Abstract
Purpose
To map the locus and identify the gene causing autosomal dominant congenital cataract (ADCC) with “snail-like” phenotype in a large Chinese family.
Methods
Clinical and ophthalmologic examinations were conducted on family members and documented by slit lamp photography. Linkage analysis was performed with an initial 41 microsatellite markers, then 3 additional markers flanking the major intrinsic protein (MIP) gene. Mutations were screened by DNA sequencing and verified by restriction fragment length polymorphism (RFLP) analysis.
Results
Significant two-point LOD scores were obtained at 5 markers flanking MIP with the highest 3.08 (θ=0.00) at marker D12S1632. Mutation screening of MIP identified a heterozygous G>A transition at the acceptor splice site of intron 3 (IVS3 −1 G>A), abolishing a BstSF I restriction site in one allele of all the affected individuals.
Conclusions
We identified a novel splice-site mutation (IVS3 −1 G>A in MIP) in a Chinese ADCC family. To our knowledge, this is the first report on an acceptor splice-site mutation in human genes associated with ADCC.
Introduction
Although surgical techniques and visual prognosis have been greatly improved in recent times, congenital cataracts remain the leading cause of visual disability in children worldwide. Without prompt treatment, cataracts can occlude clear imaging on the retina, resulting in failure to develop normal retinal-cortical synaptic connections and finally, irreversible amblyopia. Approximately 50% of congenital cataracts are inherited, with the most common being the autosomal dominant form [1]. To date, more than 25 independent loci and 17 cataract-related genes have been identified as being associated with isolated autosomal dominant congenital cataract (ADCC) [2]. These genes can be divided into 5 groups including: (1) Genes encoding crystallins: CRYAA, CRYAB, CRYBA1/A3, CRYBA4, CRYBB1, CRYBB2, CRYGC, CRYGD, and CRYGS [3-11]; (2) Genes encoding membrane transport and channel proteins: GJA3, GJA8, and MIP (also know as AQP0) [12-14]; (3) Genes encoding cytoskeletal proteins such as BFSP2 [15,16]; (4) Genes encoding transcription factors such as PITX3 and HSF4 [17,18]; and (5) Others: CHMP4B [19] and EPHA2 [20]. Most of the mutations detected in these genes are missense and nonsense mutations [21]. Few splice-site mutations have ever been reported associated with ADCC, except in CRYBA1/A3 and HSF4 [22-24]. Furthermore, to our knowledge, there is no report on an acceptor splice-site mutation in human genes associated with ADCC.
In this study, we identified MIP as the disease-causing gene in a four-generation Chinese family with ADCC by linkage analysis, and detected a novel G>A transition at the acceptor splice site of intron 3 of the MIP gene.
Methods
Family data and genomic DNA preparation
A four-generation family with ADCC was ascertained through the Eye Center of the 2nd Affiliated Hospital, Medical College of Zhejiang University, Hangzhou, China. Appropriate informed consent was obtained from all participants and the study protocol adhered to the principles of the Declaration of Helsinki. Twenty-two individuals (12 affected and 10 unaffected) from the family were enrolled in the study (Figure 1). Affected status was determined by a history of cataract extraction or ophthalmologic examination, including visual acuity, slit lamp, and fundus examination. The phenotypes were documented by slit lamp photography. Blood specimens (5 ml) from all the patients and available family members were collected in a BD Vacutainer® (BD Biosciences, San Jose, CA) containing EDTA. Genomic DNA was isolated as previously described [25]. Mutation nomenclature follows the guidelines of the Human Genome Variation Society (HGV) with the numbering based on +1 as the A of the ATG translation initiation codon in the reference sequence. The initiation codon is codon 1.
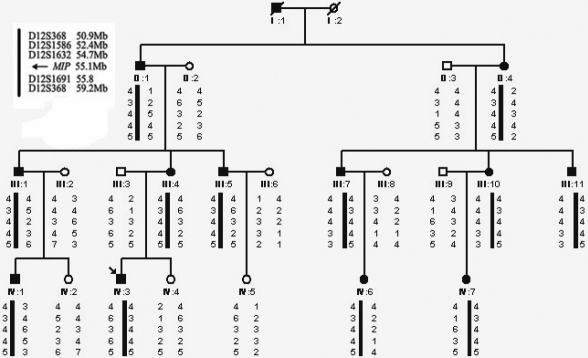
Figure 1.
