Abstract
One of the most remarkable structural aspects of Scapharca dimeric hemoglobin is the disruption of a very well-ordered water cluster at the subunit interface upon ligand binding. We have explored the role of these crystallographically observed water molecules by site-directed mutagenesis and osmotic stress techniques. The isosteric mutation of Thr-72 → Val in the interface increases oxygen affinity more than 40-fold with a surprising enhancement of cooperativity. The only significant structural effect of this mutation is to destabilize two ordered water molecules in the deoxy interface. Wild-type Scapharca hemoglobin is strongly sensitive to osmotic conditions. Upon addition of glycerol, striking changes in Raman spectrum of the deoxy form are observed that indicate a transition toward the liganded form. Increased osmotic pressure, which lowers the oxygen affinity in human hemoglobin, raises the oxygen affinity of Scapharca hemoglobin regardless of whether the solute is glycerol, glucose, or sucrose. Analysis of these results provides an estimate of six water molecules lost upon oxygen binding to the dimer, in good agreement with eight predicted from crystal structures. These experiments suggest that the observed cluster of interfacial water molecules plays a crucial role in communication between subunits.
Water plays a unique and ubiquitous role in biology. Folding, stability, and function of protein molecules are all strongly influenced by their interactions with water molecules. Despite the importance of these interactions, the precise role of water in structure and function is often difficult to elucidate. Ordered water molecules are nearly universally observed in macromolecular crystallographic analysis (1, 2), and biologically important roles for these observed water molecules have been implicated in a few cases including substrate specificity (3), catalysis (4), antigen–antibody association (5), and DNA recognition (6).
High resolution x-ray crystallography on the dimeric hemoglobin (HbI) from Scapharca inaequivalvis has revealed a striking ligand-linked rearrangement of water molecules at the subunit interface (7, 8). HbI binds oxygen cooperatively with a Hill coefficient of 1.5 and shows no change in oxygen affinity or cooperativity as pH varies from 5.5 to 9.0 (9). Quite unlike mammalian hemoglobins, where cooperativity is mediated by large quaternary structural changes (10), HbI displays minimal ligand-linked quaternary changes. The oxygen affinity of each subunit appears to depend largely on the position of the side chain of Phe-97 which packs in the heme pocket in the deoxy state, but upon heme ligation is extruded into the subunit interface where it expels several water molecules (see Fig. 1). The core of the deoxy interface includes a cluster of 17 very well-ordered water molecules, 6 of which are lost upon oxygenation. (An additional two water molecules within the heme pocket are directly displaced by the bound oxygen.)
Figure 1.
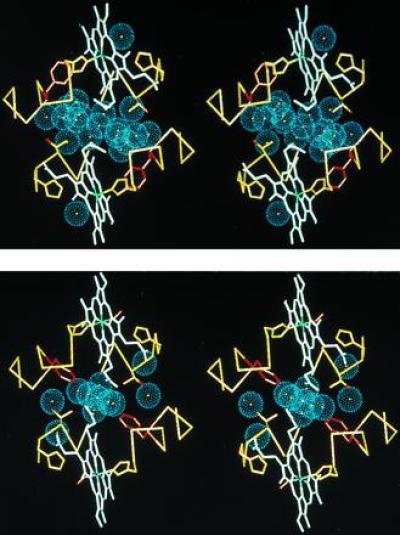
Stereo diagram depicting the interface water molecules of HbI viewed down the molecular dyad. Water molecules are shown as blue spheres representing the approximate van der Waals radii of oxygen atoms in (i) deoxy HbI (7) (Protein Data Bank code 4SDH) and (ii) oxy HbI (8) (Protein Data Bank code 1HBI). In addition to the water molecules, for both subunits an α carbon trace for residues 93–101 and 69–75 is shown in yellow, the heme groups are shown in white (with the heme iron green), side chains for the distal (residue 69) and proximal (residue 101) histidines are shown in yellow, while the side chains for Phe-97 are shown in red and the side chains for Thr-72 are shown in yellow with the hydroxyl shown in red. The deoxy water molecules are more well ordered than those of the oxy structure: the 19 deoxy water molecules shown have refined B factors that average 21.2 Å2 and average occupancy of 0.98, while the 11 shown for the oxy structure have an average B-factor of 24.3 Å2 and occupancy of 0.91. The deoxy interface water molecules form a clear cluster with five of the water molecules in position to form hydrogen bonds with other water molecules but not with protein atoms. In the oxy interface, only one water molecule has all its possible hydrogen bonds with other water molecules.
