Abstract
The salivary glands of blood sucking arthropods contain a redundant ‘magic potion’ that counteracts their vertebrate host’s hemostasis, inflammation, and immunity. We here describe the salivary transcriptome and proteomics (sialome) of the soft tick Ornithodoros coriaceus. The resulting analysis helps to consolidate the classification of common proteins found in both soft and hard ticks, such as the lipocalins, Kunitz, cystatin, basic tail, hebraein, defensin, TIL domain, metalloprotease, 5′-nucleotidase/apyrase, and phospholipase families, and also to identify protein families uniquely found in the Argasidae, such as the adrenomedullin/CGRP peptides, 7DB, 7 kDa, and the RGD containing single Kunitz proteins. Additionally, we found a protein belonging to the cytotoxin protein family that has so far only been identified in hard ticks. Three other unique families common only to the Ornithodoros genus were discovered. Edman degradation, 2D and 1D PAGE of salivary gland homogenates followed by tryptic digestion and HPLC MS/MS of results confirms the presence of several proteins. These results indicate that each genus of hematophagous arthropods studied to date evolved unique protein families that assist blood feeding, thus characterizing potentially new pharmacologically active components or antimicrobial agents.
Keywords: Ornithodoros coriaceus, Ixodidae, Argasidae, Sialotranscriptome, Salivary gland transcriptome, Sialome, Tick salivary gland, Ixolaris
1. Introduction
Soft ticks (Argasidae) feed exclusively on blood, but unlike the related hard ticks (Ixodidae) that feed for several days, soft tick meals last at most one hour. Argasidae also feed repeatedly as adults, laying smaller egg batches per meal than hard ticks. In their adaptation to blood feeding, arthropods have evolved a sophisticated cocktail of salivary components that disarm their host’s hemostatic system (consisting of platelet aggregation, vasoconstriction, and blood clotting), as well as opposing inflammatory and immunologic reactions [1]. Salivary transcriptomes (sialotranscriptomes; “sialo”; Greek: saliva, spittle, foam from the mouth; the salivary glands) of several hard tick species [2–7], as well as from the soft ticks Ornithodoros parkeri and Argas monolakensis [8–10] have been characterized. These studies indicate that the transcriptome repertoire of hard ticks have a larger complexity than those of soft ticks, probably deriving from the increased needs of hard ticks to counteract their hosts’ immune, inflammatory and angiogenic responses that occur on prolonged feeding. On the other hand, these same transcriptomes indicate a large common expansion of genes coding for several protein families, including metalloproteases, cysteine-rich proteins similar to metalloprotease domains (the ixostatins and ixodegrins), lipocalins, Kunitz-domain containing proteins, RGD-containing peptides, defensins, and many novel protein families that may include antimicrobial proteins. Argasidae-specific families have also been identified.
In the present study, we enlarge the dataset of Argasidae salivary gland transcripts by exploring that of Ornithodoros coriaceus, a vector of epizootic bovine abortion in the western U.S. [11–13]. The main protein families found in argasid and ixodid sialotranscriptomes display a low degree of similarity, which supports the idea that salivary proteins are evolving at a very fast rate compared to that of housekeeping proteins. The current dataset also contributes to expanding the tick sialotranscriptome landscape and aids in the eventual mapping of the evolutionary pathways leading to the diverse protein family expansions observed today.
2. Materials and methods
2.1. Ticks
Tick salivary gland extracts were prepared by collecting glands from O. coriaceus adult ticks. Glands were dissected by first bisecting the tick and then teasing the salivary glands away from the other internal organs and the tick exoskeleton. Glands were rinsed by immersion in PBS and added to 10 μl of distilled water (for 1D or 2D gel electrophoresis), or RNA later overnight at 4°C (for mRNA extraction), and stored frozen at −75°C until further analysis.
2.2. Chemicals
Standard laboratory chemicals were purchased from Sigma Chemicals (St. Louis, MO) if not specified otherwise. Formic acid and trifluoroacetic acid (TFA) were obtained from Fluka (Milwaukee, WI). Trypsin was purchased from Promega (Madison, WI). HPLC-grade acetonitrile was from EM Science (Darmstadt, Germany), and water was purified by a Barnstead Nanopure system (Dubuque, IA).