Pedigree of the Chinese cataract family and haplotype analysis. Squares and circles indicate males and females, respectively. Solid and open symbols denote affected and unaffected individuals, respectively. Haplotype analysis shows the segregation of five microsatellite markers on chromosome 12. The physical distance of microsatellite markers and the disease gene have been given in the top left corner. The haplotype of the disease-bearing chromosome is indicated by black bar.
Genotyping and linkage analysis
Genotyping was performed as described previously, using the initial 41 microsatellite markers, corresponding to 18 known candidate loci for ADCC [23,26], and then another 3 markers localized to 12q13. Two-point disease to marker linkage analysis was conducted by the MLINK routine of the LINKAGE software package, version 5.1. The disease locus was specified to be an autosomal dominant trait with a disease allele frequency of 0.0001. The allele frequencies for each marker were assumed to be equal as were the recombination frequencies in males and females. Genetic penetrance was assigned to be full.
PCR and DNA sequencing
Gene specific PCR primers for MIP were designed flanking each exon and intron-exon junction (Table 1). The cycling conditions for PCR were as follows: 95 °C preactivation for 5 min, 10 cycles of touchdown PCR with 0.5 °C down per cycle from 62 °C to 57 °C, followed by 25 cycles with denaturation at 94 °C for 45 s, annealing at 58 °C for 45 s and extension at 72 °C for 45 s. PCR products were isolated by electrophoresis on 3% agarose gels and sequenced using the BigDye Terminator Cycle sequencing kit V 3.1(ABI Applied Biosystems; Sangon Co., Shanghai, China) on an ABI PRISM 3730 Sequence Analyzer (ABI), according to the manufacturer’s directions.
Table 1. Primers and product sizes of MIP.
| Name | Primer | Product size (bp) |
|---|---|---|
| exon 1F |
5′-GACTGTCCACCCAGACAAGG-3′ |
492 |
| exon 1R |
5′-TCAGGGAGTCAGGGCAATAG-3′ |
|
| exon 2F |
5′-TGAAGGAGCACTGTTAGGAGATG-3′ |
500 |
| exon 2R |
5′-AGAGGGATAGGGCAGAGTTGATT-3′ |
|
| exon 3F |
5′-CCAGACAGGGCATCAGT-3′ |
373 |
| exon 3R |
5′-TGGTACAGCAGCCAACAC-3′ |
|
| exon 4F |
5′-AAGGTGTGGGATAAAGGAGT-3′ |
429 |
| exon 4R | 5′-TTCTTCATCTAGGGGCTGGC-3′ |
Restriction fragment length polymorphism (RFLP) analysis
After identifying an acceptor splice-site mutation in the intron 3-exon 4 junction, all family members and 100 unrelated control individuals were examined by RFLP analysis. The mutation abolished a BstSF I site. PCR products of exon 4 of MIP were digested for 1 h at 60 °C with BstSF I (Bio Basic Inc., Markham, Canada) and separated on a 3% agarose gel by electrophoresis.
Results
Clinical evaluation
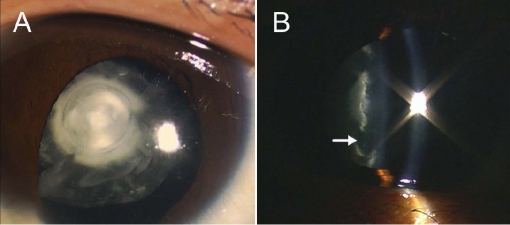
We identified a four-generation Chinese family with clear diagnosis of ADCC. Opacification of the lens was bilateral in all the affected individuals. Most of the patients had nystagmus with visual acuity ranging from hand move to 15/60 in the unoperated eyes. There was no family history of other ocular or systemic abnormalities. The lens opacification of the proband was very unique giving an appearance of a snail, with opacity density gradually increasing from the peripheral adult nucleus to the inner embryonal nucleus. The cortex remained transparent and the nuclei were separated by a transparent circle (Figure 2).
Figure 2.
Photographs of the left eye of the proband with congenital cataract. A: Diffuse illumination shows a “snail-like” cataract with opacity density gradually increased from the peripheral adult nucleus to the inner embryonal nucleus, while the cortex remains transparent. B: Slit section shows that the opacified nuclei are separated by a transparent circle (white arrow).
