical comparison of the two models may help decide which of them offers the most reliable phylogenetic tree. In addition, the observed artefacts are the symptoms of model violations, which we will analyse using a standard statistical method, the posterior predictive analysis. Note that the WAG empirical matrix is just one among the available empirical matrices, and the results obtained with WAG may not be representative of the general class of site-homogeneous models. To address this point, we also tested the general time reversible (GTR) model along with WAG and CAT.
First, based on cross-validation tests (see methods in electronic supplementary material), the CAT model was found to have a much better statistical fit than either WAG (a score of 3939±163 in favour of CAT) or GTR (2765±128). The better score of GTR relative to WAG (a difference of 1174 in favour of GTR) indicates that the dataset is big enough for the parameters of the amino acid replacement matrix to be directly inferred, rather than taken from an empirically derived empirical matrix. On the other hand, the progress in doing so is less important compared with that accomplished by the site-heterogeneous CAT model (1174 versus 2765).
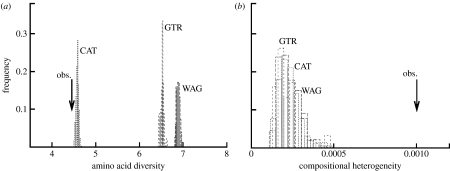
Second, we performed posterior predictive analyses (see methods in electronic supplementary material), using two test statistics: one, vertical (i.e. computed along the columns of the alignment), is the mean number of distinct amino acids per column (mean site-specific diversity; Lartillot et al. 2007); the other one, horizontal (i.e. computed along the rows of the alignment), is the chi-square compositional homogeneity test (Foster 2004). Violation of the horizontal statistic indicates that the model does not handle compositional biases correctly, whereas violation of the vertical statistic means that the model does not correctly account for site-specific biochemical patterns.
Concerning the vertical test (figure 3a), the mean number of distinct amino acids per column of the true alignment (mean observed diversity) is 4.45. Site-homogeneous models predict a much higher value (6.89±0.04 for WAG, 6.53±0.04 for GTR). Thus, the assumptions underlying the site-homogeneous models are strongly violated (p<0.001, z=62.3 for WAG and p<0.001, z=58.3 for GTR). In contrast, the CAT model performs much better (mean predicted diversity of 4.59±0.03), although it is also weakly rejected (p<0.001, z=4.6). As explained above, since overestimating the number of states per position leads to underestimating the probability of convergence (Lartillot et al. 2007), one may expect a greater risk of LBA under WAG for the present analysis. On the other hand, all models, CAT, GTR and WAG, fail the horizontal test to the same extent (compositional homogeneity; figure 3b). This is not too surprising, given that all of them are time-homogeneous amino acid replacement processes. However, the violation is strong, as measured by the z-score (z>11), which warns us that there is a risk, whichever model is used, of observing artefacts related to compositional biases.
Figure 3.
Posterior predictive tests. The observed value (arrow) of the test statistic is compared with the null distributions under CAT and WAG models. (a) Mean biochemical diversity per site. (b) Maximum compositional deviation over taxa.
3. Discussion
(a) Towards better phylogenetic analyses
Phylogenetics is still a difficult and controversial field, because no foolproof method is yet available to avoid systematic errors. In this study we have tried to combine two methods that have proven efficient in alleviating artefacts, while obtaining sufficient statistical support. First, relying on EST projects, we have tried to combine an increase of the overall amount of aligned sequence positions, so as to capture more of the primary phylogenetic signal, with an improved taxonomic sampling (Philippe & Telford 2006). Second, we have brought particular attention to the problem of probabilistic models underlying phylogenetic reconstruction. As is obvious from our statistical evaluations, the standard model used in phylogenetics today, WAG, is certainly not reliable, at least for deep-level phylogenies such as among animal phyla. Essentially, it is strongly rejected for its failure to explain either site-specific biochemical patterns or compositional differences between taxa. As indicated by our analysis of GTR, this failure is not specific to WAG and is likely to apply to all the site-homogeneous models. The alternative model used here, CAT, is significantly better, but may not be reliable enough, in particular against potential artefacts induced by compositional biases.
Interestingly, the weaknesses of the WAG model also result in an overall lack of support, which is probably due to the unstable position of some fast-evolving groups (in particular, platyhelminths). This confirms previous observations (Baurain et al. 2007), and also illustrates that improving taxonomic sampling is not in itself a sufficient response to systematic errors, but should be combined with an in-depth analysis of the probabilistic models used.