2.3. Salivary gland isolation and library construction
O. coriaceus salivary gland mRNA from 5 pairs of glands was isolated using the Micro-FastTrack mRNA isolation kit (Invitrogen, San Diego, CA). The PCR-based cDNA library was made following the instructions for the SMART cDNA library construction kit (Clontech, Palo Alto, CA). This system utilizes oligoribonucleotide (SMART IV) to attach an identical sequence at the 5′ end of each reverse-transcribed cDNA strand. This sequence is then utilized in subsequent PCR reactions and restriction digests.
First-strand synthesis was carried out using PowerScript reverse transcriptase at 42°C for 1 h in the presence of the SMART IV and CDS III (3′) primers. Second-strand synthesis was performed by a long-distance (LD) PCR-based protocol using Advantage™ Taq Polymerase (Clontech) mix in the presence of the 5′ PCR primer and the CDS III (3′) primer. The cDNA synthesis procedure resulted in the creation of SfiI A & B restriction enzyme sites at the ends of the PCR products that are used for cloning into the phage vector. The PCR conditions were: 95°C for 20 sec; 24 cycles of 95°C for 5 sec, 68°C for 6 min. A small portion of the cDNA obtained by PCR was analysed on a 1.1% agarose gel to check for the quality and range of cDNA synthesised. Double-stranded cDNA was immediately treated with proteinase K (0.8 μg/ml) at 45°C for 20 min, and the enzyme was removed by ultrafiltration though a Microcon (Amicon Inc., Beverly, CA) YM-100 centrifugal filter device. The cleaned, double-stranded cDNA was then digested with SfiI at 50°C for 2 h, followed by size fractionation on a ChromaSpin–400 column (Clontech). The profile of the fractions was checked on a 1.1% agarose gel, and fractions containing cDNAs of more than 400 bp were pooled and concentrated using a Microcon YM-100.
The cDNA mixture was ligated into the λ TriplEx2 vector (Clontech), and the resulting ligation mixture was packaged using the GigaPack® III Plus packaging extract (Stratagene, La Jolla, CA) according to the manufacturer’s instructions. The packaged library was plated by infecting log-phase XL1-Blue Escherichia coli cells (Clontech). The percentage of recombinant clones was determined by performing a blue-white selection screening on LB/MgSO4 plates containing X-gal/IPTG. Recombinants were also determined by PCR, using vector primers (5′ λ TriplEx2 and 3′ λ TriplEx2 sequencing primers) flanking the inserted cDNA and visualising the products on a 1.1% agarose/EtBr gel.
2.4. Sequencing of the O. coriaceus cDNA library
The O. coriaceus salivary gland cDNA library was plated on LB/MgSO4 plates containing X-gal/IPTG to an average of 250 plaques per 150-mm Petri plate. Recombinant (white) plaques were randomly selected and transferred to 96-well MICROTEST™ U-bottom plates (BD BioSciences, Franklin Lakes, NJ), containing 100 μl of SM buffer (0.1 M NaCl; 0.01 M MgSO4; 7 H2 O; 0.035 M Tris-HCl (pH 7.5); 0.01% gelatin) per well. The plates were covered and placed on a gyrating shaker for 30 min at room temperature. The phage suspension was either immediately used for PCR or stored at 4°C for future use.
To amplify the cDNA using a PCR reaction, 4 μl of the phage sample was used as a template. The primers were sequences from the λ TriplEx2 vector and named pTEx2 5seq (5′-TCC GAG ATC TGG ACG AGC-3′) and pTEx2 3LD (5′-ATA CGA CTC ACT ATA GGG CGA ATT GGC-3′), positioned at the 5′ end and the 3′ end of the cDNA insert, respectively. The reaction was carried out in 96-well flexible PCR plates (Fisher Scientific, Pittsburgh, PA) using TaKaRa EX Taq polymerase (TAKARA Mirus Bio, Madison, WI) on a GeneAmp® PCR system 9700 (Perkin Elmer Corp., Foster City, CA). The PCR conditions were: one hold of 95°C for 3 min; 25 cycles of 95°C for 1 min, 61°C for 30 sec; 72°C for 2 min. The amplified products were analysed on a 1.5% agarose/EtBr gel. cDNA library clones (1,100) were PCR amplified, and the ones showing single band were selected for sequencing. Approximately 200–250 ng of each PCR product was transferred to Thermo-Fast 96-well PCR plates (ABgene Corp., Epsom, Surray, UK) and frozen at −20°C before cycle sequencing using an ABI3730XL machine.
