Abstract
Motivation
Linking experimental data to mathematical models in biology is impeded by the lack of suitable software to manage and transform data. Model calibration would be facilitated and models would increase in value were it possible to preserve links to training data along with a record of all normalization, scaling, and fusion routines used to assemble the training data from primary results.
Results
We describe the implementation of DataRail, an open source MATLAB-based toolbox that stores experimental data in flexible multi-dimensional arrays, transforms arrays so as to maximize information content, and then constructs models using internal or external tools. Data integrity is maintained via a containment hierarchy for arrays, imposition of a metadata standard based on a newly proposed MIDAS format, assignment of semantically typed universal identifiers, and implementation of a procedure for storing the history of all transformations with the array. We illustrate the utility of DataRail by processing a newly collected set of ~22,000 measurements of protein activities obtained from cytokine-stimulated primary and transformed human liver cells.
1 INTRODUCTION
A fundamental goal of systems biology is constructing mathematical models that elucidate key features of biological processes as they exist in real cells. A critical step in realizing this goal is effectively calibrating models against experimental data. The challenges of model calibration are well recognized (Jaqaman and Danuser, 2006) but we have found systematizing and processing data prior to calibration to be tricky as well. This is particularly true as the volume of data or the complexity of models grows. Few information systems exist to organize, store and normalize the wide range of experimental data encountered in contemporary molecular biology in a sufficiently systematic manner to maintain provenance and meanwhile retaining the adaptability necessary to accommodate changing methods. Partly as a consequence, relatively few complex physiological processes have been modeled using a combination of theory and high throughput experimental data.
An information management system for experimental data must record data provenance and experimental conditions, maintain data integrity as various numerical transformations are performed, describe data in terms of a standardized terminology, promote data reuse, and facilitate data sharing. The most common way to achieve these requirements is via a relational database management system (RDBMS, see SBEAMS—http://www.sbeams.org —or Bioinformatics Resource Manager for relevant examples; Shah et al., 2006). Databases in biology resemble those previously developed for business and have proven spectacularly successful in managing data on DNA and protein sequences. In a relational database the subdivision of information and its subsequent storage into cross-indexed tables follows a precise, predefined schema. The granularity and stability of the schema allows an RDBMS to identify and maintain links between disparate pieces of information, even in the face of frequent read-write operations. However, this power comes at a considerable cost in terms of inflexibility. It is difficult for a relational database to accommodate frequent changes in the formats of data or metadata and to incorporate unstructured information.
Whereas the sequence of a human gene represents valuable information independent of how sequencing was performed or of the individual from whom the DNA was obtained (a statement that remains true despite the value of characterizing sequence variations) such is not the case for measures of protein activity or cellular state. Such biochemical and physiological data are highly context dependent. Data on ERK kinase activity, for example, is uninformative in the absence of information on cell type, growth conditions, etc. Moreover, a wide range of techniques are used to make biochemical and physiological measurements, and both the assays and the data they generate change over time, as new methods are developed (e.g., in imaging see Swedlow et al., 2003). Context dependence and rapidly changing data formats pose fundamental problems for databases because RDBMS schemes are not easily modified.
Moreover, even if effective metadata standards are developed to describe the context-dependence of experimental findings, data from different experiments cannot be reconciled simply by storing them in a single database. Subtle distinctions must be made about different types of data and biological insight brought to bear. Currently this is performed implicitly in the minds of individual investigators, but we envision a future in which the unique ability of mathematical models to formalize hypotheses and manage contingent information makes them the primary repositories of biological knowledge. As we work towards a model-centric future, it is our contention that information systems based solely on relational databases are unnecessarily limiting; rarely do we modify a difficult experiment simply to conform to a pre-existing database schema (whereas conformity to uniform—even arbitrary—standards is a strength for a business database). New approaches to data management that reconcile competing requirements for flexibility and structure are required.
One response to the challenges of systematizing biological data has been the creation of lightweight data standards focused on the most important metadata. Pioneered by the Microarray and Gene Expression Data Society’s Minimum Information about a Microarray Experiment (MIAME), these “minimum information” approaches typically define a simple data model that can be instantiated as an XML file, a database schema, etc. A strength of “minimum information” standards is that they specify that subset of the metadata that is relatively constant among ever-shifting and context-sensitive experiments. The philosophy is that of the Pareto principle or 80-20 rule, namely that 80% of the information can be captured with 20% of the effort whereas the final 20% requires exponentially greater effort. An underlying assumption is that a minimum information standard successfully records the information needed to make experimental data intelligible. In this paper we implement an information processing system, DataRail, intended to bridge the gap between data acquisition and modeling. A new minimum information standard (MIDAS) is part of the DataRail system, but a series of additional tools are also applied to maintain the provenance of data and ensure its integrity through multiple steps of numerical manipulation. DataRail is model- rather than data-centric in that the task of creating and transmitting knowledge is invested in mathematical models constructed using the software, rather than the data storage system itself, but it is designed to support existing modeling tools rather than serve itself as an integrated modeling environment. We illustrate this capacity in DataRail using a large set of protein measurements derived from primary and transformed hepatocytes; through the use of DataRail we derive insight both into the biology of these cells types and the optimal means by which to perform partial least squares regression (PLSR) modeling of cue-signal-response data.