In light of our model evaluation, the position of platyhelminths proposed by CAT as a sister group to annelids should be taken seriously. In this perspective, the Neotrochozoa (molluscs+annelids) found by WAG would be an artefact. This would not be too surprising, given the overall saturation of the platyhelminth sequences. Note that the vestige of artefactual attraction between platyhelminths and nematodes observed under WAG should in itself warn us that the position of platyhelminths within Lophotrochozoa may not be reliably inferred under WAG. The phylogenetic position of platyhelminths, relatively to other lophotrochozoans, is a long-standing question, whose potentially important implications have already been pointed out (Passamaneck & Halanych 2006).
The other point of disagreement concerns deuterostomes: monophyletic under WAG, they appear to be paraphyletic under CAT. This progressive emergence of deuterostome phyla is unusual. In fact, Deuterostomia sensu stricto (echinoderms, hemichordates and chordates) have long been considered as one of the most reliable phylogenetic groupings in the animal phylogeny (Adoutte et al. 2000). A possible explanation for the monophyly of deuterostomes obtained with WAG in terms of LBA would be that the fast-evolving protostomes are attracted by the out-group. Given the implications (see below), this potential artefact would certainly deserve further attention. On the other hand, caution is needed since the basal position of chordates observed under CAT does not receive a high bootstrap support (73%). It is also unstable with small variations of the taxon sampling: for instance, deuterostome monophyly is recovered with CAT if either Spadella or Xenoturbella are removed from the analysis (data not shown). Similarly, it appears that the removal of the non-bilaterian out-group leads to the non-monophyly of Xenambulacraria (Philippe et al. 2007). In addition, the fast-evolving acoels probably emerge close to the base of deuterostomes, further shortening internal branches (Philippe et al. 2007). In summary, this suggests that the signal for resolving this part of the tree is weak, all the more so that the out-group is distantly related. Additional data should be analysed with improved methods before taking sides on this issue.
(b) Implications for the evolution of Bilateria
Converging on a reliable picture of the animal phylogenetic tree is an interesting objective in itself. But more important are the implications of this phylogenetic picture for our vision of the morphological evolution of Bilateria (Telford & Budd 2003). As mentioned in §1, morphological and developmental characters were traditionally the primary source of data used to infer phylogenetic trees. Now, it has become clear that many of those characters, such as cleavage patterns or the fate of the blastopore, are not reliable phylogenetic markers. It is nevertheless interesting to map their evolution on a tree that has been inferred from independent (molecular) data, and use this to learn as much as possible about the history of morphological diversification of animal body plans. In this respect, comparative embryology or evo–devo is probably the primary customer for animal molecular phylogenetics.
Much has already been said about how the ‘new animal phylogeny’ changes our way of looking at the evolution of animals (Adoutte et al. 1999; Halanych 2004). One of the most important, and most frequent, messages has been that secondary simplifications of morphology and of developmental processes are frequent. This has been repeatedly implied by most of the successive changes brought to the animal phylogeny over the last 10 years, such as the repositioning of nematodes and platyhelminths within coelomate protostomes, or of tunicates as the sister group of vertebrates.
In the context of the present study, the position of platyhelminths between two neotrochozoan phyla, as proposed by our CAT analysis, has similar implications, specifically concerning the evolution of development. Molluscs, annelids and sipunculans have a canonical spiral development, characterized by a four quartet spiral cleavage, an invariant and evolutionary conserved cell lineage, including a single stem cell (4d mesentoblast) giving rise to the mesodermal germ bands, and a typical trochophore larva (Nielsen 2001). In contrast, platyhelminths display atypical forms of spiral cleavage, and pass through a larval stage (Müller's larva) that can only loosely be homologized to a trochophore. In this context, a basal position of platyhelminths in the lophotrochozoan group, as previously often found, is compatible with the intuitively appealing idea that evolution proceeds from simple to complex forms. Namely, platyhelminths would be ‘proto’-spiralians, outside a series of nested phyla, Trochozoa, Eutrochozoa and Neotrochozoa (Peterson & Eernisse 2001), corresponding to a graded series of increasingly complex forms of spiral development. Yet, the phylogeny favoured by CAT is at odds with the Neotrochozoan hypothesis and implies that the development of platyhelminths is a secondarily modified (and ancestrally canonical) spiral development. Further taxonomic sampling within lophotrochozoans will be important, as it may not only allow a more robust inference of the position of platyhelminths but also bring additional phyla that do not display a canonical spiral development among Eutrochozoans or Neotrochozoans, thereby leading to a completely different view of the evolution of spiral development.