2.5. Bioinformatic tools and procedures used
Expressed sequence tags (EST) were trimmed of primer and vector sequences, clusterised, and compared with other databases as described [14]. The BLAST tool [15], CAP3 assembler [16], ClustalW [17], and Treeview software [18] were used to compare, assemble, and align sequences and to visualise alignments. For functional annotation of the transcripts we used the tool BlastX [19] to compare the nucleotide sequences to the non-redundant (NR) protein database of the National Center for Biotechnology Information (NCBI, National Library of Medicine, NIH,) and to the Gene Ontology (GO) database [20]. The tool, reverse position specific Blast (RPSBLAST) [19] was used to search for conserved protein domains in the Pfam [21], SMART [22], Kog [23], and conserved domains databases (CDD) [24]. We have also compared the transcripts with other subsets of mitochondrial and rRNA nucleotide sequences downloaded from NCBI and to several organism proteomes downloaded from NCBI (yeast), Flybase (Drosophila melanogaster), or ENSEMBL (Anopheles gambiae). Segments of the three-frame translations of the EST (because the libraries were unidirectional, ix-frame translations were not used), starting with a methionine found in the first 300 predicted amino acids (AA), or the predicted protein translation in the case of complete coding sequences, were submitted to the SignalP server [25] to help identify translation products that could be secreted. O-glycosylation sites on the proteins were predicted with the program NetOGlyc [26]. Functional annotation of the transcripts was based on all the comparisons above. Following inspection of all these results, transcripts were classified as either Secretory (S), Housekeeping (H) or of Unknown (U) function, with further subdivisions based on function and/or protein families. Phylogenetic analysis and statistical neighbor-joining (NJ) bootstrap tests of the phylogenies were done with the Mega package [27]. Glycosylphosphatidylinositol (GPI) anchor predictions was made using FragAnchor [28].
2.8. Proteome characterization using Edman sequencing
Five pairs of O. coriaceus salivary glands were dissected, placed in 50 μl water and immediately frozen at −80°C. The glands were sonicated and centrifuged at 16,000 g for 10 min. The supernatant (soluble fraction, 30 μg of protein) of O. coriaceus salivary gland homogenate was mixed with LDS (lithium dodecyl sulfate) loading buffer in the presence of β-mercaptoethanol (2%, v/v) and electrophoresed in a 4% to 12% NuPAGE gel (MES buffer). The proteins in the gel were then electroblotted to PVDF membranes in 10 mM CAPS-methanol (10%, v/v) buffer, pH 11. The membrane was stained with 0.025% Coomassie blue, de-stained in 50% methanol without acetic acid, and the bands sequenced by Edman degradation. Identification of proteins was performed using in-house software. The matches can be verified in Supplemental Tables S1 and S2 where a lower case letter represents a mismatch and the upper case single letter a match. Notice that the insoluble fraction can contain otherwise soluble proteins that precipitated in the absence of ions (solubilized in water) or that had affinity for membrane fractions.