2 RESULTS
2.1 Design goals and implementation
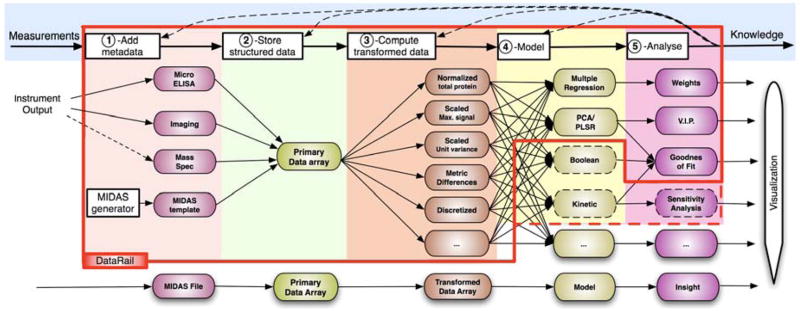
To facilitate the collection, annotation and transformation of experimental data, DataRail software is designed to meet the following specific requirements (see Fig. 1): (i) serve as a stable repository for experimental results of different types while recording key properties of the biological setting and complete information about all data processing steps; (ii) promote model development and analysis via internal visualization and modeling capabilities; (iii) interact efficiently and transparently with external modeling and mining tools; (iv) meet new requirements in data collection, annotation, and transformation as they arise; and (v) facilitate data sharing and publication through compatibility with existing bioinformatics standards. A system meeting these requirements was designed in which data is stored in a succession of regular multidimensional arrays, known as “data cubes” in information technology (Gray et al., 1997), each representing transformations of an original set of primary data. The integrity of data is maintained by tagging the primary data with metadata referenced to a controlled ontology, storing all arrays arising from the same primary data in one file structure, documenting the relationships of arrays to each other, storing algorithms used for data transformation with data arrays, and assigning each data structure a unique identifier (UID) based on a controlled semantic. DataRail was implemented as a MATLAB toolbox (http://www.mathworks.com/) with scripting and GUI-based interaction and incorporating a variety of data processing algorithms. DataRail works best as a component of a loosely coupled set of software tools including commercial data mining packages such as Spotfire (http://spotfire.tibco.com/) or public toolboxes for modeling. In addition, DataRail is designed to communicate with a semantic Wiki, to be described in a separate paper but available at the DataRail download site, that is better designed for storing textual information, such as experimental protocols, and that documents DataRail’s use of UIDs.
Fig. 1.
Process diagram for model-centric information management in DataRail. Measurements generated using one or more methods (left side of diagram) are processed to output new knowledge (right); hypothesis testing links modeling and measurement in an iterative cycle. Processes and entities within the red box have been implemented; those outside the box remain to be completed; dotted lines denote external processes that have been linked to DataRail. Experimental measurements are first converted into a MIDAS format using one or more routines (pink lozenges; see text for details) and then used to assemble a multi-dimensional primary data array (green). Alternatively, an empty MIDAS-compliant spreadsheet is generated using a Java utility and experimental values then entered. Algorithms for normalization, scaling, discretization, etc. transform the data to create new data arrays (orange) that can then be modeled using internal or external routines. Finally, analysis and visualization assist in knowledge generation. The calibration of kinetic and Boolean models is not shown explicitly, although it constitutes a critical and complicated step in the overall workflow of systems biology that is as-yet external to DataRail.
2.2 System overview
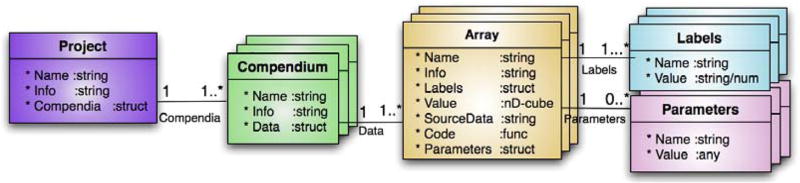
Information in DataRail arising from a single set of experiments is organized into a compendium, which consists of multiple n-dimensional data arrays, each of which contains either primary data or processed data (see Fig. 2). It is left up to users to determine the breadth of experimental data included within each compendium, but good practice is to group results with similar experimental aims, biological setting or place of publication into one compendium. DataRail also supports creation of containers for multiple compendiaknown as projects. The dimensionality of arrays containing primary data is determined by the user at the time of import, making it possible to accommodate a wide range of experimental approaches and measurement technologies. For example, measuring a few properties of many samples by flow cytometry generates an array of different dimensionality than measuring many variables in a few samples by mass spectrometry. In practice, data in our laboratory can usually be described in six dimensions: three for the experimental conditions (e.g., cell type, cytokine stimuli, and small-molecule treatment), one for time, one for experimental replicates, and one for actual measurements.