Linkage analysis
After the other candidate loci were excluded, positive two-point LOD scores were obtained at markers D12S368 (Zmax=1.66 at θ=0.0) and D12S83 (Zmax=2.01 at θ=0.0). MIP was flanked by these two markers. Therefore, three additional markers very near to MIP were subsequently used for further confirmation. All of these 5 markers received significant scores, and the maximum score was obtained with marker D12S1632 (Zmax=3.08 at θ=0.0; Table 2).
Table 2. Two-point LOD scores for linkage between autosomal dominant congenital cataract locus and chromosome 12 markers.
| Markers and MIP | Physical distance (Mbp) |
LOD scores by recombination fraction (θ) |
|||||
|---|---|---|---|---|---|---|---|
| 0 | 0.1 | 0.2 | 0.3 | 0.4 | 0.5 | ||
| D12S368 |
50.9177–50.9179 |
1.66 |
1.32 |
0.95 |
0.56 |
0.19 |
0 |
| D12S1586 |
52.4330–52.4333 |
2.78 |
2.18 |
1.57 |
0.96 |
0.42 |
0 |
| D12S1632 |
54.7016–54.7019 |
3.08 |
2.52 |
1.91 |
1.24 |
0.55 |
0 |
|
MIP |
55.1300–55.1346 |
||||||
| D12S1691 |
55.7920–55.7923 |
2.23 |
1.85 |
1.44 |
1.00 |
0.53 |
0 |
| D12S83 | 59.1756–59.1759 | 2.01 | 1.62 | 1.21 | 0.77 | 0.36 | 0 |
Mutation analysis
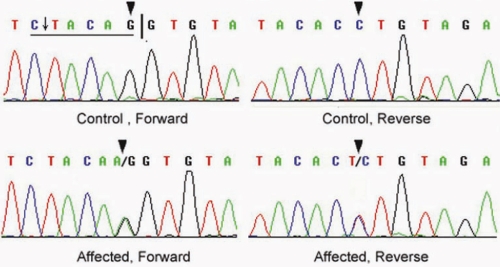
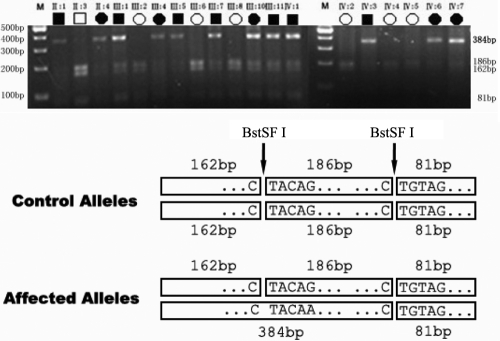
By sequencing the PCR products of MIP, we identified a single base substitution in the acceptor splice site of intron 3 (IVS3 −1 G>A) which cosegregated with all affected individuals, whereas this heterozygous mutation was not present in the unaffected family members (Figure 3). The IVS3 −1 G>A mutation changed the canonical 3′ acceptor splice site of intron 3 from AG to AA, resulting in a BstSF I restriction site abolishment. RFLP analysis verified the mutation and showed it cosegregation with all affected individuals. The mutation was not detected in unaffected family members and 100 unrelated Chinese without cataract as control (Figure 4).
Figure 3.
Forward and reverse sequence analysis of the affected and unaffected individuals in this ADCC Chinese family. It shows a heterozygous mutation (IVS3–1 G>A) in the third canonical AG sites of MIP (black triangles). The black vertical line denotes the normal intron 3-exon 4 acceptor splice site. The mutation IVS3 −1 G>A abolishes a BstSF I site (underlined) which is enzymatic cut indicated by the arrow.
Figure 4.
Restriction fragment length polymorphism (RFLP) analysis showing that the abolishment of a BstSF I site cosegregates with affected individuals. The PCR product of exon 4 with flanking sequences includes 429 bp with 2 BstSF I sites (CTACAG and CTGTAG). The unaffected has three fragments (81 bp, 162 bp, and 186 bp) after BstSF I digestion, whereas the affected has four (81 bp, 162 bp, 186 bp, and the crucial 348 bp). Only the affected allele shows the 384bp band. M means DNA ladder marker.