2.9. Proteome characterization using two-dimensional (2D) gel electrophoresis and tandem mass spectrometry
2D gel electrophoresis was performed using ZOOM IPGRunner System (Invitrogen) under manufacturer’s recommended running conditions. Briefly, approximately 90 μg of sample proteins (five pairs of salivary glands) were solubilized with 155 μl rehydration buffer (7 M urea, 2 M thiourea, 2% CHAPS (3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate), 20 mM DTT, 0.5% carrier ampholytes, pH 3–10). The samples were absorbed by rehydration ZOOM strips (7 cm; pH 3–10 NL) overnight at room temperature and then focused under manufacturer’s recommended conditions. The focused IPG strips were reduced/alkylated/equilibrated with reducing and then alkylation reagents dissolved in the sample buffer. The strips were then applied onto NuPAGE 4% to 12% Bis Tris ZOOM gels (Invitrogen). The gels were run under MOPS buffer and stained with SeeBlue staining solution (Bio Rad, Hercules, CA). A total of 60 spots were selected for tryptic digestion based on their staining intensity. Protein identification of 2D gel-separated proteins was performed on reduced and alkylated trypsin-digested samples prepared using standard mass spectrometry protocols. Tryptic digests were analyzed by coupling the Nanomate (Advion BioSciences)—an automated chip-based nano electrospray interface source—to a quadrupole time-of-flight tandem mass spectrometer (QStarXL MS/MS System, Applied Biosystems/Sciex, Foster City, CA). Computer-controlled data-dependent automated switching to tandem mass spectrometry (MS/MS) provided peptide sequence information. AnalystQS software (Applied Biosystems/Sciex) was used for data acquisition. Data processing and databank searching were performed with Mascot software (Matrix Science, Boston, MA). The NR protein database from the NCBI was used for the search analysis, as was a protein database generated during the course of this work.
2.10. Proteomic characterization using 1D gel electrophoresis and MS/MS
The soluble (S) and the insoluble (pellet, P) protein fractions (see section 2.8 above) from salivary gland homogenates from O. coriaceus corresponding to 100 μg of protein were both solubilized in reducing Laemmli gel-loading buffer. The samples were boiled for 10 min and 50 μg of protein was loaded and resolved on a NuPAGE 4% to 12% Bis-Tris precast gel with antioxidants and visualized by staining with SimplyBlue (Invitrogen). Each gel lane was sliced into approximately 30 (S)–31 (P) gel slices, de-stained, and digested overnight with trypsin at 37°C. Peptides were subsequently extracted and desalted using ZipTips (Millipore, Bedford, MA) and resuspended in 0.1% TFA prior to MS/MS analysis.
Nanoflow reversed-phase liquid chromatography (RPLC)-MS/MS was performed using a 1100 nanoflow LC system (Agilent Technologies, Palo Alto, CA) coupled online with a linear ion trap (LIT) MS (LTQ; ThermoElectron, San José, CA). NanoRPLC columns were slurry packed in-house with 5 μm 300 Å pore-size C-18 phase (Jupiter, Phenomenex, CA) in a 75 μm i.d. × 10-cm fused silica capillary (Polymicro Technologies, Phoenix, AZ) with a flame-pulled tip. After sample injection, the column was washed for 20 min with 98% mobile phase A (0.1% formic acid in water) at 0.5 μl/min, and peptides were eluted using a linear gradient of 2% mobile phase B (0.1% formic acid in acetonitrile) to 42% mobile phase B for 40 min at 0.25 μl/min, then to 98% B for an additional 10 min. The LIT-mass spectrometer was operated in a data-dependent MS/MS mode in which each full MS scan was followed by seven MS/MS scans where the seven most abundant molecular ions were dynamically selected for collision-induced dissociation (CID) using a normalized collision energy of 35%. Dynamic exclusion was applied to minimize repeated selection of peptides previously selected for CID.
Tandem mass spectra were searched using SEQUEST on a 20-node Beowulf cluster against an O. coriaceus proteome database with methionine oxidation included as dynamic modification. Only tryptic peptides with up to two missed cleavage sites meeting a specific SEQUEST scoring criteria were considered as legitimate identifications [delta correlation (ΔCn) ≥ 0.08 and charge state-dependent cross correlation (Xcorr) ≥ 1.9 for [M+H]1+, ≥ 2.2 for [M+2H]2+ and ≥ 3.1 for [M+3H]3+]. We further identified those peptides where two or more fragments were identified from a single gel fraction to make a protein call, as indicated in Figure 9, Table 4, and Supplemental Table S2.