We have undertaken the present studies to determine the functional importance of the crystallographically observed water transitions in Scapharca HbI. The results clearly indicate that the water structure is crucial for stabilizing the low affinity conformation of this molecule and appears to play a key role in its cooperative function.
MATERIALS AND METHODS
Thr-72 → Val (T72V) Mutagenesis.
Mutation of T72V was carried out by cassette mutagenesis using the unique HindIII and BclI restriction sites of an HbI expression vector (11). Recombinant protein was expressed in Escherichia coli and purified as described for wild type (11).
Crystallization and Diffraction Data Collection.
Crystals of deoxy and CO-liganded T72V isomorphous to those of wild type were grown using similar procedures (7). Diffraction data were collected from two deoxy T72V crystals at room temperature using an R-AXIS II image plate system (Molecular Structure, The Woodlands, TX). Diffraction data included 16,491 unique reflections derived from 67,578 independent observations with an R-merge of 8.39% to spacings corresponding to 1.9-Å resolution. Diffraction data were collected and processed from one CO-ligated T72V crystal yielding a data set including 32,461 unique reflections derived from 58,462 independent observations with an R-merge of 5.46% to spacings corresponding to 1.6-Å resolution.
Kinetic Ligand Binding Measurements.
Kinetic measurements of ligand binding to T72V were performed by stopped-flow, flash photolysis (12) and an oxygen pulse experiment to relate the rate of dissociation to fractional saturation with oxygen (13).
Resonance Raman Spectroscopy. Sample preparation.
CO derivatives were prepared from oxy stock solutions by passing CO gas over the surface of a 100 μl aliquot in a sealed vial. Deoxy derivatives were prepared by passing N2 over the sample and then adding five equivalents with respect to heme concentration of sodium dithionite. Both samples were ≈1 mM in heme and in 100 mM phosphate buffer (pH 7.1). The glycerol samples were prepared in a similar manner, except that a 10 mM in heme solution was made, and this was then diluted by a factor of 10 with neat glycerol to give the 90% glycerol solutions used in the experiments. The samples were then loaded in a nitrogen atmosphere into quartz sample cells with a 200 μm pathlength (P/N 124-QS; Helma, Jamaica, NY). The front window of the cell was replaced with a sapphire window, which yielded a flatter baseline in the low frequency region of the Raman spectrum. The sample cell was mounted in a custom brass holder, which was cooled to ≈10°C and rotated fast enough such that a new sample volume was interrogated with each laser shot. Photoproduct buildup and artifacts from sample spinning were found to be negligible by varying the spinning rate and by taking visible absorption spectra before and after each experiment.
Experimental apparatus.