The paraphyly of deuterostomes favoured by our CAT analysis, if confirmed, would also have deep implications concerning the way we interpret the evolution of Bilateria. First, it would result in a paraphyletic succession of three groups (Chordata, Xenambulacraria and Chaetognatha), all of which display radial cleavage, deuterostomous gastrulation and an enterocoelic mode of formation of the body cavity. Although these embryological characters are known to be evolutionarily labile (e.g. brachiopods have a deuterostomous gastrulation, and enterocoely is observed in nemerteans), this may be interpreted as phylogenetic evidence in favour of ancestral deuterostomy. Similarly, the gill slits, found in chordates and hemichordates, would also have to be considered as ancestral to all Bilateria. In addition, with respect to all other Bilateria, chordates would then be of basal emergence, which turns the traditional preconceptions radically upside down: in the perspective of this new phylogenetic hypothesis, the chordate body plan is no longer the pinnacle of a progressive evolution through a succession of body plans of increasing complexity. Rather, chordates are one of the first bilaterian offshoots. This in turn would have consequences concerning the polarization of the morphological characters. Thus far, the most chordate-specific morphological and developmental features (e.g. their unique dorsoventral polarity), with nerve cord dorsal and heart ventral, have generally been assumed to be derived (Arendt & Nubler-Jung 1994). In the context of the more traditional hypothesis of deuterostome monophyly, this assumption is justified, provided that the ancestral condition is clearly and jointly recognized in protostomes and Ambulacraria (Arendt & Nubler-Jung 1994). But the argument does not hold anymore if chordates are the sister group of all other Bilateria: in that case, it is possible that some characters of chordates, such as the dorsoventral polarity, may well have been ancestral to all bilaterally symmetrical animals.
(c) Conclusion
Several phyla, in particular brachiopods and onychophorans, are still missing in phylogenomic analyses, and some others are poorly represented (aschelminths, chaetognaths, hemichordates, among others), but the most species-rich phyla are now well sampled. Accordingly, one can be increasingly confident concerning the few robust aspects of the phylogeny of bilaterians that emerge from this and previous phylogenomic analyses. Essentially, the overall structure of protostomes (a split between lophotrochozoans and ecdysozoans, with chaetognaths at their base) seems stable, as well as the monophyly of Chordata and Ambulacraria. On the other hand, the monophyly of deuterostomes appears to be the most important point yet to be settled in order to draw a complete picture of the scaffold of the bilaterian tree. Many aspects of the detailed relationships within each supergroup (in particular, Ecdysozoa and Lophotrochozoa) remain to be investigated. Ongoing EST projects will soon bring many new species into this emerging picture, which will not only inform us about the phylogenetic position of those new species but also result in an enriched taxonomic sampling, positively impacting the overall accuracy of the phylogenetic inference. Yet, as was suggested by the present study, this will not be sufficient, and will have to be combined with a significant improvement of the underlying probabilistic models. Much work is still needed both concerning the acquisition of primary data and the methodological side, if one wants to converge towards a reliable, possibly final, picture of the bilaterian tree.
Acknowledgments
We thank Max Telford and Tim Littlewood for giving us the opportunity to write this article. The Réseau Québécois de Calcul de Haute Performance provides for computational resources. This work was supported by the Canadian Institute for Advanced Research, the Canadian Research Chair Program, the Centre National de la Recherche Scientifique (through the ACI-IMPBIO Model-Phylo funding program) and the Robert Cedergren Centre for Bioinformatics and Genomics. This work was financially supported in part by the ‘60ème commission franco-québécoise de coopération scientifique’.
Footnotes
One contribution of 17 to a Discussion Meeting Issue ‘Evolution of the animals: a Linnean tercentenary celebration’.