Discussion
In this report, we first identified an acceptor splice-site mutation (IVS3 −1 G>A in MIP) associated with ADCC in a four-generation Chinese family. To date, five other mutations in MIP have been identified from five unrelated human families (c.413C>G, c.401A>G, c.638delG, c.97C>T, and c.702G>A). Individuals with c.413C>G, c.638delG, and c.702G>A mutations have polymorphic cataracts [14,27-29]. The c.401A>G mutation causes nonprogressive lamellar cataract with sutural opacities and c.97C>T mutation causes total cataract [14,29]. In those affected with the IVS3 −1 G>A mutation, a unique cataract phenotype was observed. The proband demonstrates a “snail-like” cataract with opacity density gradually increasing from the peripheral adult nucleus to the inner embryonal nucleus, while the cortex remains transparent. It indicates that the dysfunction of AQP0 has a more severe impact on the embryonic and fetal nucleus than the adult nucleus of the lens, consistent with different stages of AQP0 expression.
Although several cases of splice sites with GT-TG, GT-CG, GC-AG, GG-AG, CT-AG, or AT-AC dinucleotides at the splice junctions were observed, the GT-AG rule is always obeyed [30]. It is reported that 87% of the 3′ splice-site mutations involved the invariant AG dinucleotide [31]. As for MIP, the canonical AG sites in intron 3 are conserved among different species (Figure 5). Splice-site mutations were reported to result in exon skipping, activation of cryptic splice sites, creation of a pseudo-exon within an intron, or intron retention, among which exon skipping is the most frequent outcome [32]. Mutations in acceptor splice sites can result either in use of the acceptor site of the next intron, with consequent loss of exon skipping, or in the utilization of a cryptic acceptor splice site upstream the mutation sites or in the next exon [33]. In our present study, the mutation occurred in the invariant AG dinucleotide of the last intron, so no existing canonical AG sites downstream are available to be used as the alternative acceptor splice site. Therefore, one or more cryptic acceptor splice sites are supposed to be used for aberrant splicing. According to the NNSPLICE program [34], we detected 8 possible cryptic acceptor splice sites scored more than 0.4 in intron 3 and exon 4. They were respectively located at +1599nt (0.81), +1758nt (0.52), +1850nt (0.52), +2588nt (0.64), +2735nt (0.92), +3040nt (0.44), +3074nt (0.88), and +3148nt (0.44) of MIP. Further study is required to confirm the above sites.
Figure 5.
Multiple-sequence alignment of acceptor splice site in intron 3 of MIP from different species. It reveals that the canonical AG sites are conserved among different species.
Several previous studies have demonstrated that the AQP0 COOH-terminus is very crucial to lens development and transparency through interactions with calmodulin, cytoskeletal proteins filensin and CP49, and connexin 45.6 [35-37]. Cleavage of the intracellular COOH-terminus decreases water permeability and enhances the adhesive properties of the extracellular surface of AQP0, indicating a conformational change in the molecule [38,39]. The possible aberrant splicing of MIP pre-mRNA may disrupt the normal COOH-terminus of AQP0, and leads to disbalance of the lens internal homeostasis, which is necessary to maintain transparency, and finally results in cataract formation. In addition, mutation analysis of AQP0 transcripts from the CatFr lens indicated that the CatFr mutation resulted in substitution of a long-terminal repeat sequence for the COOH-terminus of Mip (AQP0-LTR) [40]. AQP0-LTR in CatFr was accumulated in sub-cellular compartments and made mature fiber cells fail to stratify into uniform, concentric growth, and finally resulted in congenital cataract [41,42].
In conclusion, we first describe the identification of an acceptor splice-site mutation in human genes (IVS3 −1 G>A in MIP) associated with ADCC, characterized by “snail-like” cataract phenotype. Further investigation is required to elucidate the pathogenesis of the novel splice-site mutation of the MIP gene on cataract formation.
Acknowledgments
The authors thank the patients and their family members for their cooperation; Professor Ming Qi at Zhejiang University-Adinovo Center for Genetic & Genomic Medicine, and University of Rochester Medical Center for his critical reading of this manuscript. This work was supported by Key Projects in the National Science & Technology Pillar Program in the Eleventh Five-year Plan Period (2006BAI02B04).
