Summary
Haspin/Gsg2 is a kinase that phosphorylates Histone H3 at Thr-3 (H3T3ph) during mitosis. Its depletion by RNA interference results in failure of chromosome alignment and a block in mitosis. Haspin therefore is a novel target for development of anti-mitotic agents. We report the development of a high throughput time-resolved fluorescence resonance energy transfer (TR-FRET) kinase assay for Haspin. Histone H3 peptide was used as a substrate, and a Europium-labeled H3T3ph phosphospecific monoclonal antibody was used to detect phosphorylation. A library of 137632 small molecules was screened at Km concentrations of ATP and peptide to allow identification of diverse inhibitor types. Reconfirmation of hits and IC50 determinations were carried out with the TR-FRET assay and by a radiometric assay using recombinant Histone H3 as the substrate. A preliminary assessment of specificity was made by testing inhibition of two unrelated kinases. EC50 values in cells were determined using a cell-based ELISA assay of H3T3ph. Five compounds were selected as leads based on potency and chemical structure considerations. These leads form the basis for the development of specific inhibitors of Haspin that will have clear utility in basic research and possible use as starting points for development of anti-mitotic anticancer therapeutics.
Keywords: anti-mitotics, chromatin, Haspin, kinase, time-resolved resonance energy transfer
Introduction
Agents that target mitosis are used widely and successfully for therapy of a variety of cancers. Docetaxel, for example, is a taxane that binds tubulin and stabilizes mitotic spindle microtubules, likely inhibiting mitosis by activating the spindle checkpoint. Microtubules have important functions outside mitosis, however, and peripheral neurotoxicity is a known side effect of microtubule-targeting agents, for example.1 Because of this, great efforts are being made to identify more selective drug targets in mitosis. For at least two reasons, protein kinases have been the focus of much work in this area. First, kinases play central roles in regulation of cellular events, including mitosis. Second, enzymes such as kinases are excellent targets for inhibitor drug development because their active sites can be effectively blocked by the binding of small molecules. Recently, mitotic kinases such as the Aurora kinases and Polo like kinase-1 (Plk1) have received considerable attention as molecular targets for anti-cancer therapy (for review see refs 1-4).
In this article we report our efforts to identify inhibitors of the chromatin-modifying protein kinase Haspin/Gsg2 by high throughput screening of a small molecule library. Haspin is a serine-threonine kinase that phosphorylates Thr-3 in the amino terminal tail of Histone H3 during mitosis.5-7 A number of factors indicate that the identification of specific Haspin inhibitors might be feasible and useful for basic biological studies and as candidate agents for cancer treatment. First, and most important, Haspin plays a significant role in the regulation of mitosis. Haspin RNAi in tumor cell lines prevents chromosome alignment and completion of normal mitosis,5,7 suggesting that Haspin inhibitors might be novel anti-mitotic agents capable of preventing cancer cell proliferation.8,9 Second, Haspin mRNA is expressed in proliferating cells but not in non-cycling cells10 indicating that Haspin inhibition is a viable strategy for cancer treatment. Third, based on its primary sequence, Haspin is a structurally divergent kinase that does not fall into any of the known eukaryotic protein kinase families, and humans have a single Haspin homologue.11-13 Compared with kinases that are members of large families or have closely related homologues (e.g. the three Aurora kinases), these factors increase the likelihood of identifying specific inhibitors of Haspin that may result in fewer off-target effects. Finally, fast-acting Haspin inhibitors would circumvent many of the problems inherent in using slow-acting methods such as RNAi to study the rapid successive events in mitosis.14
To identify Haspin inhibitors by high throughput screening we have used a homogeneous kinase assay based on time-resolved fluorescence resonance energy transfer (TR-FRET; Figure 1). Mathis first described the application of TR-FRET to assay kinase activity,15 which has emerged as one of the preferred fluorescent assay formats in drug discovery. Such TR-FRET assays use a lanthanide donor species conjugated to a phospho-specific antibody that binds specifically to the product of kinase reaction labeled with an acceptor fluorophore. This induced proximity of the donor and acceptor fluorophores leads to resonance energy transfer, resulting in a detectable increase of TR-FRET signal. In the assay described here, we use a Europium chelate, conjugated to an anti-Histone H3T3ph antibody, as the donor species. The acceptor fluorophore, allophycocyanin (APC) is used as a streptavidin conjugate that can bind to a biotinylated Histone H3 peptide substrate. The TR-FRET read-out is a dimensionless number calculated as a ratio of acceptor specific fluorescence signal to the donor signal, which provides a robust internal standard to compensate for compound interference and variations in assay volume.16,17 Lanthanide ions like Europium have a much longer emission lifetime, often measured in hundreds of microseconds, compared with traditional organic reagents that have lifetimes measured on the scale of hundreds of nanoseconds. TR-FRET assays are thus less susceptible to compound interference generated by short-lived compound or matrix component fluorescence. Furthermore, TR-FRET can be carried out in a homogeneous format that avoids time-consuming separation steps that introduce variability. Based on these properties TR-FRET based assay kinases have been widely used in high throughput screening.
Figure 1.
Assay principles for TR-FRET detection of Haspin activity. Kinase action in presence of ATP results in phosphorylation of an H3(1-21)-biotin substrate peptide. Phosphospecific Europium-labeled anti-Histone H3T3ph antibodies can bind the phosphorylated product. The biotinylated peptide is also captured by Streptavidin molecules conjugated to allophycocyanin (APC). Donor Europium excitation at 337 nm results in fluorescence resonance energy transfer to the acceptor APC which fluoresces at 665 nm. The TR-FRET data readout is the ratio of acceptor-specific fluorescence at 665 nm to donor-specific fluorescence at 620 nm (ratio of 665/620 nm).
