Abstract
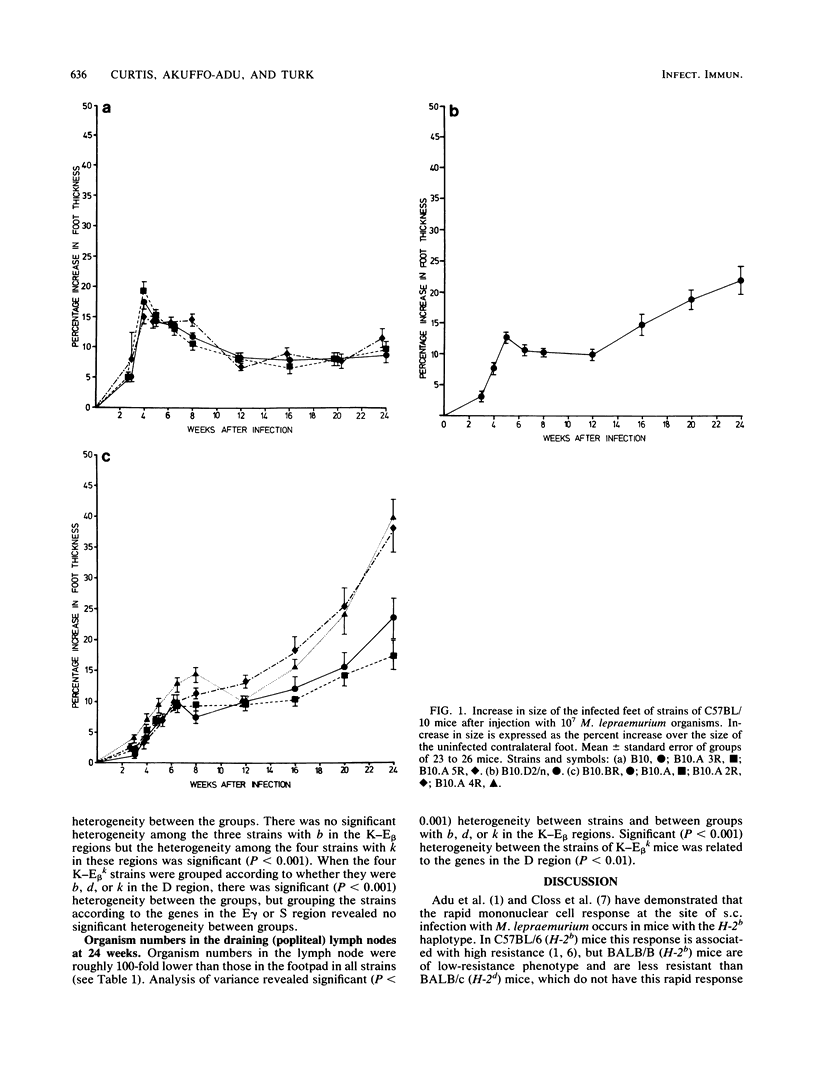
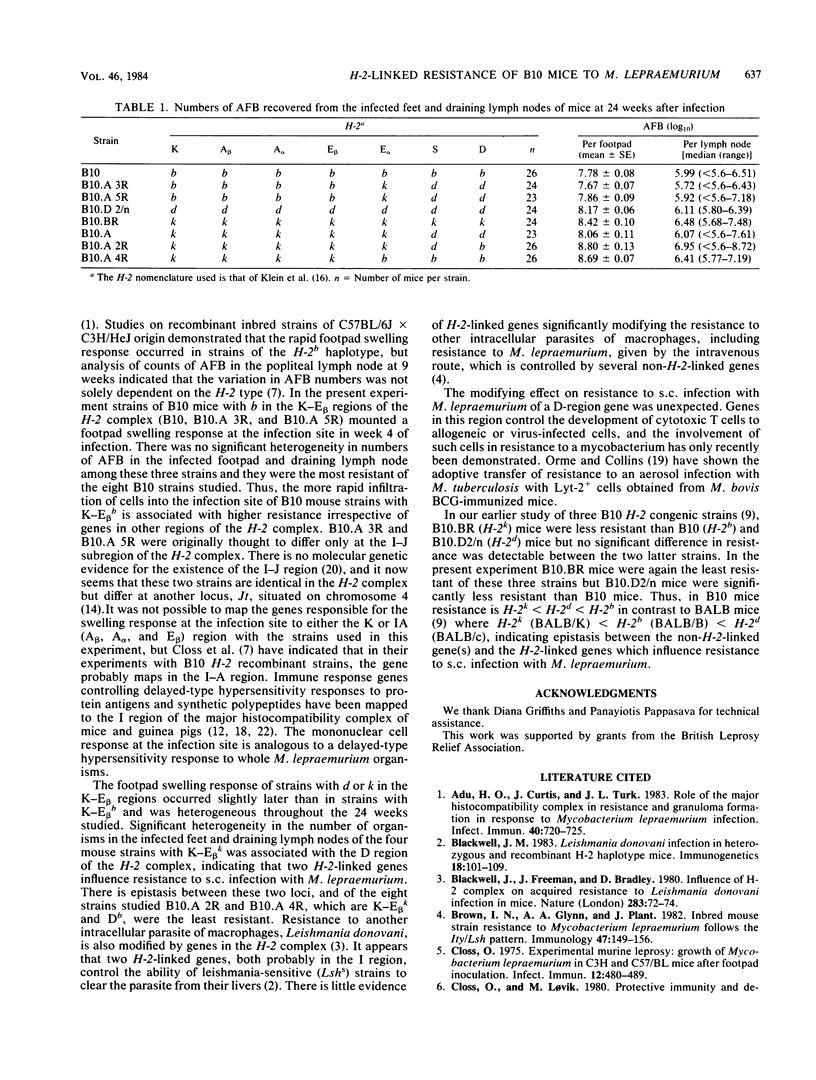
Strains of C57BL/10 mice with recombinants within the H-2 complex were used to map the genes which control the mononuclear cell response at the infection site and modify resistance to subcutaneous infection with Mycobacterium lepraemurium. Strains with b in the K-E beta regions of the H-2 complex mounted a more rapid cellular response in the infected footpad and were more resistant than mice with d or k in the K-E beta regions. Significant differences between strains with k in the K-E beta regions appeared to be controlled by a gene in the D region.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adu H. O., Curtis J., Turk J. L. Role of the major histocompatibility complex in resistance and granuloma formation in response to Mycobacterium lepraemurium infection. Infect Immun. 1983 May;40(2):720–725. doi: 10.1128/iai.40.2.720-725.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell J. M. Leishmania donovani infection in heterozygous and recombinant H-2 haplotype mice. Immunogenetics. 1983;18(2):101–109. doi: 10.1007/BF00368537. [DOI] [PubMed] [Google Scholar]
- Blackwell J., Freeman J., Bradley D. Influence of H-2 complex on acquired resistance to Leishmania donovani infection in mice. Nature. 1980 Jan 3;283(5742):72–74. doi: 10.1038/283072a0. [DOI] [PubMed] [Google Scholar]
- Brown I. N., Glynn A. A., Plant J. Inbred mouse strain resistance to Mycobacterium lepraemurium follows the Ity/Lsh pattern. Immunology. 1982 Sep;47(1):149–156. [PMC free article] [PubMed] [Google Scholar]
- Closs O. Experimental murine leprosy: growth of Mycobacterium lepraemurium in C3H and C57/BL mice after footpad inoculation. Infect Immun. 1975 Sep;12(3):480–489. doi: 10.1128/iai.12.3.480-489.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Closs O., Løvik M., Wigzell H., Taylor B. A. H-2-linked gene(s) influence the granulomatous reaction to viable Mycobacterium lepraemurium in the mouse. Scand J Immunol. 1983 Jul;18(1):59–63. doi: 10.1111/j.1365-3083.1983.tb00836.x. [DOI] [PubMed] [Google Scholar]