Visible time resolved resonance Raman spectra were obtained by using an 8-ns 435.8-nm pulse to both photodissociate the ligand and Raman scatter off the sample. The laser used was a neodymium: yttrium aluminum garnet (Nd:YAG) laser (model NY81C-20; Continuum, Santa Clara, CA), which produced 450 mJ pulses at 20 Hz in the second harmonic output at 532 nm. Four watts of the 532 nm beam was focused into a homemade 90-cm-long cell filled with 120 psi of hydrogen to Raman shift the laser to 435.8 nm. Neutral density filters were used to reduce the energy of the 435.8 nm pulses to 150 μJ and these were focused with a 150 mm plano-convex lens on the sample at an incidence angle of 45°. The Raman scattered light was collected at normal incidence to the sample (135° to the laser) with a 50 mm Nikon F/1.4 camera lens and focused with an f-matching lens onto the 50 μm × 5 mm slit of a 0.64 m single monochromator utilizing an 1800 grooves/mm grating (model HR640; Instruments S.A., Metuchen, NJ). The Rayleigh line was reduced in intensity with a holographic notch filter (P/N HNF-1171 centered at 442 nm; Kaiser, Ann Arbor, MI) and a depolarizer (P/N DPL-10; CVI, Putnam, CT) was used to scramble the polarization of the collected light and thus eliminate intensity artifacts created by polarization dependent grating reflectivity. The detector was an intensified diode array run in the ungated mode (P/N IRY-1024S/B; Princeton Instruments, Trenton, NJ). The total accumulation time per spectrum was typically 30 min. The spectral bandwidth of the monochromator was ≈2.5 cm−1, and the discretization on the detector face was ≈0.9 cm−1 per pixel. Raman spectra were calibrated by using solvent spectra with previously determined peak assignments. A least squares fit was used to map pixel number into relative wavenumbers (Raman shift). The Raman spectra were baselined by using a polynomial fitting routine in lab calc and are presented without smoothing (Galactic, Salem, NH).
Equilibrium Oxygen Binding Measurements.
Oxygen binding measurements on HbI and mutant T72V were performed in 0.1 M Tris/1 mM EDTA (pH 7.2) at 23°C primarily by tonometric methods (14) with a few measurements performed on wild-type HbI with a Hem-O-Scan Oxygen Dissociation Analyzer (SLM–Aminco, Silver Spring, MD). All measurements were carried out in the presence of an enzymatic reduction system to reduce oxidation (15). Tonometer measurements were carried out at a concentration of about 50 μM heme following absorbance changes at nine wavelengths in the visible range. Hem-O-Scan measurements were performed on samples at a concentration of about 4 mM heme. [The dimer to monomer dissociation constant is about 10−8 M for liganded HbI, and much lower for deoxy HbI (W.E.R., R. Fox, F. Smith, D. Zhu, and E. Braswell, unpublished data).]
Osmotic Pressure Determination.
The water activity for hemoglobin solutions was measured with a two-step method devised by Wiebe (16) using a modified C52 sample chamber and an HR33T dewpoint microvoltmeter (Wescor, Logan, UT). This two-step procedure was required due to the high osmotic pressures (low water activity) of the solutions used. Osmotic pressure (Π) is related to water activity (aw) by the equation Π = −(RT/V1°)ln(aw), where V1° is the molar volume of water (17).
RESULTS
The T72V Mutant.
The high resolution crystal structures of Scapharca HbI in the deoxy, oxy, and carbon monoxide liganded forms (7, 8) allow identification of likely hydrogen bonds between interface water molecules and a number of protein and heme atoms. While several interactions between main chain atoms and water molecules are disrupted upon heme ligation, Thr-72 has the only side chain whose apparent hydrogen bonding with water is lost upon ligation. Each Thr-72 is strategically positioned to form a hydrogen bond with a water molecule at the periphery of the water cluster in deoxy HbI (Fig. 1). To determine the importance of this interaction, we have mutated Thr-72 to the nearly isosteric residue Val (T72V).
Ligand binding properties are altered dramatically as a result of the T72V mutation, with remarkable increases in both affinity and cooperativity. Equilibrium oxygen binding measurements at 23°C show a p50 of 0.2 Torr with a Hill coefficient of 1.7, whereas wild-type HbI has values of 9.8 Torr and 1.5 at 25°C (18). Kinetic studies reveal that increased oxygen affinity of T72V relative to wild type results largely from reductions in the oxygen dissociation rates, as shown in Table 1. A remarkable aspect of these results is an increase in kinetic cooperativity, as reflected in a comparison of ligand combination rates for the first and second steps. The association rate constant for the binding of the first oxygen molecule to T72V is identical to that observed for the binding of the first molecule to the wild-type protein. However, the rate constant for the binding of the second oxygen to the mutant is 2.2 times greater than that for wild-type HbI (20, 21). These differences are much more pronounced for CO binding. The rate constants for association of the first CO molecule are 0.9 and 0.09 μM−1 s−1 for T72V and wild-type HbI, respectively, and the corresponding rate constants for the second step are 7.0 and 0.2 μM−1 s−1, respectively (Table 1). These results indicate that cooperativity is not only preserved but significantly enhanced in the mutant.