Fig. 2.
Containment hierarchy for DataRail. Individual arrays of primary or transformed data are gathered together into a MATLAB structure we call a compendium; multiple compendia are linked together into a project. Each compendium contains a unique name (UID), a short textual documentation, and a set of multi-dimensional arrays. Each array is stored together with simple metadata (name, free-text information, source, algorithm, and free parameters used in array creation). The representation follows the conventions of UML (Unified Modeling Language) format, indicating that a compendium contains one or more arrays, which contain one or more labels and zero or more parameters.
Arrays of transformed data are generated from primary data by applying numerical algorithms that normalize, scale, or otherwise increase accuracy and information content. Algorithms used during data processing, along with the values of all free parameters, are stored with each arrayto maintain a complete record of all transformations performed prior to data mining or modeling.
2.3 Test cases
We have tested DataRail on seven sets of recent data available in our laboratories, containing between 5×103 and ~1.6×106 data points. Each set had a unique structure and gave rise to arrayswith 4–6 dimensions (see Supplementary Table S1). Here we discuss the analysis of a “CSR Liver compendium,” a cue-signal-response dataset (Gaudet et al., 2005) comprising 22,512 measurements in primary human hepatocytes and a hepatocarcinoma cell line (HepG2 cells; L.A. et al., unpublished data). In this compendium, cells were exposed to 11 cytokine treatments and 8 small-molecule drugs upon which the states of phosphorylation of 17 signaling proteins (at 30 minutes and 3 hr) and the concentrations of 50 extracellular cytokines (at 12 and 24 hr) were measured using bead-based micro-ELISA assays.
2.4 Storing primary data and metadata
Tagging primary data with metadata is essential to its utility and involves two aspects of DataRail: a new metadata standard and a process for actually collecting the metadata. The metadata standard is based on our proposed MIDAS format (Minimum Information for Data Analysis in Systems Biology) that is itself based on pre-existing minimum-information standards such as MIACA (Minimum Information About a Cellular Assay, http://miaca.sourceforge.net/). MIDAS is a tabular (or spreadsheet) format that specifies the layout of experimental data files that gives rise, upon import into DataRail, to an n-dimensional data array. The MIDAS format was derived from the “experimental module” concept in MIACA, with modifications required for model-centric data management (see Fig. 3). Typically a MIDAS file is used to input information from instruments into DataRail and to export information from DataRail into other software that uses spreadsheets. However, export from a data array to a MIDAS file entails loss of information about data provenance and prior processing steps. We are therefore in the process of implementing a standardized format for exchanging DataRail files that does not depend on the use of MATLAB files (see discussion for details). Each row in a MIDAS table represents a single experimental sample; each column represents one sample attribute, such as identity (e.g., multi-well plate name or well coordinate), treatment condition, or value obtained from an experimental assay. A column header consists of two values: (i) a two-letter code defining the type of column, (e.g., TR for treatment, DV for data value), and (ii) a short column name (e.g., a small molecule inhibitor added or a protein assayed). The body of each column stores the corresponding value for each row (sample) such as a plate/well name, reagent concentration, time point, or data value (see supplementary information for details and example MIDAS spreadsheets). MIDAS is designed to fulfill the need for data exchange and analysis within a closely knit research group. It is not a stand-alone solution for archival storage or publication and should be implemented in conjunction with MIAME, MIACA, or DataRail itself..
Fig. 3.