- Curtis J., Adu H. O., Turk J. L. A lack of correlation between antigen-specific cellular reactions and resistance to Mycobacterium lepraemurium infection in mice. Immunology. 1981 Jun;43(2):293–301. [PMC free article] [PubMed] [Google Scholar]
- Curtis J., Adu H. O., Turk J. L. H-2 linkage control of resistance to subcutaneous infection with Mycobacterium lepraemurium. Infect Immun. 1982 Nov;38(2):434–439. doi: 10.1128/iai.38.2.434-439.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis J., Turk J. L. Resistance to subcutaneous infection with Mycobacterium lepraemurium is controlled by more than one gene. Infect Immun. 1984 Mar;43(3):925–930. doi: 10.1128/iai.43.3.925-930.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper P. The walls of Mycobacterium lepraemurium: chemistry and ultrastructure. J Gen Microbiol. 1971 Dec;69(3):313–324. doi: 10.1099/00221287-69-3-313. [DOI] [PubMed] [Google Scholar]
- Green I., Paul W. E., Benacerraf B. A study of the passive transfer of delayed hypersensitivity to DNP-poly-L-lysine and DNP-GL in responder and nonresponder guinea pigs. J Exp Med. 1967 Nov 1;126(5):959–967. doi: 10.1084/jem.126.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HART P. D., REES R. J. Effect of macrocyclon in acute and chronic pulmonary tuberculous infection in mice as shown by viable and total bacterial counts. Br J Exp Pathol. 1960 Aug;41:414–421. [PMC free article] [PubMed] [Google Scholar]
- Hayes C. E., Klyczek K. K., Krum D. P., Whitcomb R. M., Hullett D. A., Cantor H. Chromosome 4 Jt gene controls murine T cell surface I-J expression. Science. 1984 Feb 10;223(4636):559–563. doi: 10.1126/science.6607530. [DOI] [PubMed] [Google Scholar]