We describe here the development of a high throughput TR-FRET assay and secondary assays suitable for the identification and initial validation of Haspin inhibitors. We have used the TR-FRET assay to screen a small molecule library of approximately 140000 compounds. Primary hits were re-tested by TR-FRET assay using the peptide substrate and then revalidated by assaying the compounds in a radiometric assay using full-length Histone H3 as a protein substrate. Finally, candidate compounds were evaluated in a cellular assay of Haspin activity to select lead compounds for further development.
Materials and Methods
Expression and Purification of Recombinant Haspin
A synthetic codon-optimized human Haspin cDNA was made in vector pUC57 at GenScript Corporation (Piscataway, NJ) to facilitate bacterial expression. This full-length haspin gene was cloned into the pMALc2E vector (New England Biolabs, Ipswich, MA) using EcoR I and Sal I sites. Haspin was expressed and purified as an N-terminal MBP fusion protein from E. coli Rosetta™2(DE3)pLysS cells (Novagen, Madison, WI). A freshly transformed colony was used to initiate a small volume liquid culture in LB medium with 2 g/l glucose, 34 μg/ml chloramphenicol and 100 μg/ml ampicillin. This culture was used to inoculate a large volume of the same medium and grown until an absorbance at 600 nm of 0.5 was reached. Protein expression was induced by adding 0.3 mM isopropyl thiogalactoside and growth with shaking at room temperature for 14 hours. Affinity column chromatography was carried out using amylose resin following the manufacturer’s instructions (New England Biolabs). The fusion protein was eluted in 50 mM Tris, pH 7.5, 200 mM NaCl, 10 mM maltose and dialyzed into 50 mM Tris, pH 7.5, 200 mM NaCl, 2 mM DTT and 50% glycerol. The purity and yield of intact fusion protein was determined by SDS-PAGE and Coomassie Blue staining, in comparison with known quantities of bovine serum albumin.
Reagents and Substrates
A synthetic peptide representing the first 21 amino acid residues of human Histone H3 was designated H3(1-21)-biotin peptide (ARTKQTARKSTGGKAPRKQLA-GGK-biotin) and a peptide representing the expected product of kinase reaction was designated H3(1-21)T3ph-biotin (ARTphKQTARKSTGGKAPRKQLA-GGK-biotin), and were synthesized at Abgent (San Diego, CA). These peptides carried biotin on the side chain of the C-terminal lysine. Recombinant full-length human histone H3 was from New England Biolabs. ATP and Staurosporine were purchased from Sigma-Aldrich (St Louis, MO). Rabbit monoclonal anti-Histone H3T3ph antibody (clone JY325) from Millipore (Billerica, MA) was directly labeled by PerkinElmer (Waltham, MA) with LANCE Eu W1024. For indirect detection, LANCE Eu W1024 labeled anti-rabbit IgG antibody was used (PerkinElmer). Streptavidin conjugated to SureLight-Allophycocyanin (PerkinElmer) was used as the acceptor fluorophore.
Compound Library
The compound library consisted of approximately 140,000 small molecules, including compounds approved by the Food and Drug Administration (FDA), a purified natural products library, compounds purchased from Peakdale (High Peak, UK), Maybridge plc. (Cornwall, UK), Cerep (Paris, France), Bionet Research Ltd. (Cornwall, UK), Prestwick (Ilkirch, France), Specs and Biospecs (CP Rijswijk, the Netherlands), ENAMINE (Kiev, Ukraine), Life Chemicals, Inc. (Burlington, Canada), MicroSource Diversity System’s NINDS custom collection (Gaylordsville, CT) and ChemDiv (San Diego, CA), and from various academic institutions. Compounds were selected from the different vendors by applying a series of filters, including for clogP and predicted solubility. All of the small molecules generally adhere to Lipinski’s rules (i.e. molecular weight < 500, H-bond donors ≤ 5, H-bond acceptors ≤ 10 and logP < 5) and contain a low proportion of known toxicophores (i.e. Michael acceptors and alkylating agents) and unwanted functionalities (i.e. imines, thiols, and quaternary amines), and have been optimized to maximize molecular diversity.
TR-FRET assays
A CRS CataLyst Express robotic arm (Thermo Fisher Scientific, Waltham, MA) and a Cybi-well 384 channel simultaneous pipettor (CyBio AG, Jena, Germany) were used to carry out the high throughput screening of a small molecule library. Kinase reactions were performed in 50 mM Tris, pH 7.5, 5 mM MgCl2, 1 mM DTT, 0.01% Brij-35 using Proxiplate 384 Plus white assay plates (PerkinElmer). In the final HTS conditions, 0.17 nM enzyme (0.05 nM MBP-Haspin final) and 0.33 μM biotinylated H3(1-21) peptide (0.1 μM peptide final, at the Km) in a volume of 3 μl kinase buffer were added to 2 μl solutions of 50 μM compound (10 μM final) and pre-incubated for 20 minutes. The kinase reaction was initiated by addition of 5 μl of 400 μM ATP per reaction (200 μM ATP final, at the Km). The reaction was incubated for 10 minutes at room temperature. Reaction was terminated by addition of 10 μl 50 mM EDTA, 2 nM Europium labeled anti-Histone H3T3ph antibody, 40 nM Streptavidin-APC. After a two hour incubation at room temperature, TR-FRET measurements were performed using a PHERAstar HTS microplate reader (BMG Labtech, Offenberg, Germany), and were expressed as ratios of acceptor fluorescence at 665 nm over donor fluorescence at 620 nm.