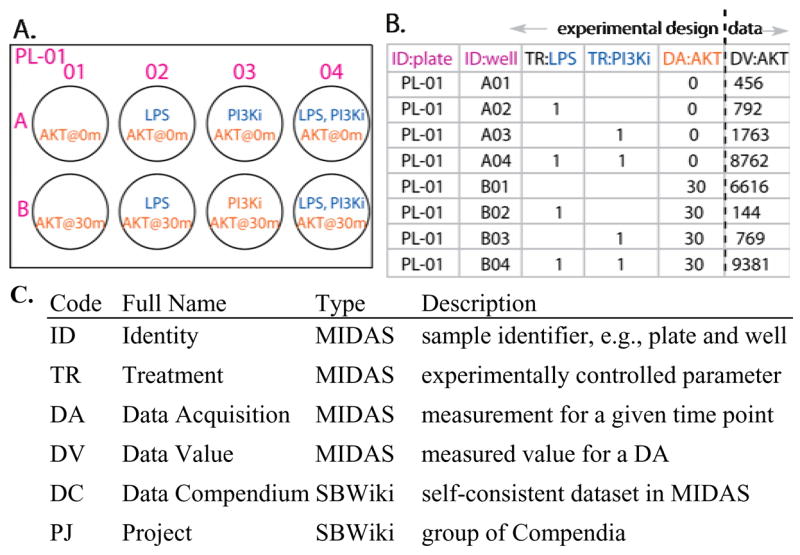
Minimum Information for Data Analysis in Systems Biology (MIDAS). A: A simplified map of a multi-well experiment in which Akt phosphorylation is to be assayed at 0 and 30 min in extracts from cells treated, or not, with lipo-polysacharide (LPS) and a PI3-kinase inhibitor (PI3Ki). B: MIDAS representation of the experiment. A column header consists of a two-letter code defining the type of column and a short column name. For clarity headers are color-coded to match the corresponding values on the plate map. The leftmost five columns (codes ID: identity, TR: treatment, and DA: data acquisition) are experimental design parameters and would be filled in before bench work begins. The rightmost column holds measured data values (DV) that are appended as data acquisition is performed. See Supplementary Table S2 for a larger example. C: A list of the type codes used for MIDAS columns and a few relevant SBWiki types.
The sequence of steps involved in entering metadata into DataRail is designed to accommodate the rhythm of a typical laboratory in which simple annotation is possible while experiments are in progress, but detailed data analysis is performed subsequently. As an experiment is being designed, a MIDAS file specifying the dimensionality and format of the data (treatments, time points, readouts, etc.) is created, and scripts specialized to different instruments or experimental methodologies are then used to add results to the “empty” MIDAS file. Thus far we have written a script to import bead-based micro-ELISA data generated by a Luminex reader running BioRad software (Bio-Plex). We have also implemented a general purpose Java program for MIDAS file creation that can be used to import data into DataRail, used as a stand-alone application, or integrated into other software. Within the MIDAS layout utility, wells that will be treated in a similar or sequential manner are selected via a GUI and appropriate descriptions of the samples added via pop-up tabs (see Supplementary Fig. S1). When layout is complete, a correctly formatted MIDAS file is generated, ready for the addition of data. Lists that assist experimentation are also created (these lists typically specify times of reagent addition, sample withdrawal, etc.). We invite instrument manufacturers to incorporate this utility into their software so that creation of MIDAS-compliant files is automatic; the code is therefore distributed under a non-viral caBIG open source license developed by the National Cancer Institute. If a MIDAS file has not been generated at the outset of an experiment, it is possible to convert experimental data at any point prior to import, but in this case MIDAS-associated support tools are not available to help with experiments.
As mentioned above, DataRail can but need not be used in combination with SBWiki, a wiki based on semantic web technology (Berners-Lee, 2001). For the current discussion, four features of SBWiki are important. First, a web form used for upload prompts users to enter the metadata such as user name, date, cell type, etc., required for full MIDAS compliance. Because continuous web access is easy to arrange, even for geographically dispersed instruments, users record metadata when files are first saved to a central location. This is very important in practice because metadata is rarely added when the process is cumbersome or separated in time from data collection. Second, use of semantic web forms makes it possible to create simple, familiar, and easily modified interfaces while collecting structured information. In contrast, tools for accessing metadata in traditional databases or XML files are more difficult to use and require considerable expertise to modify. Third, as data is imported it is assigned a UID by SBWiki itself, which directly encodes, among other things, the type of data and the person who created it (see supplementary materials). The assignment of a UID makes it possible to track the origin of all data in DataRail, independent of the array-compendium-projectstructure. Fourth, although metadata describing specific details of experiments are stored internal to the MIDAS file, protocols and reagents are stored in SBWiki. Storage external to the MIDAS file allows complex textual information to be modified and reused more easily. Links from data arrays to external files are made via URLs that follow the UID scheme described above and can use the revision history in SBWiki to reference a specific version of a protocol or reagent..
When constructing the CSR Liver compendium, a spreadsheet generated by Bio-Plex software was appended to a MIDAS file, and a second MIDAS file containing data on total protein concentrations was generated using a plate reader. Overall three primary data arrayswere created from CSR Liver data: one recording phosphorylation states of 17 proteins at three time points (0, 30 min, 3 hr), one recording extracellular cytokine concentrations, also at three time points (0, 12 hr, 24 hr), and one recording total protein concentration. In principle these arrayscould be combined to bundle data together, but the resulting single arraywould be sparsely populated. In addition, bundling data into a single arrayis not the same as fusing different types of data. The fusion of flow cytometry, Western blot and live-cell imaging data (J.A. et al., unpublished data) is facilitated by DataRail but also requires biological insight and problem-specific modeling.