- Jarnagin J. L., Luchsinger D. W. The use of fluorescein diacetate and ethidium bromide as a stain for evaluating viability of mycobacteria. Stain Technol. 1980 Jul;55(4):253–258. doi: 10.3109/10520298009067249. [DOI] [PubMed] [Google Scholar]
- Klein J., Figueroa F., David C. S. H-2 haplotypes, genes and antigens: second listing. II. The H-2 complex. Immunogenetics. 1983;17(6):553–596. doi: 10.1007/BF00366126. [DOI] [PubMed] [Google Scholar]
- Lagrange P. H., Hurtrel B. Local immune response to Mycobacterium lepraemurium in C3H and C57Bl/6 mice. Clin Exp Immunol. 1979 Dec;38(3):461–474. [PMC free article] [PubMed] [Google Scholar]
- Miller J. F., Vadas M. A., Whitelaw A., Gamble J., Bernard C. Histocompatibility linked immune responsiveness and restrictions imposed on sensitized lymphocytes. J Exp Med. 1977 Jun 1;145(6):1623–1628. doi: 10.1084/jem.145.6.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orme I. M., Collins F. M. Adoptive protection of the Mycobacterium tuberculosis-infected lung. Dissociation between cells that passively transfer protective immunity and those that transfer delayed-type hypersensitivity to tuberculin. Cell Immunol. 1984 Mar;84(1):113–120. doi: 10.1016/0008-8749(84)90082-0. [DOI] [PubMed] [Google Scholar]
- Steinmetz M., Minard K., Horvath S., McNicholas J., Srelinger J., Wake C., Long E., Mach B., Hood L. A molecular map of the immune response region from the major histocompatibility complex of the mouse. Nature. 1982 Nov 4;300(5887):35–42. doi: 10.1038/300035a0. [DOI] [PubMed] [Google Scholar]
- Turcotte R. Influence of route of Mycobacterium lepraemurium injection on susceptibility to mouse leprosy and on lymphoblastic transformation. Infect Immun. 1980 Jun;28(3):660–668. doi: 10.1128/iai.28.3.660-668.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]