References
- 1.Hejtmancik JF, Smaoui N. Molecular genetics of cataract. Dev Ophthalmol. 2003;37:67–82. doi: 10.1159/000072039. [DOI] [PubMed] [Google Scholar]
- 2.Shiels A, Hejtmancik JF. Genetic origins of cataract. Arch Ophthalmol. 2007;125:165–73. doi: 10.1001/archopht.125.2.165. [DOI] [PubMed] [Google Scholar]
- 3.Litt M, Kramer P, LaMorticella DM, Murphey W, Lovrien EW, Weleber RG. Autosomal dominant congenital cataract associated with a missense mutation in the human alpha crystallin gene CRYAA. Hum Mol Genet. 1998;7:471–4. doi: 10.1093/hmg/7.3.471. [DOI] [PubMed] [Google Scholar]
- 4.Berry V, Francis P, Reddy MA, Collyer D, Vithana E, MacKay I, Dawson G, Carey AH, Moore A, Bhattacharya SS, Quinlan RA. Alpha-B crystallin gene (CRYAB) mutation causes dominant congenital posterior polar cataract in humans. Am J Hum Genet. 2001;69:1141–5. doi: 10.1086/324158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kmoch S, Brynda J, Asfaw B, Bezouska K, Novak P, Rezacova P, Ondrova L, Filipec M, Sedlacek J, Elleder M. Link between a novel human gammaD-crystallin allele and a unique cataract phenotype explained by protein crystallography. Hum Mol Genet. 2000;9:1779–86. doi: 10.1093/hmg/9.12.1779. [DOI] [PubMed] [Google Scholar]
- 6.Billingsley G, Santhiya ST, Paterson AD, Ogata K, Wodak S, Hosseini SM, Manisastry SM, Vijayalakshmi P, Gopinath PM, Graw J, Heon E. CRYBA4, a novel human cataract gene, is also involved in microphthalmia. Am J Hum Genet. 2006;79:702–9. doi: 10.1086/507712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun H, Ma Z, Li Y, Liu B, Li Z, Ding X, Gao Y, Ma W, Tang X, Li X, Shen Y. Gamma-S crystallin gene (CRYGS) mutation causes dominant progressive cortical cataract in humans. J Med Genet. 2005;42:706–10. doi: 10.1136/jmg.2004.028274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Padma T, Ayyagari R, Murty JS, Basti S, Fletcher T, Rao GN, Kaiser-Kupfer M, Hejtmancik JF. Autosomal dominant zonular cataract with sutural opacities localized to chromosome 17q11–12. Am J Hum Genet. 1995;57:840–5. [PMC free article] [PubMed] [Google Scholar]
- 9.Mackay DS, Boskovska OB, Knopf HL, Lampi KJ, Shiels A. A nonsense mutation in CRYBB1 associated with autosomal dominant cataract linked to human chromosome 22q. Am J Hum Genet. 2002;71:1216–21. doi: 10.1086/344212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Litt M, Carrero-Valenzuela R, LaMorticella DM, Schultz DW, Mitchell TN, Kramer P, Maumenee IH. Autosomal dominant cerulean cataract is associated with a chain termination mutation in the human beta-crystallin gene CRYBB2. Hum Mol Genet. 1997;6:665–8. doi: 10.1093/hmg/6.5.665. [DOI] [PubMed] [Google Scholar]
- 11.Heon E, Priston M, Schorderet DF, Billingsley GD, Girard PO, Lubsen N, Munier FL. The gamma-crystallins and human cataracts: a puzzle made clearer. Am J Hum Genet. 1999;65:1261–7. doi: 10.1086/302619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mackay D, Ionides A, Kibar Z, Rouleau G, Berry V, Moore A, Shiels A, Bhattacharya S. Connexin46 mutations in autosomal dominant congenital cataract. Am J Hum Genet. 1999;64:1357–64. doi: 10.1086/302383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shiels A, Mackay D, Ionides A, Berry V, Moore A, Bhattacharya S. A missense mutation in the human connexin50 gene (GJA8) underlies autosomal dominant “zonular pulverulent” cataract, on chromosome 1q. Am J Hum Genet. 1998;62:526–32. doi: 10.1086/301762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berry V, Francis P, Kaushal S, Moore A, Bhattacharya S. Missense mutations in MIP underlie autosomal dominant 'polymorphic' and lamellar cataracts linked to 12q. Nat Genet. 2000;25:15–7. doi: 10.1038/75538. [DOI] [PubMed] [Google Scholar]