2.5 Adding transformed arrays to compendia
Once primary data is imported into a new compendium, it is then transformed by one or more algorithms internal to DataRail, by user-specified algorithms, or by external programs, to create a new transformed data array. Transformations can change numerical values within an arrayor can expand or collapse the dimensionality of arrays. A long time series, for example, can be transformed into a shorter series involving selected times, time-averaged data, or integrated values. When a transformation is performed on an array, the code used for the transformation and the values of free parameters are stored, along with a reference to the input data (in the current implementation, the algorithms themselves are recorded as the text of MATLAB functions), so that the compendiumis a self-documenting entity, in which the provenance of data can be tracked.
Overall, DataRail can perform a diversity of transformations falling into several general categories. Simple arithmetic operations include subtracting background from primary data, or dividing one type of data by another (see Supplementary Fig. S2). For example, Bio-Plex-based measures of protein phosphorylation in CSR Liver data were divided by total protein concentration to correct for differences in cell number and extraction efficiency. In a second type of transformation, metrics such as “area under the curve,” maximal value of a variable in a series, standard deviation of a series, and relative values are computed. Third, complex data transformations are performed, including mean-centering and variance-scaling, both of which are helpful in performing principal component analysis (PCA) or assembling models using PLSR (Gaudet et al., 2005). Finally, computations specific to particular modeling methods are performed, including transformation of continuous variables into discrete values for the purpose of Boolean or discrete data modeling. For example, to support Boolean modeling, a discretization routine assigns a value of “1” to a variable if and only if (i) it is above a typical background value for the assay, as determined by the user or extracted automatically from primary data, (ii) it is above a user-supplied threshold, and (iii) it is high with respect to the values of the same signal under other conditions in the data set.
2.6 Data mining and visualization
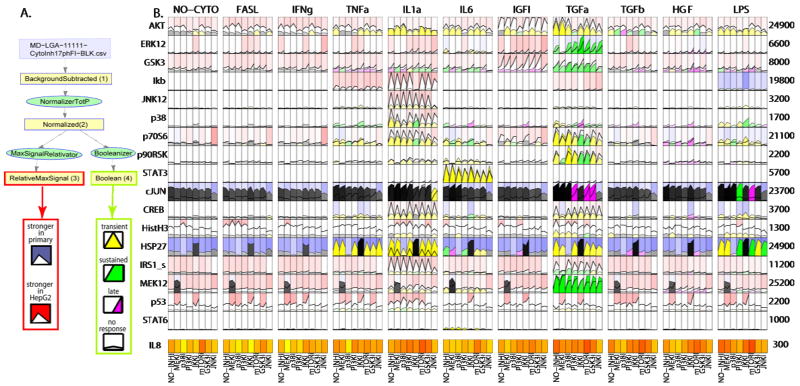
Visualization can involve data export directly to an external application such as Spotfire, or it can be performed within the pipeline. Internal visualization routines that make use of transformations performed by DataRail are often an effective means to create thumbnails of time-courses, heat maps, etc. For example, the data viewer in Fig. 4 was developed to display time courses of protein modification in the CSR Liver compendium, corrected for background and protein concentration and scaled to a common vertical axis. Data from primary hepatocytes and HepG2 cells was compared, and the difference between the integrated activities in the two lines then computed and displayed in the background as a red-blue heat map. Discretization was then used to score responses as transient, sustained, or invariant, each of which was assigned a different color. Finally, a heat map of the phenotypic responses was generated to facilitate comparison of signals and outcomes (see Fig. 4). Importantly, efficient generation of plots such as this relies on the inclusion in DataRail of multiple data transformation routines.
Fig. 4.
Visualizing data in DataRail by exploiting data in transformed arrays. A. Structure of the compendium used to generate this plot and the relationship of each feature to data in a transformed array. This structural map was generated using routines internal to DataRail. B. Time courses for the phosphorylation of 17 key proteins (rows) in primary hepatocytes under 11 different conditions of cytokine stimulation (columns) and treated with 7 different small molecule drugs (subpanels within each cytokine-signal block). Curves are colored according to their dynamics (green=sustained, yellow=transient, magenta=late activation, grey=no significant signal). The intensity of the signal determines the intensity of the color. The corresponding signals from HepG2 tumor cells are plotted behind without color coding. The background is blue if the mean signal is stronger for primary cells and red if it is stronger for HepG2 cells; larger differences lead to stronger coloring. In addition, the levels of IL8 at 24 hr, a measure of cellular response, are added as a heat map.