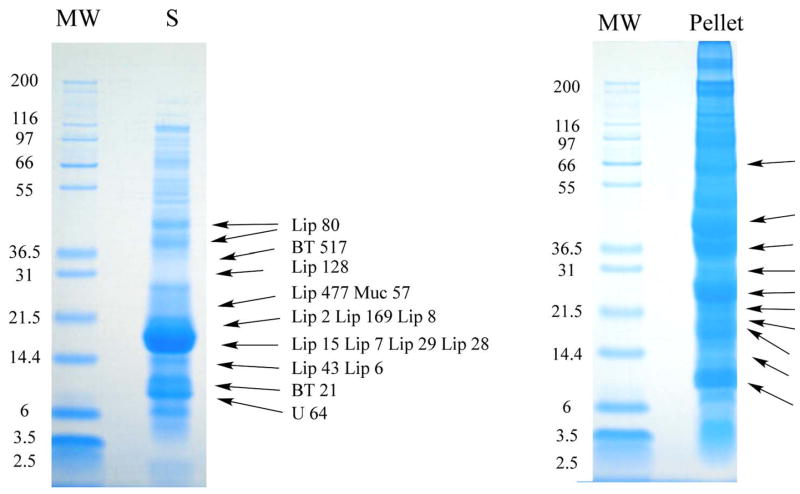
Fig. 9.
1D gel electrophoresis of O. coriaceus salivary gland homogenates followed by tryptic digestion and HPLC MS/MS of results. Soluble (S) and insoluble (Pellet) salivary gland homogenates of O. coriaceus were dissolved in reducing SDS buffer and applied to a polyacrylamide gel. After electrophoresis, each gel was sliced into 28 (S) or 31 (Pellet) fractions and submitted to tryptic digest and reverse-phase HPLC MS/MS. Numbers on the left indicate molecular weight marker positions in the gel. The bands labeled with Lip X correspond to lipocalins where X is the number of the OC-X protein found in Supplemental Table S2. Similarly, BT is for basic tail proteins, Muc for mucin, Cy for cystatin, DK for double Kunitz, Pl for phospholipase, Mt for metalloprotease, Ctx for cytotoxin-like, 5Nuc for 5′-nucleotidase/apyrase, GR for glycine rich, and U for Unknown protein. Other protein matches can be found in Supplemental Table S2.
Table 4.
Description of salivary secreted polypeptides deposited in GenBank, with summary of proteomic experiments
| Seq name | GenBank accession number and link to NCBI | Status | Description | Match to 1D gel Edman (Fig. 7) | Match to 2D gel MS/MS results (Fig. 8) | Match to 1D PAGE HPLC MS/MS results (Fig. 9) |
|---|---|---|---|---|---|---|
|
Lipocalin family
| ||||||
| OC-15 | 172051078 | Complete | Salivary lipocalin | Y | Y | Y |
| OC-43 | 172051154 | Complete | Salivary lipocalin | Y | ||
| OC-41 | 172051146 | Complete | Salivary lipocalin | Y | ||
| OC-2 | 172051090 | Complete | Salivary lipocalin | Y | Y | |
| OC-3 | 172051116 | Complete | Salivary lipocalin | Y | Y | |
| OC-62 | 172051210 | Complete | Salivary lipocalin | Y | ||
| OC-80 | 172051236 | Complete | Salivary lipocalin | Y | ||
| OC-169 | 172051084 | Complete | Moubatin-like lipocalin | Y | ||
| OC-677 | 172051218 | Complete | Salivary lipocalin | |||
| OC-477 | 172051166 | Complete | Salivary secreted lipocalin | Y | ||
| OC-6 | 172051204 | Complete | Salivary lipocalin | Y | Y | |
| OC-7 | 172051222 | Truncated | Salivary lipocalin | Y | Y | |
| OC-8 | 172051234 | Complete | Salivary lipocalin | Y | ||
| OC-29 | 172051114 | Complete | Salivary lipocalin | Y | Y | |
| OC-28 | 172051112 | Complete | Salivary lipocalin | Y | Y | |
| OC-60 | 172051206 | Complete | Moubatin-like lipocalin | Y | ||
| OC-481 | 172051168 | Complete | Salivary lipocalin | Y | ||
| OC-128 | 172051062 | Complete | Salivary lipocalin | Y | ||
|
Single-Kunitz protease inhibitors | ||||||
| OC-51 | 172051178 | Complete | Savignygrin-like | |||
| OC-52 | 172051182 | Complete | Savignygrin-like RGD containing salivary peptide | |||
| OC-53 | 172051186 | Complete | Savignygrin-like peptide | |||
|
Dual-Kunitz protease inhibitors | ||||||
| OC-24 | 172051102 | Complete | Dual-Kunitz salivary protein | Y | ||
| OC-25 | 172051106 | Complete | Dual-Kunitz salivary protein | Y | ||
| OC-26 | 172051110 | Complete | Dual-Kunitz salivary protein | Y | ||
|
Cystatin | ||||||
| OC-38 | 172051140 | Complete | Salivary cystatin | Y | ||
| OC-37 | 172051136 | Complete | Salivary cystatin 2 | Y | ||
|
Calcitonin/adrenomedulin family peptide | ||||||
| OC-39 | 172051142 | Complete | Calcitonin-related polypeptide | |||
| OC-40 | 172051144 | Complete | Similar to calcitonin/calcitonin-related polypeptide | |||
| OC-49 | 172051174 | Complete | Calcitonin/calcitonin-related polypeptide | |||
| OC-50 | 172051176 | Complete | Calcitonin/calcitonin-related polypeptide | |||
|
Possible mucin | ||||||