Table 1.
Kinetic ligand binding parameters to Scapharca HbI and T72V
Despite these large functional changes, crystallographic analysis indicates that the accompanying structural changes are quite small. Difference Fourier maps show very little difference between CO-liganded HbI and T72V, with evidence of only a slight change in orientation of side-chain 72. Difference Fourier maps comparing crystals of deoxy T72V and native HbI reveal that the only significant difference in deoxy structures is the loss, or disordering, of the water molecule normally in a position to form a hydrogen bond with each Thr-72 (Fig. 2). Thus, the destabilization of these two water molecules, at the periphery of the water cluster, is sufficient to dramatically alter the balance between the liganded and unliganded form of this protein.
Figure 2.
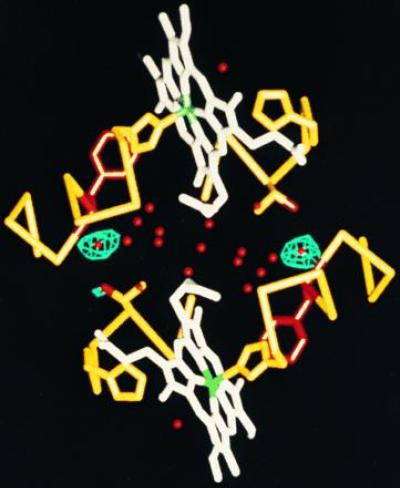
Difference Fourier map comparing wild-type HbI and T72V deoxy crystals. Atomic structure is shown as in Fig. 1, but with water molecules in red. Contours are drawn at 6σ for a map calculated between T72V and wild-type deoxy data (Protein Data Bank code R4SDHSF) using 10,578 reflections corresponding to spacings between 10- and 2.0-Å resolution. This indicates the only significant differences between the structures is the loss, or disordering, in T72V of the two water molecules normally in a position to hydrogen bond to the hydroxyl group of Thr-72 in each subunit.
Destabilization of the Deoxy State in Glycerol.
An indication that osmotic forces could alter the conformation of HbI by destabilizing the water structure has been obtained by resonance Raman measurements of deoxy HbI in the presence of high concentrations of glycerol. In an earlier study (22), it was shown that the low frequency resonance Raman spectra of deoxy HbI and HbI* (the 10-ns photoproduct of HbICO) reflect the low and high affinity structures of HbI, respectively. Fig. 3 shows changes in the low frequency resonance Raman spectra of deoxy HbI upon addition of glycerol (90% by volume) to aqueous buffer. Also shown are the corresponding spectra of HbI*, which do not show a glycerol effect. It can be seen that with the addition of glycerol, the resulting deoxy spectrum closely resembles the photoproduct spectrum. The results indicate that glycerol induces a structural transition from the low affinity deoxy state toward the conformation of the ligated state even in the absence of ligand.
Figure 3.
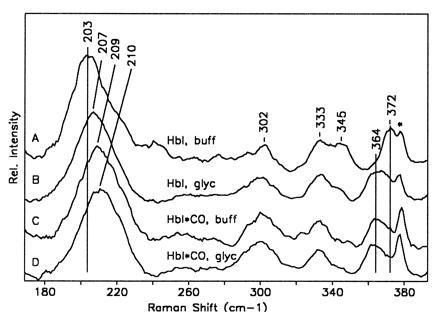
Plot showing the low frequency resonance Raman spectra of deoxy HbI in aqueous buffer (pH 7.1, 100 mM phosphate) (line A); deoxy HbI in a glycerol/water solution (90% glycerol by volume) (line B); HbI*, the 10-ns photoproduct of HbICO in aqueous buffer (line C); and HbI* in a 90% glycerol/water solution (line D). The low frequency band occurring between 203 and 210 cm−1 is the iron-proximal histidine stretching mode. The bands occurring between 330 and 372 cm−1 are all sensitive to the propionate stretching motions. The shift in the propionate bands in going from deoxy HbI to HbI* in buffer is ascribed in this case to the large shift in the heme upon ligation (E.S.P., W.E.R., and J.M.F., unpublished data). It can be seen that the addition of glycerol to the deoxy sample produces a population that has almost the same iron-proximal histidine stretching and propionate frequencies as HbI*. The spectral feature marked by ∗ is from the sapphire window.