2.7 Constructing and evaluating models
DataRail supports three approaches to modeling. First, several routines that create statistical models, such as PLSR, have been integrated directly into the code. Second, efficient links have been created to other MATLAB toolboxes such as CellNetAnalyzer (Klamt et al., 2007), which performs Boolean modeling, and the differential-equation based modeling package PottersWheel (http://www.PottersWheel.de/). Third, export of primary or transformed data from DataRail as vectors, matrices or n-dimensional arrayshas been implemented to facilitate links to other modeling tools. In this case, users need to ensure continuing compliance with the MIDAS data standard so as to preserve the integrity of metadata. Thus far we have implemented export into a MIDAS file, which can be read by Spotfire, and formats compatible with either PottersWheel or CellNetAnalyzer.
It is well recognized that modeling in biology is an iterative process in which modeling, hypotheses generation, and experiments alternate. Less obvious is that the relationship between models and data can be very complex. We have previously shown that the quality of statistical models can be improved by various preprocessing algorithms that mean-center data or scale it to unit variance (Gaudet et al., 2005). Moreover, metrics derived from time course data such as area under the curve, maximum slope, and mean value can be more informative than primary data because they implicitly account for time in different ways. However, it is rarely known a priori which data transformations will yield the best model. Instead, multiple models must be computed and the choice among them made using an objective function such as least squares fit to experimental data. From the point of view of work-flow, the key point is that a single primary data array can give rise to multiple transformed arrays, and each of these to multiple models that differ in their underlying assumptions. As a consequence, a very large number of models are generated, each of which needs to be referenced correctly to underlying data and data processing algorithms. DataRail excels at maintaining these links between model and data.
For example, data in the Liver CSR Compendiumwere processed to account for variation in experimental protocol. PCA was then used to reduce the dimensionality of the cytokine data, and k-means clustering applied to identify relevant cytokine subsets. PLSR was then performed, taking as an input phosphorylation data (signals) and as an output a PCA-derived cluster of important mediators of the inflammatory response, namely the pro-inflammatory cytokine IL1β and several activators of granulocytes (MIP1α/CCL3, MIP1β/CCL4, RANTES/CCL5, GCSF). We could have chosen a different response set, but this cluster served to demonstrate key steps in statistical modeling by PLSR. Next, 24 transformed data arrays were created for signals and responses based on different scaling (mean-centering or variance-scaling) or metrics (area under the curve, slope, and mean activation; see Fig. 5). PLSR was performed on pairs of signal-response arrays, generating 576 models that were then ranked by goodness of fit to data (a least squares fit based on R2, see Supplementary Table S3). To prevent overfitting, the number of components for each model was determined using 7-fold cross-validation (Wold et al., 2004). Importantly, the whole process of creating and evaluating models ran in DataRail in a matter of minutes, and every model could be traced back to the transformed data from which it was derived.
Fig. 5.
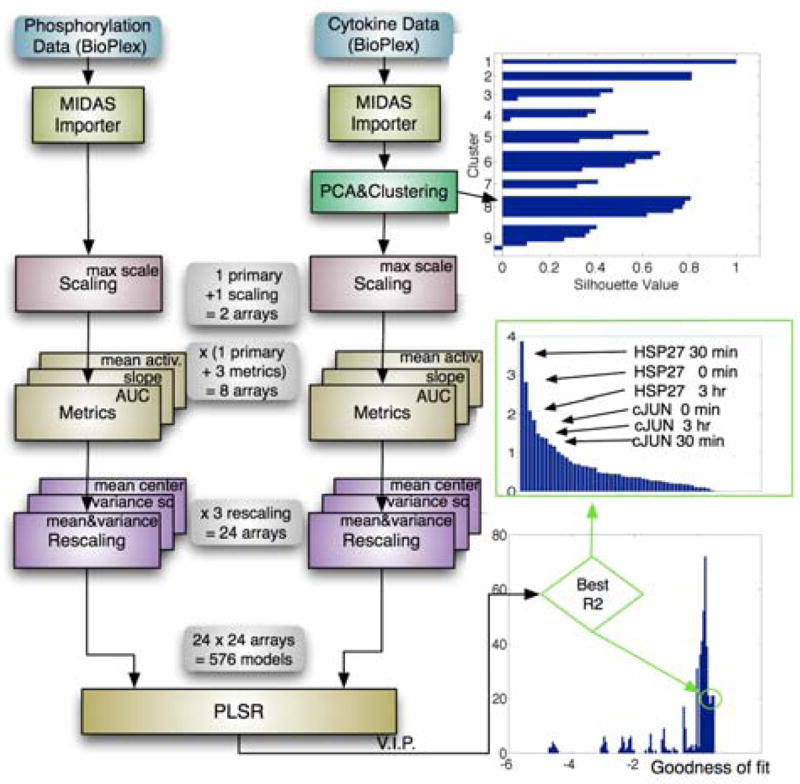
PLSR analysis in DataRail. Liver CSR data was imported to DataRail and values for protein phosphorylation designated as inputs and levels of secreted cytokine as outputs. The data was not normalized with respect to total protein concentration, to not introduce additional experimental error. The extent of cytokine co-expression was determined using internal PCA and k-means clustering routines. This yielded as set of 5 tightly clustered cytokines that were used as outputs for modeling (see row 1 of Table S1 for information about the dimensionality of the data). Primary data and data scaled with respect to maximum signal were then analyzed to compute area under the curve, slope, and mean change; this generated 8 transformed arrays for both input and output data. The resulting arrays were rescaled using routines for mean-centering, variance-scaling, or both combined (auto-scaling). The resulting 24 input cubes and 24 output cubes gave rise to 576 PLSR models, which were ranked according to their goodness of fit. For the best model, the variable importance of projection (VIP) is shown as a way to assess the relative importance of different inputs for cytokine secretion.