Radiometric filter binding assay
In radiometric assays, 10 μM test compound was incubated with 4 nM MBP-Haspin in a 25 μl enzyme reaction containing 0.3 μM Histone H3 (slightly above the apparent Km value of 0.18 μM for Histone H3 in this assay) and 11 μM ATP (apparent Km value), 0.73 μCi γ33P-ATP (PerkinElmer), 50 mM Tris-HCl, 5 mM MgCl2, pH 7.5. The reaction was stopped after 10 minutes by directly spotting 10 μl of reaction mix on P81 phosphocellulose filters (Whatman plc, Maidstone, UK). P81 filter discs were subsequently washed thrice with 0.2 M ammonium bicarbonate (5 ml/circle) and air dried. The dried P81 filter discs were transferred to a 6 ml scintillation vial (Pony-Vial, PerkinElmer) and, following addition of 3 ml of scintillation fluid, were read using an LS5801 liquid scintillation counter (Beckman Coulter, Fullerton, CA). Background 33P incorporation was defined from similar reactions carried out in the absence of enzyme.
Cell Based ELISA
For cell-based ELISA, myc-Haspin overexpressing HeLa Tet-on cells5 were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% Tet-system approved fetal bovine serum (Clontech, Mountain View, CA). Approximately 15000 cells per well were seeded in a 96 well Nunclon™Δ surface plates (Thermo Fisher Scientific). Following 16 h growth in the presence of 1 μM doxycycline to induce myc-Haspin expression, cells were treated for 2 hours with various inhibitor concentrations. The cells were then fixed with 4% formaldehyde in PBS for 2 h, followed by washing thrice with 200 μl Wash Buffer per well (PBS, 0.1% Triton X-100, pH 7.4). The wells of assay plate were subsequently treated with quench buffer (0.1% NaN3, 1% H2O2 in Wash Buffer) for 1 h. Then the plates were again washed thrice with Wash Buffer and incubated overnight at 4°C with rabbit anti-Histone H3T3ph affinity-purified polyclonal antibody B86345 in 3% BSA in Wash Buffer. The plates were warmed to room temperature, washed thrice with Wash Buffer and incubated with 1:3000 anti-rabbit IgG-HRP (Jackson Immunoresearch, West Grove, PA) in Wash Buffer for 1 h. After washing thrice with Wash Buffer, a 1:1 mix of chemiluminescent substrate and hydrogen peroxide was added to each well (SuperSignal ELISA Pico Chemiluminescent Substrate, Thermo Fisher Scientific). Chemiluminescence was measured after five minute incubation on a Victor2 Plate Reader (PerkinElmer). To control for cell viability, we assayed duplicate plates using CellTiter-Glo® (Promega, Madison, WI), following themanufacturer’s protocol, which uses luciferase to measure ATP as an indicator of metabolically active viable cells. Significant loss of viable cell numbers was detected only at doses above 100 μM after a two hour treatment with the 5 lead compounds.
Data analysis
Data were analyzed using GraphPad Prism Version 4 (GraphPad Software Inc, La Jolla, CA). No inhibitor (“MAX”) and Staurosporine inhibitor (“MIN”) controls were used to calculate Z’ values18 and signal to background ratios during the high throughput screen. Percentage inhibition of enzyme activity was calculated according to the following equation: % inhibition = 100 x (average of MAX controls - test compound value) / (average of MAX controls - average of MIN controls). For determination of IC50 and EC50 concentrations, mean % inhibition dose response curves were fitted to the sigmoidal dose response equation: Y = Bottom+(Top-Bottom)/(1+10logEC50-X) where X is log(compound concentration), Y is % inhibition, and Bottom and Top are the lower and upper plateaux. Km concentrations were also determined by non-linear regression.
Results and Discussion
Selection and Optimization of TR-FRET Detection Reagents
To ensure a reproducible source of phospho-specific detection antibody for TR-FRET, we made use of a rabbit monoclonal anti-Histone H3T3ph antibody (clone JY325, Millipore). By immunoblotting of synthetic peptides representing nonphosphorylated and phosphorylated portions of Histone H3, we confirmed that this antibody recognized a peptide containing H3T3ph, and not an equivalent unphosphorylated peptide or H3T11ph. Weaker cross-reactivity with peptides containing H3T22ph was observed (data not shown), but because residue 22 was excluded from the peptide substrates used in all subsequent experiments, we considered the antibody suitable for further assay development.
Initially, we compared the detection of a synthetic H3(1-21)T3ph-biotin peptide (in which Thr-3 was phosphorylated) using direct Eu-labeled anti-Histone H3T3ph antibody or a combination of primary anti-Histone H3T3ph and a secondary LANCE Eu W-1024 labeled anti-rabbit IgG antibody (PerkinElmer). Directly labeled antibodies were found to yield higher TR-FRET signal strength and were selected for assay development (data not shown). These and similar experiments also confirmed that the detection system responded in a dose-dependent manner to the expected product of Haspin kinase activity, Histone H3 peptides phosphorylated at Thr-3 (data not shown). It should be noted that the N-terminal tail of histone H3 is thought to be relatively unstructured19 and therefore peptides are likely to be a reasonable mimic of the in vivo substrate in this case.