Oxygen Binding as a Function of Osmotic Pressure.
The resonance Raman results imply that high glycerol concentrations destabilize the deoxy conformation either directly or through an indirect osmotic effect that could be probed with osmotic methods (23, 24). We have performed equilibrium oxygen binding experiments on HbI in the presence of glycerol, glucose, and sucrose, each of which significantly raises oxygen affinity of HbI while maintaining cooperativity. The increased oxygen affinity is consistent with the idea that the deoxy state is destabilized relative to the higher affinity oxygenated form. The effect on oxygen affinity is dependent on osmotic pressure in the same manner for all three solutes (Fig. 4).
Figure 4.
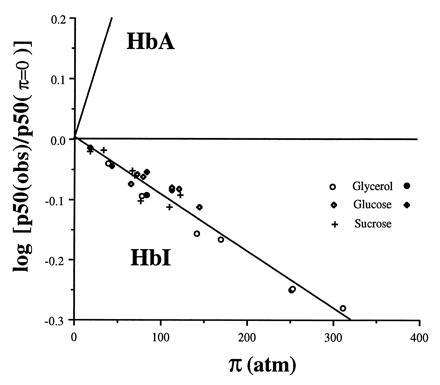
Plot showing the dependence of oxygen affinity with osmotic pressure for HbI and human hemoglobin (HbA). The value of p50 at π = 0 was estimated from measurements in buffer alone. The line showing dependence for human hemoglobin is obtained from Colombo et al. (25). Data points are shown for tonometer measurements with glycerol (○), glucose (⋄), and sucrose (+) solutions and Hem-O-Scan measurements for glycerol (•) and glucose (♦). Note the opposite direction for HbI, in which osmotic pressure increases oxygen affinity, compared with HbA, for which osmotic pressure decreases oxygen affinity. The slope of the least squares fit of the HbI data is −9.5 × 10−4 ± 0.5 atm−1, which indicates that an additional 6.2 ± 0.3 water molecules are bound to the deoxy state relative to the oxy state.
Colombo et al. (25, 26) have investigated the effect of osmotic pressure on oxygen binding to human hemoglobin and developed an analysis to estimate the linkage between binding of water and oxygen using relationships first derived by Wyman (17, 27). With the approximation of p50 (partial pressure of oxygen at half saturation) for the mean ligand activity:
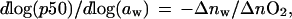 |
where aw is the chemical potential of water, Δnw is the difference in the number of water molecules bound between oxy and deoxy hemoglobin, and ΔnO2 is the number of oxygen binding sites. For human hemoglobin the observed dependence suggests that oxygen uptake results in the binding of an additional 65–80 water molecules per tetramer (26). As shown in Fig. 4, the dependence of oxygen affinity on osmotic pressure is quite different for HbI. Least squares fit of the data shown for all three solutes suggests that 6.2 ± 0.3 water molecules are released upon oxygen binding, in good agreement with the eight expected based on the crystal structures.