A variety of input arrays gave rise to top scoring models, but area under the curve was clearly the best measure of output (Supplementary Table S4). Models based on unit variance scaling of input data and area under the curve, which constituted the best form for the input in an earlier PLSR study (Gaudet et al., 2005), scored no better than 218 out of the 576 models and had R2 values 4-fold worse than the best model. Had we simply assumed our previous findings to be universally applicable, we would have generated models with very poor performance. When the best performing model (whose scores and loading plots can be found in Fig. S3) was examined by variable importance of projection (VIP; Gaudet et al., 2005) to see which signals were most predictive of cytokine secretion, the levels of phosphorylation of Hsp27 and cJun (each at 0, 30 min and 3 hr) comprised six of the ten highest scoring variables. Phospho-Hsp27 is an integrated measure of p38 kinase activity and cJun of JNK kinase activity; intriguingly, the levels of activating phosphorylation on p38 and JNK kinases were considerably less informative. Thus, the steady-state activities of p38 and JNK (captured by t=0 data) appear to play a key role in determining the extracellular concentrations of five cytokines and growth factors involved in epithelia-immune cell interactions. Consistent with this idea, it has previously been described that RANTES secretion is positively regulated by p38 MAPK and JNK in intestinal and airway epithelial cells (Yan et al., 2006; Pazdrak et al., 2002), as it is in liver.
2.8 Facilitating data sharing and publication
The fact that DataRail packages primary and transformed data arrays and their provenance together makes it a good means to share data among laboratories. However, knowledge transfer would be greatly facilitated by including figures, particularly those destined for publication or public presentation, within DataRail in a manner that maintained the analysis itself, the provenance of the data, and the identities of all algorithms and free parameters. Users could then interact with published figures in a dynamic fashion that would go far beyond what is available in today’s journals, while also discovering new ways in which the data could be viewed or put to use. We have implemented a special category of project whose UID can contain a Pubmed ID and in which figures are saved as structured variables, “pseudo-arrays,” that are embedded in compendia in the same manner as other arrays. We are currently working on an additional feature in which all of the relevant data in a linked SBWiki are stored as a wiki-book (e.g., a PDF file), thereby ensuring a complete description of all experimental procedures, reagents, etc. In the case of open-source publication, the actual manuscript could also be embedded; otherwise, a link would be provided to the publisher.
3 DISCUSSION
We describe the implementation of DataRail, a flexible toolbox for storing and manipulating experimental data for the purpose of numerical modeling. Metadata in DataRail is based on a “minimum information” MIDAS standard closely related to standards that have already proven their utility in the analysis of DNA microarray and other types of high-throughput data. Because MIDAS is a simplified version of the MIACA standard, export from DataRail into a MIACA-compliant file is straightforward. Based on several use cases with up to 1.5×106 data points (see Table S1), DataRail appears to be scalable and broadly useful, thanks to its efficient reuse of primary data and data processing algorithms. Compared to traditional relational databases, DataRail is significantly easier to deploy and modify, and it can accommodate a wider range of data formats since its internal arrays can have any dimensionality. Careful management of arrays via semantically typed identifiers (which also serve as URLs), use of a strict containment hierarchy, and imposition of metadata standard take the place of the rigid tabular structure found in relational databases. However, in cases in which data formats stabilize, or greater transactional capacity is desired, all or part of a DataRail data model can be implemented in an RDBMS.