Supplementary Material
Description of the data set assembly and of the phylogenetic methods
References
- Abouheif E, Zardoya R, Meyer A. Limitations of metazoan 18S rRNA sequence data: implications for reconstructing a phylogeny of the animal kingdom and inferring the reality of the Cambrian explosion. J. Mol. Evol. 1998;47:394–405. doi: 10.1007/pl00006397. doi:10.1007/PL00006397 [DOI] [PubMed] [Google Scholar]
- Adoutte A, Balavoine G, Lartillot N, de Rosa R. Animal evolution. The end of the intermediate taxa? Trends Genet. 1999;15:104–108. doi: 10.1016/s0168-9525(98)01671-0. doi:10.1016/S0168-9525(98)01671-0 [DOI] [PubMed] [Google Scholar]
- Adoutte A, Balavoine G, Lartillot N, Lespinet O, Prud'homme B, de Rosa R. The new animal phylogeny: reliability and implications. Proc. Natl Acad. Sci. USA. 2000;97:4453–4456. doi: 10.1073/pnas.97.9.4453. doi:10.1073/pnas.97.9.4453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguinaldo A.M, Turbeville J.M, Linford L.S, Rivera M.C, Garey J.R, Raff R.A, Lake J.A. Evidence for a clade of nematodes, arthropods and other moulting animals. Nature. 1997;387:489–493. doi: 10.1038/387489a0. doi:10.1038/387489a0 [DOI] [PubMed] [Google Scholar]
- Arendt D, Nubler-Jung K. Inversion of dorsoventral axis? Nature. 1994;371:26. doi: 10.1038/371026a0. doi:10.1038/371026a0 [DOI] [PubMed] [Google Scholar]
- Baurain D, Brinkmann H, Philippe H. Lack of resolution in the animal phylogeny: closely spaced cladogeneses or undetected systematic errors? Mol. Biol. Evol. 2007;24:6–9. doi: 10.1093/molbev/msl137. doi:10.1093/molbev/msl137 [DOI] [PubMed] [Google Scholar]
- Blair J.E, Ikeo K, Gojobori T, Hedges S.B. The evolutionary position of nematodes. BMC Evol. Biol. 2002;2:7. doi: 10.1186/1471-2148-2-7. doi:10.1186/1471-2148-2-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore J.L. The use of genome-level characters for phylogenetic reconstruction. Trends Ecol. Evol. 2006;21:439–446. doi: 10.1016/j.tree.2006.05.009. doi:10.1016/j.tree.2006.05.009 [DOI] [PubMed] [Google Scholar]
- Bourlat S.J, et al. Deuterostome phylogeny reveals monophyletic chordates and the new phylum Xenoturbellida. Nature. 2006;444:85–88. doi: 10.1038/nature05241. doi:10.1038/nature05241 [DOI] [PubMed] [Google Scholar]
- Brinkmann H, Philippe H. Archaea sister group of Bacteria? Indications from tree reconstruction artifacts in ancient phylogenies. Mol. Biol. Evol. 1999;16:817–825. doi: 10.1093/oxfordjournals.molbev.a026166. [DOI] [PubMed] [Google Scholar]
- Brusca R.C, Brusca G.J. Sinauer Associates; Sunderland, MA: 1990. Invertebrates. [Google Scholar]
- Conway Morris S. The fossil record and the early evolution of the Metazoa. Nature. 1993;361:219–225. doi:10.1038/361219a0 [Google Scholar]
- Delsuc F, Brinkmann H, Philippe H. Phylogenomics and the reconstruction of the tree of life. Nat. Rev. Genet. 2005;6:361–375. doi: 10.1038/nrg1603. doi:10.1038/nrg1603 [DOI] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. doi:10.1038/nature04336 [DOI] [PubMed] [Google Scholar]
- Dopazo H, Santoyo J, Dopazo J. Phylogenomics and the number of characters required for obtaining an accurate phylogeny of eukaryote model species. Bioinformatics. 2004;20:i116–i121. doi: 10.1093/bioinformatics/bth902. doi:10.1093/bioinformatics/bth902 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Cases in which parsimony or compatibility methods will be positively misleading. Syst. Zool. 1978;27:401–410. doi:10.2307/2412923 [Google Scholar]
- Felsenstein J. Sinauer Associates, Inc; Sunderland, MA: 2004. Inferring phylogenies. [Google Scholar]