DISCUSSION
The experiments presented here reveal that Scapharca HbI is quite sensitive to the stability of bound water molecules. These results reflect the effect of water in modulating the intricate balance between alternate allosteric conformations that underlie cooperativity. An important feature of the ligand-linked structural transitions in Scapharca HbI is the absence of large quaternary changes. Thus, the functional effects may plausibly be attributed to the observed structural changes localized at the subunit interface. The interfacial water cluster in deoxy HbI evidently provides an important contribution to the stability of the low affinity state, such that significant increases in oxygen affinity are observed as water activity is lowered. In the mutant T72V, selective removal of just two hydrogen bonds from the deoxy water cluster and the concomitant loss of two water molecules is sufficient to drastically tip the balance away from the low affinity form toward the high affinity conformation. As the two water molecules directly influenced by this mutation are at the periphery of the water cluster (Figs. 1 and 2), rather than in a position to stabilize the deoxy conformation through direct interactions with the hemes, the observed functional effects appear to reflect the importance of an intact water cluster in the deoxy state.
An important and unexpected aspect of our results is the enhancement of cooperativity as ligand affinity increases upon mutation of Thr-72 → Val. More often, increasing oxygen affinity has been coupled with a decrease in cooperativity as, for instance, in an earlier mutation in HbI of Thr-72 → Ile (28). The observed effects with the T72V mutation may be rationalized within a two-state model (29). It should be noted first that values of kinetic or equilibrium parameters obtained by experiment are not the same as those assigned to the R and T allosteric states in a two-state model. For example, in deoxy hemoglobin, the proportions of the R and T forms are 1/(1 + L) and L/(1 + L), respectively, where L is the allosteric constant (defined as the ratio of T to R molecules in the deoxy state), and the observable rate constant for binding the first ligand is kR′/(1 + L) + kT′ L/(1 + L) where kR′ and kT′ are the intrinsic rates for oxygen binding to the R and T states. The observed rate will be greater than kT′, although if L is large it closely approximates kT′. The analogous expression for the dissociation of the first ligand molecule from a dimer is kR/(1 + Lc2) + kTLc2/(1 + Lc2), where c is the ratio KT/KR of the affinities of the allosteric states (statistical factors omitted). These considerations are often of little significance in work with mammalian hemoglobins where the distribution between R- and T-states in liganded and deoxy forms may differ by a factor of 108 and experimental values come close to allosteric parameters.
Although the situation is not so simple for dimers in which significant populations of both R and T forms may be present in either deoxy or fully liganded hemoglobin, this possibility offers a means for rationalizing our experimental data. Given the structural similarities between native and T72V and the plausible assumption that this mutation does not affect intrinsic subunit reactivity, the functional behavior of deoxy native HbI may be taken to represent T-states in general, and liganded T72V to represent R-states. It is then possible to simulate the behavior of both native and mutant molecules with a single set of kinetic parameters, varying only the allosteric parameter L and taking the T-state parameters from native HbI and R state parameters from T72V. As Table 2 shows, a surprisingly good representation of experimental values is achieved by this procedure. In the native hemoglobin the equilibrium is weighted heavily toward the low affinity conformation such that in the singly liganded species a low affinity form still predominates. In T72V, this equilibrium is shifted dramatically, with the singly liganded form chiefly in a higher affinity conformation.
Table 2.
Fit of HbI ligand binding data to allosteric model
From a physiological point of view, it is interesting to note that with the ligand binding parameters of Table 2 the value of L does not lead to optimal cooperativity in oxygen binding, which would be the case when L equals 1/c. This may be because maximum cooperativity would be accompanied by an oxygen affinity too great to permit efficient oxygen transport.