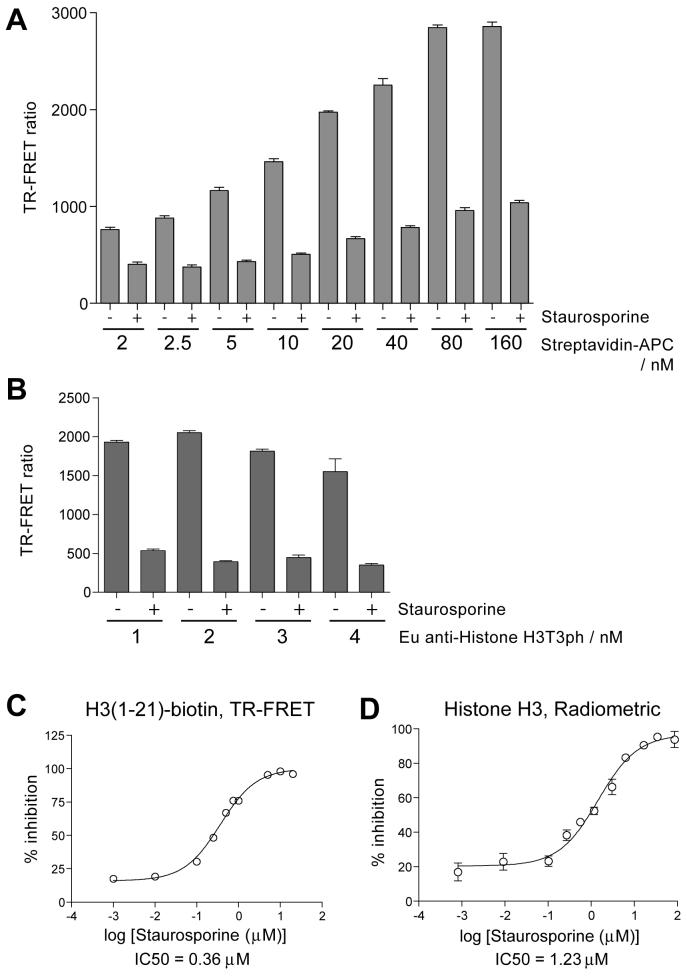
Optimization of detection conditions was then carried out in kinase reactions with H3(1-21)-biotin peptide in near-screening conditions in 384 well plates. The parameters tested were primarily the effect of streptavidin-APC:biotin ratio and concentration of detection antibody necessary for a robust signal. Using an H3(1-21)-biotin peptide concentration of 100 nM, we tested streptavidin-APC concentrations between 2 and 160 nM (Figure 2A). Staurosporine was used as an inhibitor control (see “Enzyme stability and Staurosporine inhibition”, below). We subsequently used 40 nM Streptavidin-APC in high throughput screening to provide a balance of high signal-to-background and cost-effectiveness, and which corresponds to a 1:2.5 streptavidin-APC to biotin peptide ratio (note that each streptavidin-APC molecule is assumed to bind 2 biotinylated peptides). We also tested Eu-labeled anti-Histone H3T3ph antibody concentrations of 1 nM to 4 nM using 40 nM Streptavidin-APC (Figure 2B). Use of 2 nM antibody resulted in efficient signal detection and Z’ values18 obtained in test plates were always above 0.6.
Figure 2.
Optimization of detection conditions for TR-FRET and determination of IC50 for Staurosporine. A. Optimization of H3(1-21)-biotin peptide to Streptavidin-APC ratio in high throughput screening conditions. Means plus standard deviation are shown, n=16. B. Effect of Eu-labeled Histone H3T3ph antibody concentration on TR-FRET detection. Means plus standard deviation are shown, n=16. C. Determination IC50 of Staurosporine in the TR-FRET assay with H3(1-21)-biotin as substrate. Means plus standard deviation are shown, n=3. D. As for C, but in a radiometric assay with Histone H3 protein as a substrate (see text for details).
Enzyme Stability and Staurosporine Inhibition
We used both radiometric and TR-FRET assays to test reaction conditions for the activity of recombinant MBP-Haspin using both an H3(1-21)-biotin peptide and recombinant Histone H3. The enzyme activity was stable in a range of pH from 6.5 to 9.0, and the optimum pH was 7.5 (data not shown). The optimum reaction temperature for MBP-Haspin was 30°C (data not shown), however for convenience all enzymatic reactions were carried out at room temperature. We tested the stability of the enzyme by leaving stock solutions at room temperature for 2 h and overnight. The enzyme showed little decrease in activity when left at room temperature for 2 h. When incubated overnight at room temperature, however, there was a 20% decrease in enzymatic activity. Subsequently, working solutions of MBP-Haspin were used within 30 min. DMSO tolerance was tested using both the peptide and recombinant Histone H3 as substrates. MBP-Haspin was remarkably stable in up to 5% DMSO and did not show any significant decrease in activity. At 20% DMSO, a 25% decrease in enzymatic activity was observed. DMSO was subsequently used at a concentration of 1% or lower.
We found that the broad-spectrum kinase inhibitor Staurosporine was an effective inhibitor of MBP-Haspin, with IC50 concentrations of 0.36 μM and 1.2 μM in TR-FRET and radiometric assays respectively (Figure 2C, D). Staurosporine at 2 μM in TR-FRET assays and at 10 μM in radiometric assays was used in subsequent experiments to define 100% inhibition of enzyme activity.