- Field K.G, Olsen G.J, Lane D.J, Giovannoni S.J, Ghiselin M.T, Raff E.C, Pace N.R, Raff R.A. Molecular phylogeny of the animal kingdom. Science. 1988;239:748–753. doi: 10.1126/science.3277277. doi:10.1126/science.3277277 [DOI] [PubMed] [Google Scholar]
- Foster P.G. Modeling compositional heterogeneity. Syst. Biol. 2004;53:485–495. doi: 10.1080/10635150490445779. doi:10.1080/10635150490445779 [DOI] [PubMed] [Google Scholar]
- Foster P.G, Hickey D.A. Compositional bias may affect both DNA-based and protein-based phylogenetic reconstructions. J. Mol. Evol. 1999;48:284–290. doi: 10.1007/pl00006471. doi:10.1007/PL00006471 [DOI] [PubMed] [Google Scholar]
- Halanych K.M. The new view of animal phylogeny. Annu. Rev. Ecol. Evol. Syst. 2004;35:229–256. doi:10.1146/annurev.ecolsys.35.112202.130124 [Google Scholar]
- Halanych K.M, Bacheller J.D, Aguinaldo A.M, Liva S.M, Hillis D.M, Lake J.A. Evidence from 18S ribosomal DNA that the lophophorates are protostome animals. Science. 1995;267:1641–1643. doi: 10.1126/science.7886451. doi:10.1126/science.7886451 [DOI] [PubMed] [Google Scholar]
- Hendy M.D, Penny D. A framework for the quantitative study of evolutionary trees. Syst. Zool. 1989;38:297–309. doi:10.2307/2992396 [Google Scholar]
- Hordijk W, Gascuel O. Improving the efficiency of SPR moves in phylogenetic tree search methods based on maximum likelihood. Bioinformatics. 2005;21:4338–4347. doi: 10.1093/bioinformatics/bti713. doi:10.1093/bioinformatics/bti713 [DOI] [PubMed] [Google Scholar]
- Irimia M, Maeso I, Penny D, Garcia-Fernandez J, Roy S.W. Rare coding sequence changes are consistent with Ecdysozoa, not Coelomata. Mol. Biol. Evol. 2007;24:1604–1607. doi: 10.1093/molbev/msm105. doi:10.1093/molbev/msm105 [DOI] [PubMed] [Google Scholar]
- Jenner R.A. Bilaterian phylogeny and uncritical recycling of morphological data sets. Syst. Biol. 2001;50:730–742. doi: 10.1080/106351501753328857. doi:10.1080/106351501753328857 [DOI] [PubMed] [Google Scholar]
- Jobb G, von Haeseler A, Strimmer K. Treefinder: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol. Biol. 2004;4:18. doi: 10.1186/1471-2148-4-18. doi:10.1186/1471-2148-4-18 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. doi:10.1093/molbev/msh112 [DOI] [PubMed] [Google Scholar]
- Lartillot N, Brinkmann H, Philippe H. Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol. Biol. 2007;7(Suppl. 1):S4. doi: 10.1186/1471-2148-7-S1-S4. doi:10.1186/1471-2148-7-S1-S4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavrov D.V, Lang B.F. Poriferan mtDNA and animal phylogeny based on mitochondrial gene arrangements. Syst. Biol. 2005;54:651–659. doi: 10.1080/10635150500221044. doi:10.1080/10635150500221044 [DOI] [PubMed] [Google Scholar]
- Marletaz F, et al. Chaetognath phylogenomics: a protostome with deuterostome-like development. Curr. Biol. 2006;16:R577–R578. doi: 10.1016/j.cub.2006.07.016. doi:10.1016/j.cub.2006.07.016 [DOI] [PubMed] [Google Scholar]
- Matus D.Q, Copley R.R, Dunn C.W, Hejnol A, Eccleston H, Halanych K.M, Martindale M.Q, Telford M.J. Broad taxon and gene sampling indicate that chaetognaths are protostomes. Curr. Biol. 2006;16:R575–R576. doi: 10.1016/j.cub.2006.07.017. doi:10.1016/j.cub.2006.07.017 [DOI] [PubMed] [Google Scholar]
- Nielsen C. Oxford University Press; Oxford, UK: 2001. Animal evolution, interrelationships of the living phyla. [Google Scholar]
- Olsen G. Earliest phylogenetic branching: comparing rRNA-based evolutionary trees inferred with various techniques. Cold Spring Harb. Symp. Quant. Biol. 1987;LII:825–837. doi: 10.1101/sqb.1987.052.01.090. [DOI] [PubMed] [Google Scholar]