- 15.Conley YP, Erturk D, Keverline A, Mah TS, Keravala A, Barnes LR, Bruchis A, Hess JF, FitzGerald PG, Weeks DE, Ferrell RE, Gorin MB. A juvenile-onset, progressive cataract locus on chromosome 3q21-q22 is associated with a missense mutation in the beaded filament structural protein-2. Am J Hum Genet. 2000;66:1426–31. doi: 10.1086/302871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jakobs PM, Hess JF, FitzGerald PG, Kramer P, Weleber RG, Litt M. Autosomal-dominant congenital cataract associated with a deletion mutation in the human beaded filament protein gene BFSP2. Am J Hum Genet. 2000;66:1432–6. doi: 10.1086/302872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Semina EV, Ferrell RE, Mintz-Hittner HA, Bitoun P, Alward WL, Reiter RS, Funkhauser C, Daack-Hirsch S, Murray JC. A novel homeobox gene PITX3 is mutated in families with autosomal-dominant cataracts and ASMD. Nat Genet. 1998;19:167–70. doi: 10.1038/527. [DOI] [PubMed] [Google Scholar]
- 18.Bu L, Jin Y, Shi Y, Chu R, Ban A, Eiberg H, Andres L, Jiang H, Zheng G, Qian M, Cui B, Xia Y, Liu J, Hu L, Zhao G, Hayden MR, Kong X. Mutant DNA-binding domain of HSF4 is associated with autosomal dominant lamellar and Marner cataract. Nat Genet. 2002;31:276–8. doi: 10.1038/ng921. [DOI] [PubMed] [Google Scholar]
- 19.Shiels A, Bennett TM, Knopf HL, Yamada K, Yoshiura K, Niikawa N, Shim S, Hanson PI. CHMP4B, a novel gene for autosomal dominant cataracts linked to chromosome 20q. Am J Hum Genet. 2007;81:596–606. doi: 10.1086/519980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shiels A, Bennett TM, Knopf HL, Maraini G, Li A, Jiao X, Hejtmancik JF. The EPHA2 gene is associated with cataracts linked to chromosome 1p. Mol Vis. 2008;14:2042–55. [PMC free article] [PubMed] [Google Scholar]
- 21.Hejtmancik JF. Congenital cataracts and their molecular genetics. Semin Cell Dev Biol. 2008;19:134–49. doi: 10.1016/j.semcdb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bateman JB, Geyer DD, Flodman P, Johannes M, Sikela J, Walter N, Moreira AT, Clancy K, Spence MA. A new betaA1-crystallin splice junction mutation in autosomal dominant cataract. Invest Ophthalmol Vis Sci. 2000;41:3278–85. [PubMed] [Google Scholar]
- 23.Kannabiran C, Rogan PK, Olmos L, Basti S, Rao GN, Kaiser-Kupfer M, Hejtmancik JF. Autosomal dominant zonular cataract with sutural opacities is associated with a splice mutation in the betaA3/A1-crystallin gene. Mol Vis. 1998;4:21. [PubMed] [Google Scholar]
- 24.Smaoui N, Beltaief O, BenHamed S, M'Rad R, Maazoul F, Ouertani A, Chaabouni H, Hejtmancik JF. A homozygous splice mutation in the HSF4 gene is associated with an autosomal recessive congenital cataract. Invest Ophthalmol Vis Sci. 2004;45:2716–21. doi: 10.1167/iovs.03-1370. [DOI] [PubMed] [Google Scholar]
- 25.Jin C, Yao K, Jiang J, Tang X, Shentu X, Wu R. Novel FBN1 mutations associated with predominant ectopia lentis and marfanoid habitus in Chinese patients. Mol Vis. 2007;13:1280–4. [PubMed] [Google Scholar]
- 26.Yao K, Tang X, Shentu X, Wang K, Rao H, Xia K. Progressive polymorphic congenital cataract caused by a CRYBB2 mutation in a Chinese family. Mol Vis. 2005;11:758–63. [PubMed] [Google Scholar]