| OC-57 | 172051196 | Complete | Salivary mucin | Y | ||
|
Antimicrobial-hebraein family | ||||||
| OC-32 | 172051122 | Truncated | Microplusin-like antimicrobial | |||
| OC-31 | 172051120 | Complete | Hebraein-like | |||
| OC-33 | 172051124 | Complete | Hebraein-like | |||
| OC-696 | 172051220 | Complete | Hebraein-like | |||
|
Defensin | ||||||
| OC-487 | 172051172 | Truncated | Defensin | |||
|
TIL domain-containing peptides | ||||||
| OC-1000 | 172051048 | Complete | Ixodidin | |||
| OC-86 | 172051240 | Complete | Hypothetical conserved secreted protein | |||
|
Salivary enzymes with possible blood-feeding function | ||||||
|
Metalloproteases | ||||||
| OC-372 | 172051138 | Truncated | Metalloprotease | Y | ||
| OC-676 | 172051216 | Truncated | Salivary metalloprotease | |||
|
Phospholipase A2 fragments | ||||||
| OC-420 | 172051150 | Truncated | Phospholipase A2 | |||
| OC-92 | 172051246 | Truncated | Phospholipase A2 | Y | ||
| OC-96 | 172051250 | Truncated | Phospholipase A2 | Y | ||
|
5′-nucleotidase/apyrase precursor fragment | ||||||
| OC-79 | 172051232 | Truncated | 5′-nucleotidase/putative apyrase precursor | Y | ||
|
Tick families of unknown function | ||||||
|
Basic tail family | ||||||
| OC-21 | 172051094 | Complete | BTSP | Y | Y | |
| OC-20 | 172051092 | Complete | BTSP | Y | Y | |
| OC-22 | 172051096 | Complete | BTSP | Y | ||
| OC-54 | 172051188 | Complete | Acid tail salivary protein | Y | ||
| OC-56 | 172051192 | Complete | Acid tail salivary protein | Y | ||
| OC-84 | 172051238 | Truncated | BTSP | |||
| OC-139 | 172051076 | Complete | Salivary secreted basic tail protein | |||
| OC-23 | 172051098 | Complete | BTSP | |||
| OC-517 | 172051180 | Complete | BTSP | Y | ||
| OC-133 | 172051070 | Complete | Hypothetical secreted peptide precursor | Y | ||
|
Cytotoxin family | ||||||
| OC-574 | 172051198 | Truncated | Putative salivary secreted protein | Y | ||
|
Glycine-rich protein family | ||||||
| OC-130 | 172051066 | Complete | Putative secreted glycine-rich salivary protein | Y | ||
|
7DB family unique to Argasidae | ||||||
| OC-104 | 172051050 | Complete | 7DB family | |||
| OC-120 | 172051060 | Complete | 7DB family | Y | ||
| OC-106 | 172051054 | Complete | 7DB family | Y | ||
|
Peptides of the 8-kDa family | ||||||
| OC-98 | 172051254 | Complete | Hypothetical secreted peptide precursor | |||
| OC-194 | 172051088 | Complete | Hypothetical secreted peptide precursor | |||
|
Unique Ornithodoros families | ||||||
| OC-64 | 172051214 | Complete | Hypothetical secreted peptide precursor | Y | ||
|
Disulphide-rich 5-kDa family | ||||||
| OC-93 | 172051248 | Complete | Putative salivary secreted peptide | |||
| OC-129 | 172051064 | Complete | Hypothetical secreted peptide precursor | |||
|
Novel families | ||||||
| OC-724 | 172051226 | Complete | Hypothetical secreted peptide precursor | |||
| OC-458 | 172051158 | Complete | Hypothetical secreted peptide precursor | |||
| OC-46 | 172051160 | Complete | Hypothetical secreted peptide precursor | |||
| OC-520 | 172051184 | Complete | Hypothetical secreted peptide precursor | |||
| OC-639 | 172051212 | Complete | Hypothetical secreted peptide precursor | |||
| OC-73 | 172051228 | Complete | Hypothetical secreted peptide precursor | |||
| OC-97 | 172051252 | Complete | Hypothetical secreted peptide precursor | |||
3. Results and Discussion
3.1. cDNA library characteristics