Optimization of Enzyme Concentration and Km Estimation for High Throughput Screening
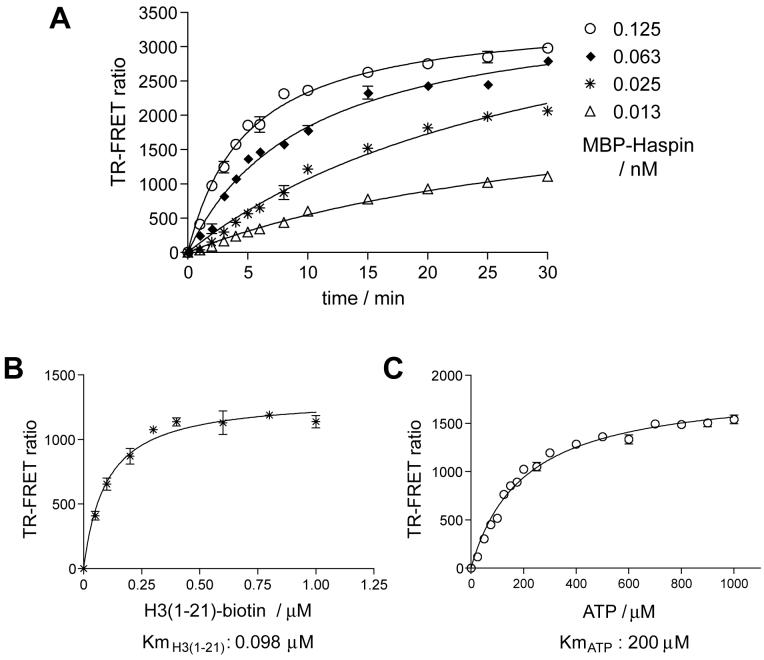
For high throughput screening it was necessary to screen at an enzyme concentration that would yield a near-linear dependency of substrate conversion with respect to reaction time. Using the TR-FRET assay, reaction progression curves were generated at several enzyme concentrations (Figure 3A). Reaction conditions using 0.05 nM MBP-Haspin for 5 minutes were then used to determine the apparent Km of ATP and H3(1-21)-biotin substrate peptide. The ATP Km is one of the most important parameters in drug discovery efforts involving protein kinases because, while uncompetitive inhibitors might be more specific on theoretical grounds, in practice the vast majority of kinase inhibitors that have been identified are ATP competitive. Therefore, to allow identification of diverse inhibitor types, we aimed to use conditions in which an equal amount of free and substrate-bound enzyme are present and the reaction was “balanced” (i.e. at Km for both substrates20). Initially, we determined the Km for ATP utilizing a saturating concentration of 1 μM H3(1-21)-biotin peptide in a radiometric assay (data not shown). Using this Km of ATP, the apparent Km of peptide was determined in the TR-FRET assay (Km H3(1-21) = 0.1 μM; Figure 3B). The apparent Km of ATP was then re-estimated at the Km concentration of peptide and was found to be 200 μM (Figure 3C).
Figure 3.
Optimization of the kinase reaction conditions and determination of apparent Km of substrates. A. Kinase reaction progress was monitored as an increase in TR-FRET ratio. Reactions were performed at Km concentrations of both the substrate peptide (100 nM) and ATP (200 μM), at different MBP-Haspin concentrations. B. The apparent Km for H3(1-21)-biotin peptide was determined by titration at a fixed concentration of 200 μM ATP. C. The apparent Km for ATP was determined by titration at a fixed concentration of 0.1 μM H3(1-21)-biotin. Means +/- standard deviations of are shown, n=3.
High Throughput Screening and Data Analysis
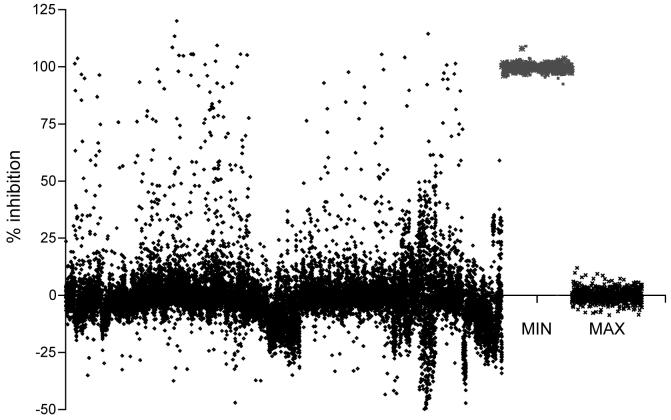
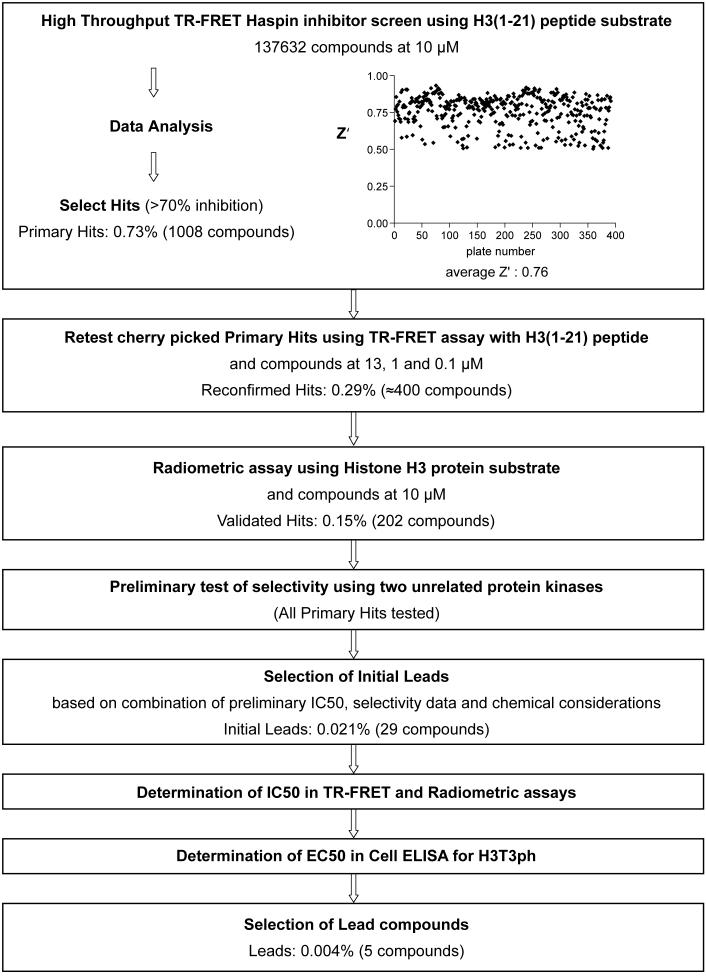
The finalized high throughput screen was carried out in 384-well plate format with a 10 μl reaction volume containing 0.05 nM MBP-Haspin, 0.1 μM H3(1-21)-biotin peptide and 200 μM ATP, incubated for 10 min at 25°C. Test compounds were included in single wells at a final concentration of 10 μM and 0.65% DMSO. In each plate, 16 wells contained 0.65% DMSO alone as the “MAX” control, and 16 wells contained 2 μM Staurosporine as the “MIN” control. Percentage inhibition was calculated as described in Materials and Methods. Compounds showing greater than 70% inhibition (1008 compounds) were defined as primary hits from a screen