- 27.Geyer DD, Spence MA, Johannes M, Flodman P, Clancy KP, Berry R, Sparkes RS, Jonsen MD, Isenberg SJ, Bateman JB. Novel single-base deletional mutation in major intrinsic protein (MIP) in autosomal dominant cataract. Am J Ophthalmol. 2006;141:761–3. doi: 10.1016/j.ajo.2005.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin H, Hejtmancik JF, Qi Y. A substitution of arginine to lysine at the COOH-terminus of MIP caused a different binocular phenotype in a congenital cataract family. Mol Vis. 2007;13:1822–7. [PubMed] [Google Scholar]
- 29.Gu F, Zhai H, Li D, Zhao L, Li C, Huang S, Ma X. A novel mutation in major intrinsic protein of the lens gene (MIP) underlies autosomal dominant cataract in a Chinese family. Mol Vis. 2007;13:1651–6. [PubMed] [Google Scholar]
- 30.Burset M, Seledtsov IA, Solovyev VV. Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Res. 2000;28:4364–75. doi: 10.1093/nar/28.21.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krawczak M, Reiss J, Cooper DN. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum Genet. 1992;90:41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- 32.Nakai K, Sakamoto H. Construction of a novel database containing aberrant splicing mutations of mammalian genes. Gene. 1994;141:171–7. doi: 10.1016/0378-1119(94)90567-3. [DOI] [PubMed] [Google Scholar]
- 33.Sappal BS, Ghosh SS, Shneider B, Kadakol A, Chowdhury JR, Chowdhury NR. A novel intronic mutation results in the use of a cryptic splice acceptor site within the coding region of UGT1A1, causing Crigler-Najjar syndrome type 1. Mol Genet Metab. 2002;75:134–42. doi: 10.1006/mgme.2001.3284. [DOI] [PubMed] [Google Scholar]
- 34.Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J Comput Biol. 1997;4:311–23. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 35.Rose KM, Wang Z, Magrath GN, Hazard ES, Hildebrandt JD, Schey KL. Aquaporin 0-calmodulin interaction and the effect of aquaporin 0 phosphorylation. Biochemistry. 2008;47:339–47. doi: 10.1021/bi701980t. [DOI] [PubMed] [Google Scholar]
- 36.Lindsey Rose KM, Gourdie RG, Prescott AR, Quinlan RA, Crouch RK, Schey KL. The C-terminus of lens aquaporin 0 interacts with the cytoskeletal proteins filensin and CP49. Invest Ophthalmol Vis Sci. 2006;47:1562–70. doi: 10.1167/iovs.05-1313. [DOI] [PubMed] [Google Scholar]
- 37.Yu XS, Yin X, Lafer EM, Jiang JX. Developmental regulation of the direct interaction between the intracellular loop of connexin 45.6 and the C-terminus of major intrinsic protein (aquaporin-0). J Biol Chem. 2005;280:22081–90. doi: 10.1074/jbc.M414377200. [DOI] [PubMed] [Google Scholar]
- 38.Ball LE, Little M, Nowak MW, Garland DL, Crouch RK, Schey KL. Water permeability of C-terminally truncated aquaporin 0 (AQP0 1–243) observed in the aging human lens. Invest Ophthalmol Vis Sci. 2003;44:4820–8. doi: 10.1167/iovs.02-1317. [DOI] [PubMed] [Google Scholar]
- 39.Gonen T, Cheng Y, Kistler J, Walz T. Aquaporin-0 membrane junctions form upon proteolytic cleavage. J Mol Biol. 2004;342:1337–45. doi: 10.1016/j.jmb.2004.07.076. [DOI] [PubMed] [Google Scholar]
- 40.Shiels A, Bassnett S. Mutations in the founder of the MIP gene family underlie cataract development in the mouse. Nat Genet. 1996;12:212–5. doi: 10.1038/ng0296-212. [DOI] [PubMed] [Google Scholar]
- 41.Shiels A, Mackay D, Bassnett S, Al-Ghoul K, Kuszak J. Disruption of lens fiber cell architecture in mice expressing a chimeric AQP0-LTR protein. FASEB J. 2000;14:2207–12. doi: 10.1096/fj.99-1071com. [DOI] [PubMed] [Google Scholar]
- 42.Kalman K, Nemeth-Cahalan KL, Froger A, Hall JE. AQP0-LTR of the Cat Fr mouse alters water permeability and calcium regulation of wild type AQP0. Biochim Biophys Acta. 2006;1758:1094–9. doi: 10.1016/j.bbamem.2006.01.015. [DOI] [PubMed] [Google Scholar]