A total of 1,089 clones were sequenced and used to assemble a database (see Supplemental Table1) yielding 726 clusters of related sequences, 628 of which contained only one EST. As in our previous papers [2, 7, 29], the consensus sequence of each cluster is named either a contig (deriving from two or more sequences) or a singleton (deriving from a single sequence). For simplicity sake, this manuscript uses ‘cluster’ to denote sequences deriving both from consensus sequences and from singletons. The 726 clusters were compared using the program BlastX, BlastN, or RPSBLAST [19] to the NR protein database of the NCBI, to a gene ontology database [20], to the CDD of the NCBI [24], and to a custom-prepared subset of the NCBI nucleotide database containing either mitochondrial or rRNA sequences.
Because the libraries used are unidirectional, three-frame translations of the dataset were also derived, and open reading frames starting with a methionine and longer than 40 AA residues were submitted to SignalP server [25] to help identify putative-secreted proteins. The EST assembly, BLAST, and signal peptide results were loaded into an Excel spreadsheet for manual annotation and can be browsed as Supplemental Table S1.
Four categories of expressed genes derived from the manual annotation of the contigs (Table 1). The putatively secreted (S) category contained 17.5% of the clusters and 31.5% of the sequences, with an average number of 2.7 sequences per cluster. The housekeeping (H) category had 23% and 22% of the clusters and sequences, respectively, and an average of 1.45 sequences per cluster. Fifty-nine percent of the clusters, containing 46% of all sequences, were classified as unknown (U), because no functional assignment could be made. This category had an average of 1.2 sequences per cluster. A good proportion of these transcripts could derive from 3′ or 5′ untranslated regions of genes of the above two categories, as was recently indicated for a sialotranscriptome of An. gambiae [30]. We also found three transcripts possibly related to transposable elements (TE class), a common occurrence in sialotranscriptomes.
Table 1.
Transcript abundance according to functional class
| Clusters
|
Sequences
|
||||
|---|---|---|---|---|---|
| Class | Number | (Percent of total) | Number | (Percent of total) | Sequences/Cluster |
| Housekeeping | 167 | (23.0) | 242 | (22.2) | 1.45 |
| Secreted | 127 | (17.5) | 343 | (31.5) | 2.70 |
| Unknown | 429 | (59.1) | 501 | (46.0) | 1.17 |
| Transposable elements | 3 | (0.4) | 3 | (0.3) | 1.00 |
| Total | 726 | 1089 | |||
3.2. Housekeeping (H) genes
The 167 clusters (comprising 242 EST) attributed to H genes expressed in the salivary glands of O. coriaceus were further characterized into 14 subgroups according to function (Table 2). Not surprisingly for an organ specialized in secreting polypeptides, the two larger sets were associated with protein synthesis machinery (68 EST in 51 clusters) and energy metabolism (80 EST ion 37 clusters), a pattern also observed in other sialotranscriptomes [8–10, 29, 31, 32]. We have arbitrarily included in the H category a group of 28 EST that grouped into 26 clusters, representing highly conserved proteins of unknown function, presumably associated with cellular function, but still uncharacterized. They are named ‘conserved proteins of unknown function’ in Supplemental Table S1, immediately preceding the clusters of the TE class. These sets may help functional identification of the ‘conserved hypothetical’ proteins as previously reviewed [33]. The complete list of all gene clusters, along with further information about each, is given in Supplemental Table S1.