Our results suggest a new mechanism for cooperativity in which water molecules are key mediators for information transfer between hemes. Oxygen affinity in Scapharca HbI has been postulated to be primarily regulated by the disposition of the side chain of Phe-97 (7). In the deoxy form, Phe-97 is packed in the heme pocket which appears to lower oxygen affinity by sterically restricting movement of the iron atom into the heme plane and lengthening a hydrogen bond involving Nδ of the proximal histidine (His-101). In the liganded structures, Phe-97 is extruded from the heme pocket, permitting movement of the iron atom into the heme plane and a stronger hydrogen bond to be formed between Nδ of His-101 and the main-chain carbonyl oxygen of Phe-97. The crystal structures of deoxy HbI and CO-liganded HbI (7) implicated the interface water cluster in a number of specific interactions that stabilize the low affinity state observed in deoxy HbI. These include forming hydrogen bonds that compensate for lost main-chain hydrogen bonding when Phe-97 packs in the heme pocket and interactions with the heme propionates. Upon oxygen binding, extrusion of Phe-97 from the heme pocket into the interface disrupts the interface water network, with the expulsion of three water molecules. This may lower the stability of other water molecules in the cluster sufficiently to encourage the second subunit to attain a higher affinity conformation (with its Phe-97 extruded) prior to ligand binding. The exquisite sensitivity of Scapharca HbI to water, as illustrated by osmotic agents and mutagenesis, strongly supports such a direct role.
An important area of investigation in molecular biology is the role of water in biological function (30, 31). In the case of allosteric interactions, water may play a direct role in the intricate stability between alternate protein conformations. For instance, in phosphofructokinase the insertion of a layer of water molecules between β-strands replaces β-sheet hydrogen bonding lost in the T to R transition (32). In human hemoglobin, preferential hydration appears essential for stabilization of the R-state (25, 26). Our present study illustrates that a crystallographically observed water cluster is crucial in the cooperative functioning of Scapharca dimeric hemoglobin. These examples demonstrate how proteins can evolve specific conformations to utilize the binding of individual water molecules in biological function.
Acknowledgments
We thank R. Briscoe and T. Boyle for advice and assistance with water activity measurements; S. Suzuka, M. Fabrey, and R. Nagel for use of and assistance with Hem-O-Scan oxygen binding measurements; and F. Smith and J. Olson for helpful comments on the manuscript. This work was supported by National Institutes of Health Grants DK43323 (W.E.R.), GM14276 (Q.H.G.), and GM44343 (J.M.F.), and the W. M. Keck Foundation (J.M.F.). Work was done during the tenure of an Established Investigatorship from the American Heart Association to W.E.R.
Footnotes
Abbreviations: HbI, Scapharca dimeric hemoglobin; T72V, Thr-72 → Val mutant of HbI.
References
- 1.Karplus P A, Faerman C. Curr Opin Struct Biol. 1994;4:770–776. [Google Scholar]
- 2.Badger J, Caspar D L D. Proc Natl Acad Sci USA. 1991;88:622–626. doi: 10.1073/pnas.88.2.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quiocho F A, Wilson D K, Vyas N K. Nature (London) 1989;340:404–407. doi: 10.1038/340404a0. [DOI] [PubMed] [Google Scholar]
- 4.Singer P T, Smalås A, Carty R P, Mangel W F. Science. 1993;259:669–673. doi: 10.1126/science.8430314. [DOI] [PubMed] [Google Scholar]