performed on a diverse chemical library of 137632 compounds (see Materials and Methods). Examples of the results from a representative one eighth of all screening plates are shown in Figure 4. Z’ values for each plate were calculated from the 16 MAX and 16 MIN readings according to Zhang et al.,18 to provide a statistical measure of assay robustness. An assay with a Z’ over 0.5 is usually considered suitable for high throughput screening, and screening of compound plates with Z’ values below 0.5 was repeated to yield Z’ values above 0.5 before they were included in the analysis. The average Z’ of the screen was 0.76 (see Figure 5).
Figure 4.
Primary TR-FRET screening data from fifty representative 384-well plates. The first 800 columns contain % inhibition data for 17,600 library compounds (only one eighth of the total is shown to maintain visual clarity). The final two columns show aggregated “MIN” controls (2 μM Staurosporine, defined as 100% inhibition) and “MAX” controls (DMSO alone; 0% inhibition) from the 50 plates.
Figure 5.
Flow chart of Haspin inhibitor lead discovery process also showing the distribution of Z’ values for plates in the TR-FRET high throughput screen.
Hit Confirmation and Characterization by TR-FRET and Radiometric Assay
The primary hits were cherry picked from 10 mM stock compound plates (see Figure 5 for a chart of the screening workflow). TR-FRET Haspin assays were then repeated at three concentrations of test compound (12.9 μM, 1.1 μM and 0.1 μM) using H3(1-21)-biotin peptide substrate to confirm inhibition and to obtain a preliminary estimate of potency. Approximately 400 of the compounds inhibited by 60% or more at 12.9 μM. Among the potential drawbacks of the homogeneous TR-FRET assay are the continuous presence of test compounds that can lead to compound interference in the detection step, and the use of histone peptides rather than protein substrate. We therefore assayed all 1008 primary hits at a compound concentration of 10 μM using a radiometric filter binding assay with recombinant Histone H3 as a protein substrate (see Materials and Methods). Compounds that resulted in greater then 60% inhibition of enzyme activity were considered positive (202 compounds) and further tested at 1 μM inhibitor concentration. All 1008 primary hit compounds were also tested at 10 μM concentration in TR-FRET assays for their inhibitory effect on two unrelated protein kinases (one tyrosine and one serine-threonine kinase) using appropriate substrates. Compounds that strongly inhibited these unrelated kinases were considered likely to be broad-spectrum kinase inhibitors and were not chosen for further evaluation. Based on a combination of these results we selected 29 confirmed leads to pursue. The number of initial leads constitutes 0.021% of the compound library.