The current DataRail implementation meets our original design goals in the following ways: (i) data provenance is maintained through the containment hierarchy, the record of processing steps, and the assignment of UIDs; (ii) visualization and modeling are possible with internal tools specialized to PCA and PLSR; (iii) interaction with external software such as CellNetAnalyzer, PottersWheel, and Spotfire is implemented, and export routines are available to expand this list; (iv) flexibility is provided by the use of data arrays with user-determined dimensionality and a simple interface for adding new analysis routines; (v) data sharing and publication are facilitated by a special category of project that packages together transformed arrays and figures describing key analyses, including those in published papers. Future developments in DataRail include the creation of utilities for managing image and mass spectrometry data, importers for a range of common laboratory instruments, and support for the HDF5 file format (http://hdf.ncsa.uiuc.edu/HDF5/). HDF is a widely supported, open-source format used in many fields dealing with large data sets, such as earth imaging or astronomy. HDF files are self-describing and permit access to binary data in manner that is much more efficient that with XML rules. Moreover, integration of DataRail with Gaggle and similar interoperability standards is a high priority (Shannon et al., 2006). Gaggle coordinates multiple analysis tools, among them the R/Bioconductor statistical environment (Gentleman et al., 2004), thereby providing access to tools for the statistical analysis of high-throughput data. Finally, versions of DataRail based on the open-source languages R or Python are in development, as are discussions with instrument vendors to create direct export routines for MIDAS-compatible files. In the context of commercial use, we are discussing, with a commercial partner, the implementation of granular access control functionality.
A model-centric approach explicitly encodes specific hypotheses about data and its meaning, and can therefore merge data not only at the level of information but at the more useful level of knowledge. Even in the database dependent world of business, knowledge is usually derived from information in specialized databases (data warehouses, which are static representations of transactional databases processed to ensure data consistency) using business intelligence tools. Business intelligence is, in essence, an approach to modeling business and financial processes mathematically and then testing the models on data. In the case of biological models, data plays an even more central role because many model parameters can be estimated only by induction from experimental observations. Thus, for mathematical models of biology to realize their full potential, a tight link between model and experiment is necessary. This involves not only an effective means to calibrate models, but also reliable information on data provenance. Only then can model-based predictions be evaluated in light of assumptions and uncertainties. DataRail therefore represents a step forward in the complex task of designing software that supports model-driven knowledge creation in biomedicine.
Supplementary Material
Supplementary information: accompanies this paper.
Acknowledgments
We thank S. Gaudet and J. Albeck for helpful discussions and B. Hendriks, C. Espelin, and M. Zhang for testing DataRail.
FUNDING
This work was funded by NIH grant P50-GM68762 and by a grant from Pfizer Inc. to P.K.S. and D.A.L.
Footnotes
Availability: DataRail is distributed under the GNU General Public License and available at http://code.google.com/p/sbpipeline/
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/2.0/uk/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
References
- Berners-Lee T, et al. The Semantic Web—A new form of Web content that is meaningful to computers will unleash a revolution of new possibilities. Scientific American. 2001;284:34. [Google Scholar]
- Gaudet S, et al. A compendium of signals and responses triggered by prodeath and prosurvival cytokines. Mol Cell Proteomics. 2005;4:1569–1590. doi: 10.1074/mcp.M500158-MCP200. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J, et al. Data cube: A relational aggregation operator generalizing group-by, cross-tab, and sub-totals. Data Mining and Knowledge Discovery. 1997;1:29–53. [Google Scholar]
- Jaqaman K, Danuser G. Linking data to models: data regression. Nat Rev Mol Cell Biol. 2006;7:813–819. doi: 10.1038/nrm2030. [DOI] [PubMed] [Google Scholar]
- Klamt S, et al. Structural and functional analysis of cellular networks with CellNetAnalyzer. BMC Systems Biology. 2007;1:2–14. doi: 10.1186/1752-0509-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazdrak K, et al. MAPK activation is involved in posttranscriptional regulation of RSV-induced RANTES gene expression. Am J Physiol Lung Cell Mol Physiol. 2002;283:L364–L372. doi: 10.1152/ajplung.00331.2001. [DOI] [PubMed] [Google Scholar]
- Shah AR, et al. Enabling high-throughput data management for systems biology: the Bioinformatics Resource Manager. Bioinformatics. 2006;23:906–909. doi: 10.1093/bioinformatics/btm031. [DOI] [PubMed] [Google Scholar]
- Shannon PT, et al. The Gaggle: An open-source software system for integrating bioinformatics software and data sources. BMC Bioinformatics. 2006;7:176. doi: 10.1186/1471-2105-7-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swedlow JR, et al. Informatics and Quantitative Analysis in Biological Imaging. Science. 2003;300:100–102. doi: 10.1126/science.1082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold S, et al. The PLS method and its applications in industrial RDP (research, development, and production) [Accessed 2007 Dec 2];2004 http://umetrics.com/default.asp/pagename/news_pastevents/c/2.
- Yan SR, et al. Differential Pattern of Inflammatory Molecule Regulation in Intestinal Epithelial Cells Stimulated with IL-1. J Immunology. 2006;177:5604–5611. doi: 10.4049/jimmunol.177.8.5604. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information: accompanies this paper.