- 5.Bhat T N, Bentley B A, Boulot G, Breene M I, Tello D, Dall’Acqua W, Souchon H, Schwarz F P, Mariuzza R A, Poljak R J. Proc Natl Acad Sci USA. 1994;91:1089–1093. doi: 10.1073/pnas.91.3.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joachimiak A, Haran T E, Sigler P B. EMBO J. 1994;13:367–372. doi: 10.1002/j.1460-2075.1994.tb06270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Royer W E., Jr J Mol Biol. 1994;235:657–681. doi: 10.1006/jmbi.1994.1019. [DOI] [PubMed] [Google Scholar]
- 8.Condon P J, Royer W E., Jr J Biol Chem. 1994;269:25259–25267. doi: 10.2210/pdb1hbi/pdb. [DOI] [PubMed] [Google Scholar]
- 9.Chiancone E, Vecchini P, Verzili D, Ascoli F, Antonini E. J Mol Biol. 1981;152:577–592. doi: 10.1016/0022-2836(81)90270-9. [DOI] [PubMed] [Google Scholar]
- 10.Perutz M F, Fermi G, Luisi B, Shaanan B, Liddington R C. Acc Chem Res. 1987;20:309–321. doi: 10.1101/sqb.1987.052.01.063. [DOI] [PubMed] [Google Scholar]
- 11.Summerford C M, Pardanani A, Betts A H, Poteete A R, Colotti G, Royer W E., Jr Protein Eng. 1995;8:593–599. doi: 10.1093/protein/8.6.593. [DOI] [PubMed] [Google Scholar]
- 12.Sawicki C A, Gibson Q H. J Biol Chem. 1977;252:7538–7547. [PubMed] [Google Scholar]
- 13.Gibson Q H. Proc Natl Acad Sci USA. 1973;70:1–4. doi: 10.1073/pnas.70.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rossi-Fanelli A, Antonini E. Arch Biochem Biophys. 1958;77:478–492. doi: 10.1016/0003-9861(58)90094-8. [DOI] [PubMed] [Google Scholar]
- 15.Hayashi A, Suzuki T, Shin M. Biochim Biophys Acta. 1973;310:309–316. doi: 10.1016/0005-2795(73)90110-4. [DOI] [PubMed] [Google Scholar]
- 16.Wiebe H H. Plant Physiol. 1981;68:1218–1221. doi: 10.1104/pp.68.6.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wyman J, Gill S J. Binding and Linkage: Functional Chemistry of Biological Macromolecules. Mill Valley, CA: University Science Books; 1990. [Google Scholar]
- 18.Ikeda-Saito M, Yonetani T, Chiancone E, Ascoli F, Verzili D, Antonini E. J Mol Biol. 1983;170:1009–1018. doi: 10.1016/s0022-2836(83)80200-9. [DOI] [PubMed] [Google Scholar]
- 19.Adair G S. J Biol Chem. 1925;63:529–545. [Google Scholar]
- 20.Chiancone E, Elber R, Royer W E, Jr, Regan R, Gibson Q H. J Biol Chem. 1993;268:5711–5718. [PubMed] [Google Scholar]
- 21.Antonini E, Ascoli F, Brunori M, Chiancone E, Verzili D, Morris R J, Gibson Q H. J Biol Chem. 1984;259:6730–6738. [PubMed] [Google Scholar]
- 22.Rousseau D, Song S, Friedman J M, Boffi A, Chiancone E. J Biol Chem. 1993;268:5719–5723. [PubMed] [Google Scholar]
- 23.Parsegian V A, Rand R P, Rau D C. Methods Enzymol. 1995;259:43–94. doi: 10.1016/0076-6879(95)59039-0. [DOI] [PubMed] [Google Scholar]
- 24.Robinson C R, Sligar S G. Methods Enzymol. 1995;259:395–426. doi: 10.1016/0076-6879(95)59054-4. [DOI] [PubMed] [Google Scholar]
- 25.Colombo M F, Rau D C, Parsegian V A. Science. 1992;256:655–659. doi: 10.1126/science.1585178. [DOI] [PubMed] [Google Scholar]
- 26.Colombo M F, Bonilla-Rodriguez G O. J Biol Chem. 1996;271:4895–4899. doi: 10.1074/jbc.271.9.4895. [DOI] [PubMed] [Google Scholar]
- 27.Wyman J. Adv Prot Chem. 1964;19:223–286. doi: 10.1016/s0065-3233(08)60190-4. [DOI] [PubMed] [Google Scholar]
- 28.Gambacurta A, Piro M C, Coletta M, Clementi M E, Polizio F, Desideri A, Santucci R, Ascoli F. J Mol Biol. 1995;248:910–917. doi: 10.1006/jmbi.1995.0271. [DOI] [PubMed] [Google Scholar]
- 29.Monod J, Wyman J, Changeux J-P. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 30.Westhoff E. Water and Biological Macromolecules. Boca Raton, FL: CRC; 1993. [Google Scholar]
- 31.Gregory R B. Protein–Solvent Interactions. New York: Dekker; 1995. [Google Scholar]
- 32.Schirmer T, Evans P R. Nature (London) 1990;343:140–145. doi: 10.1038/343140a0. [DOI] [PubMed] [Google Scholar]