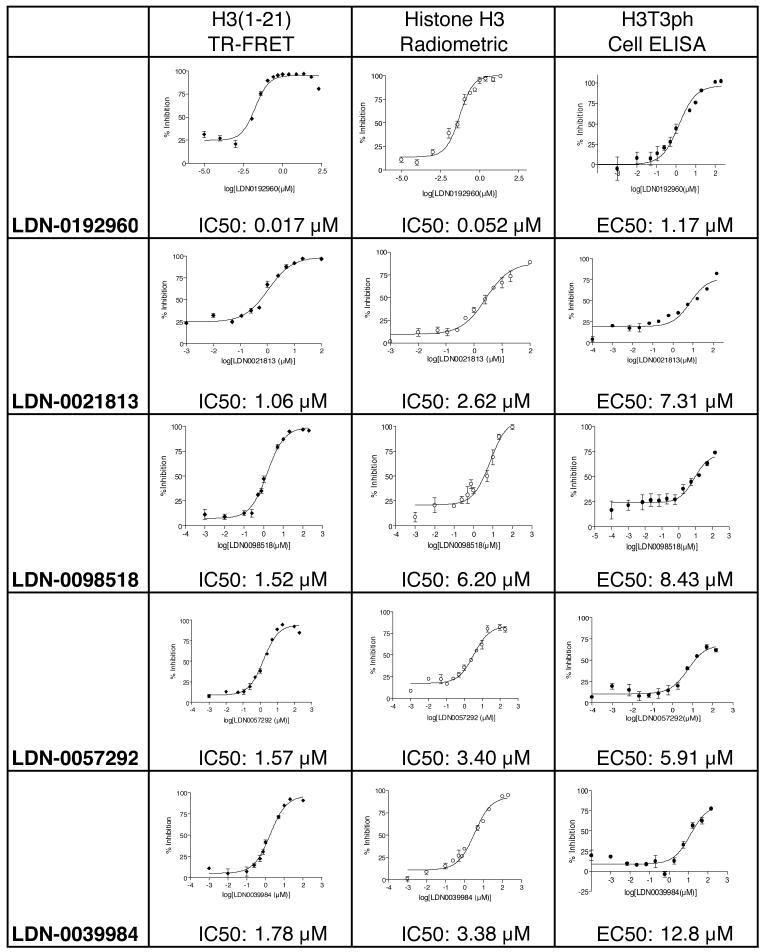
The 29 initial lead compounds were freshly obtained from their respective suppliers prior to further analysis. The dose dependence of MBP-Haspin inhibition for each compound was then determined in triplicate utilizing both TR-FRET and radiometric assays. Several compounds had IC50 values below 5 μM, and one lead compound (LDN-0192960) had IC50 values of 17 nM and 52 nM in the TR-FRET and radiometric assays respectively (Figure 6). In vitro selectivity testing revealed that 10 μM LDN-0192960 inhibited only 9 of 270 kinases by more than 90% when assayed at approximately Km concentrations of ATP. Of these, only 5 kinases were inhibited with an IC50 below 1 μM, indicating that LDN-0192960 is a relatively selective inhibitor.
Figure 6.
List of final lead compounds showing determination of IC50 concentrations in TR-FRET assays with H3(1-21) peptide substrate and in radiometric assays with Histone H3 protein substrate, and showing EC50 concentration determination in myc-Haspin-expressing HeLa Cell ELISA for H3T3ph. Means +/- standard deviations are shown, n=3.
Activity in a Cellular Assay
So far, all inhibitors had been tested solely on the isolated enzyme. To determine if the compounds could enter cells and inhibit Haspin, we wished to develop a medium throughput cellular assay for Haspin activity. Haspin phosphorylates Histone H3 at Thr-3 during mitosis. Because of this, assays of total cellular H3T3ph are limited in sensitivity by the low mitotic index of most cell lines. Furthermore, the generation of H3T3ph in cells is normally dependent upon entry into mitosis. Therefore, compounds that directly or indirectly prevent entry into mitosis may give “false-positive” results in assays for loss of H3T3ph, even if those compounds are incapable of inhibiting Haspin, limiting the selectivity of such approaches. To circumvent these problems, we made use of a stable HeLa cell line over-expressing myc-Haspin under an inducible promoter that we previously generated. Under conditions of Haspin over-expression, H3T3ph is seen throughout the cell cycle in these cells,5 yielding a robust signal that is independent of cell cycle stage. We therefore developed a 96-well plate cell-based ELISA for H3T3ph using these cells (see Materials and Methods). This assay allowed us to determine lead compound effectiveness in a physiologically relevant environment that simultaneously provided an indirect assessment of compound solubility, membrane permeability and cell retention. We identified 5 lead compounds that have EC50 values less than 10 μM when Haspin over-expressing cells were treated for 2 h. One compound, LDN-0192960 demonstrated a nanomolar IC50 value and had an EC50 of approximately 1 μM in the cell-based assay (Figure 6).
Conclusions
We have developed and utilized a high throughput TR-FRET assay with a peptide substrate to screen a small molecule library of nearly 140000 compounds for identifying Haspin kinase inhibitors. Primary hits (a total of 1008 compounds) that showed significant inhibition in a second TR-FRET assay (approximately 400 compounds) were validated in a radiometric assay using recombinant Histone H3 as the substrate (202 compounds). Both radiometric and TR-FRET assays were then used to determine accurate IC50 values, and cellular activity was assessed in a cell-based ELISA for phosphorylation of H3 at Thr-3. Twenty-nine initial lead compounds were chosen based on their potency in these three assay formats, and based on their failure to inhibit two unrelated kinases. Finally five lead compounds have been selected based on their potency and chemical tractability for medicinal chemistry optimization. These compounds belong to four different chemical classes, including two structurally diverse fused pyridine derivatives, a pyrrole derivative, an alkaloid natural product and a synthetic derivative of this alkaloid. The average molecular weight of the five compounds was 296 (range: 198-372) and the average clogP was 3.86 (range: 2.6-4.8). All of the lead compounds lacked reactive functional groups and were predicted to be reversible inhibitors. It may therefore be possible to develop structurally diverse Haspin inhibitors, increasing the likelihood of producing compounds with advantageous pharmacological profiles, and allowing increased confidence that observations made in cellular assays are due to the intended kinase target. Preliminary data indicates that at least one of the compounds is ATP-competitive and it seems likely that all of the compounds are similar in this respect. The lead compounds show a similar ratio of IC50 in the TR-FRET/H3 peptide assay to IC50 in the radiometric assay/H3 protein assay (IC50 TR-FRET/IC50 radiometric = 2.8 +/-0.9) that may simply reflect minor kinetic differences in these endpoint assays. There is more variation in ratio of IC50 in vitro versus EC50 in cells. Such variations may be expected, however, based on the likelihood that the compounds enter, accumulate, and are metabolized and transported out of cells with different efficiencies. Further development of these compounds will including testing for selectivity on a large panel of kinases, analysis of metabolic stability and of the persistence of H3T3ph suppression in cells, assessment of effects on cell proliferation and the cell cycle, determination of structure-activity relationships and chemical optimization for potency, specificity and ADMET parameters. This work has the potential to yield specific inhibitors of Haspin that will have clear utility for basic research and possible use as anti-mitotic anticancer therapeutics.