- Passamaneck Y, Halanych K.M. Lophotrochozoan phylogeny assessed with LSU and SSU data: evidence of lophophorate polyphyly. Mol. Phylogenet. Evol. 2006;40:20–28. doi: 10.1016/j.ympev.2006.02.001. doi:10.1016/j.ympev.2006.02.001 [DOI] [PubMed] [Google Scholar]
- Peterson K.J, Eernisse D.J. Animal phylogeny and the ancestry of bilaterians: inferences from morphology and 18S rDNA gene sequences. Evol. Dev. 2001;3:170–205. doi: 10.1046/j.1525-142x.2001.003003170.x. doi:10.1046/j.1525-142x.2001.003003170.x [DOI] [PubMed] [Google Scholar]
- Philippe H, Laurent J. How good are deep phylogenetic trees? Curr. Opin. Genet. Dev. 1998;8:616–623. doi: 10.1016/s0959-437x(98)80028-2. doi:10.1016/S0959-437X(98)80028-2 [DOI] [PubMed] [Google Scholar]
- Philippe H, Telford M.J. Large-scale sequencing and the new animal phylogeny. Trends Ecol. Evol. 2006;21:614–620. doi: 10.1016/j.tree.2006.08.004. doi:10.1016/j.tree.2006.08.004 [DOI] [PubMed] [Google Scholar]
- Philippe H, Chenuil A, Adoutte A. Can the Cambrian explosion be inferred through molecular phylogeny? Development. 1994;120:S15–S25. [Google Scholar]
- Philippe H, Delsuc F, Brinkmann H, Lartillot N. Phylogenomics. Annu. Rev. Ecol. Evol. Syst. 2005a;36:541–562. doi:10.1146/annurev.ecolsys.35.112202.130205 [Google Scholar]
- Philippe H, Lartillot N, Brinkmann H. Multigene analyses of bilaterian animals corroborate the monophyly of Ecdysozoa, Lophotrochozoa, and Protostomia. Mol. Biol. Evol. 2005b;22:1246–1253. doi: 10.1093/molbev/msi111. doi:10.1093/molbev/msi111 [DOI] [PubMed] [Google Scholar]
- Philippe H, Brinkmann H, Martinez P, Riutort M, Baguñà J. Acoel flatworms are not Platyhelminthes: evidence from phylogenomics. PLoS ONE. 2007;2:e717. doi: 10.1371/journal.pone.0000717. doi:10.1371/journal.pone.0000717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Ezpeleta N, Brinkmann H, Roure B, Lartillot N, Lang B.F, Philippe H. Detecting and overcoming systematic errors in genome-scale phylogenies. Syst. Biol. 2007;56:389–399. doi: 10.1080/10635150701397643. doi:10.1080/10635150701397643 [DOI] [PubMed] [Google Scholar]
- Rogozin I.B, Wolf Y.I, Carmel L, Koonin E.V. Ecdysozoan clade rejected by genome-wide analysis of rare amino acid replacements. Mol. Biol. Evol. 2007;24:1080–1090. doi: 10.1093/molbev/msm029. doi:10.1093/molbev/msm029 [DOI] [PubMed] [Google Scholar]
- Rokas A, Kruger D, Carroll S.B. Animal evolution and the molecular signature of radiations compressed in time. Science. 2005;310:1933–1938. doi: 10.1126/science.1116759. doi:10.1126/science.1116759 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. doi:10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Ludwig T, Meier H. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–463. doi: 10.1093/bioinformatics/bti191. doi:10.1093/bioinformatics/bti191 [DOI] [PubMed] [Google Scholar]
- Telford M.J, Budd G.E. The place of phylogeny and cladistics in evo–devo research. Int. J. Dev. Biol. 2003;47:479–490. [PubMed] [Google Scholar]
- Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- Wolf Y.I, Rogozin I.B, Koonin E.V. Coelomata and not Ecdysozoa: evidence from genome-wide phylogenetic analysis. Genome Res. 2004;14:29–36. doi: 10.1101/gr.1347404. doi:10.1101/gr.1347404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L. Molecules as documents of evolutionary history. J. Theor. Biol. 1965;8:357–366. doi: 10.1016/0022-5193(65)90083-4. doi:10.1016/0022-5193(65)90083-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of the data set assembly and of the phylogenetic methods