Acknowledgements
We thank Dr Simon Ng and Kevin Giroux of PerkinElmer for their assistance in setting up the TR-FRET assay and Sudeepa Sanyal of the Partners Center for Drug Discovery for preparing the compound plates for this screen. This work was supported by NIH/NCI R01CA122608 (to JMGH) and the Partners Center for Drug Discovery.
References
- 1.Jackson JR, Patrick DR, Dar MM, Huang PS. Targeted anti-mitotic therapies: can we improve on tubulin agents? Nat Rev Cancer. 2007;7:107–117. doi: 10.1038/nrc2049. [DOI] [PubMed] [Google Scholar]
- 2.Keen N, Taylor S. Aurora-kinase inhibitors as anticancer agents. Nat Rev Cancer. 2004;4:927–936. doi: 10.1038/nrc1502. [DOI] [PubMed] [Google Scholar]
- 3.Mountzios G, Terpos E, Dimopoulos MA. Aurora kinases as targets for cancer therapy. Cancer Treat Rev. 2008;34:175–182. doi: 10.1016/j.ctrv.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Strebhardt K, Ullrich A. Targeting polo-like kinase 1 for cancer therapy. Nat Rev Cancer. 2006;6:321–330. doi: 10.1038/nrc1841. [DOI] [PubMed] [Google Scholar]
- 5.Dai J, Sultan S, Taylor SS, Higgins JMG. The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes Dev. 2005;19:472–488. doi: 10.1101/gad.1267105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dai J, Higgins JMG. Haspin: a mitotic histone kinase required for metaphase chromosome alignment. Cell Cycle. 2005;4:665–668. doi: 10.4161/cc.4.5.1683. [DOI] [PubMed] [Google Scholar]
- 7.Dai J, Sullivan BA, Higgins JMG. Regulation of mitotic chromosome cohesion by Haspin and Aurora B. Dev Cell. 2006;11:741–750. doi: 10.1016/j.devcel.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 8.de Cárcer G, de Castro IP, Malumbres M. Targeting cell cycle kinases for cancer therapy. Curr Med Chem. 2007;14:969–985. doi: 10.2174/092986707780362925. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt M, Bastians H. Mitotic drug targets and the development of novel anti-mitotic anticancer drugs. Drug Resist Updat. 2007;10:162–181. doi: 10.1016/j.drup.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 10.Higgins JMG. The Haspin gene: location in an intron of the Integrin αE gene, associated transcription of an Integrin αE-derived RNA and expression in diploid as well as haploid cells. Gene. 2001;267:55–69. doi: 10.1016/s0378-1119(01)00387-0. [DOI] [PubMed] [Google Scholar]
- 11.Higgins JMG. Haspin-like proteins: A new family of evolutionarily conserved putative eukaryotic protein kinases. Prot Sci. 2001;10:1677–1684. doi: 10.1110/ps.49901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higgins JMG. Structure, function and evolution of haspin and haspin-related proteins, a distinctive group of eukaryotic protein kinases. Cell Mol Life Sci. 2003;60:446–462. doi: 10.1007/s000180300038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kannan N, Taylor SS, Zhai Y, Venter JC, Manning G. Structural and functional diversity of the microbial kinome. PLoS Biol. 2007;5:e17. doi: 10.1371/journal.pbio.0050017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor S, Peters JM. Polo and Aurora kinases: lessons derived from chemical biology. Curr Opin Cell Biol. 2008;20:77–84. doi: 10.1016/j.ceb.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 15.Mathis G. Probing molecular interactions with homogeneous techniques based on rare earth cryptates and fluorescence energy transfer. Clin Chem. 1995;41:1391–1397. [PubMed] [Google Scholar]
- 16.Hemmilä I. LANCE™: Homogeneous Assay Platform for HTS. J Biomol Screen. 1999;4:303–308. doi: 10.1177/108705719900400604. [DOI] [PubMed] [Google Scholar]
- 17.Mathis G. HTRF® Technology. J Biomol Screen. 1999;4:309–314. doi: 10.1177/108705719900400605. [DOI] [PubMed] [Google Scholar]
- 18.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 19.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 20.Case A, Ni J, Yeh LA, Stein RL. Development of a mechanism-based assay for tissue transglutaminase-results of a high-throughput screen and discovery of inhibitors. Anal Biochem. 2005;338:237–244. doi: 10.1016/j.ab.2004.09.047. [DOI] [PubMed] [Google Scholar]