Table 2.
Functional classification of housekeeping transcripts
| Function | Clusters | Sequences | Sequences/Cluster |
|---|---|---|---|
| Energy metabolism | 37 | 80 | 2.16 |
| Protein synthesis | 51 | 68 | 1.33 |
| Unknown conserved | 26 | 28 | 1.08 |
| Cytoskeletal | 7 | 11 | 1.57 |
| Transcription machinery | 11 | 11 | 1.00 |
| Transporters/Storage | 5 | 11 | 2.20 |
| Nuclear regulation | 8 | 9 | 1.13 |
| Proteasome machinery | 6 | 7 | 1.17 |
| Protein modification | 5 | 5 | 1.00 |
| Lipid, amino acid and carbohydrate metabolism | 3 | 3 | 1.00 |
| Protein export | 3 | 3 | 1.00 |
| Detoxication metabolism | 3 | 3 | 1.00 |
| Immunity | 1 | 2 | 2.00 |
| Transcription factors | 1 | 1 | 1.00 |
| Total | 167 | 242 |
3.3. Possibly secreted (S) class of expressed genes
Inspection of Supplemental Table S1 indicates the expression of several expanded gene families, including those coding for lipocalins, Kunitz domain-containing polypeptides and mucins, among others (Table 3). Lipocalin members alone represent 30 clusters and 137 EST, or 13% of the total number of sequenced clones, a pattern observed previously in both hard and soft ticks ticks [2, 8, 9]. Similarly, Kunitz domain- and basic tail-containing peptides account for 20 clusters and 65 EST. Several other peptide families, described in greater detail below, were also observed.
Table 3.
Functional classification of transcripts coding for secreted proteins
| Family | Clusters | Sequences | Sequences/Cluster |
|---|---|---|---|
| Lipocalins | 30 | 137 | 4.57 |
| Basic tail | 14 | 46 | 3.29 |
| Other | 29 | 40 | 1.38 |
| Antimicrobials | 13 | 28 | 2.15 |
| Kunitz | 6 | 19 | 3.17 |
| Similar to various Argasidae proteins | 9 | 16 | 1.78 |
| 7 DB Argasidae family | 6 | 9 | 1.50 |
| CGRP related peptide | 4 | 14 | 3.50 |
| Cystatins | 2 | 11 | 5.50 |
| Metalloproteases | 7 | 7 | 1.00 |
| RGD containing disagregins | 3 | 7 | 2.33 |
| Lipases | 3 | 6 | 2.00 |
| 5′ nucleotidase/apyrase | 1 | 3 | 3.00 |
| Total | 127 | 343 |
3.4. Analysis of the O. coriaceus sialotranscriptome
Several clusters of sequences coding for H and S polypeptides indicated in Supplemental Table S1 are abundant and complete enough to extract consensus sequences of novel sequences. Additionally, we have performed primer extension studies in several clones to obtain full- or near-full-length sequences of products of interest. A total of 104 novel sequences, 74 of which code for S proteins, are grouped together in Supplemental Table S2. Table 4 has a summary of the secreted subset, with links to GenBank.
Following is a detailed description of the full-length or near-full-length transcripts found in the salivary glands of O. coriaceus:
3.5 Putative salivary secreted proteins having ubiquitous domains or with known function
3.5.1. Lipocalins
Ticks and triatomine bugs, among the remaining blood-sucking arthropod orders studied so far, uniquely and abundantly express many members of the lipocalin family in their salivary glands [2, 8, 9, 31, 34–37], clearly an example of convergent evolution. The function of only a few of these proteins are known, and they vary from nitric oxide carriers in Rhodnius [38], to small ligand binders such as serotonin in both Rhodnius and ticks [39, 40] and as anticlotting and anticomplement found in Rhodnius [41, 42] and soft ticks [43], respectively. A recent review of tick lipocalins revealed several hundreds such sequences found in diverse tick sialotranscriptomes, indicating the large expansion and divergence of this protein family in ticks [44].
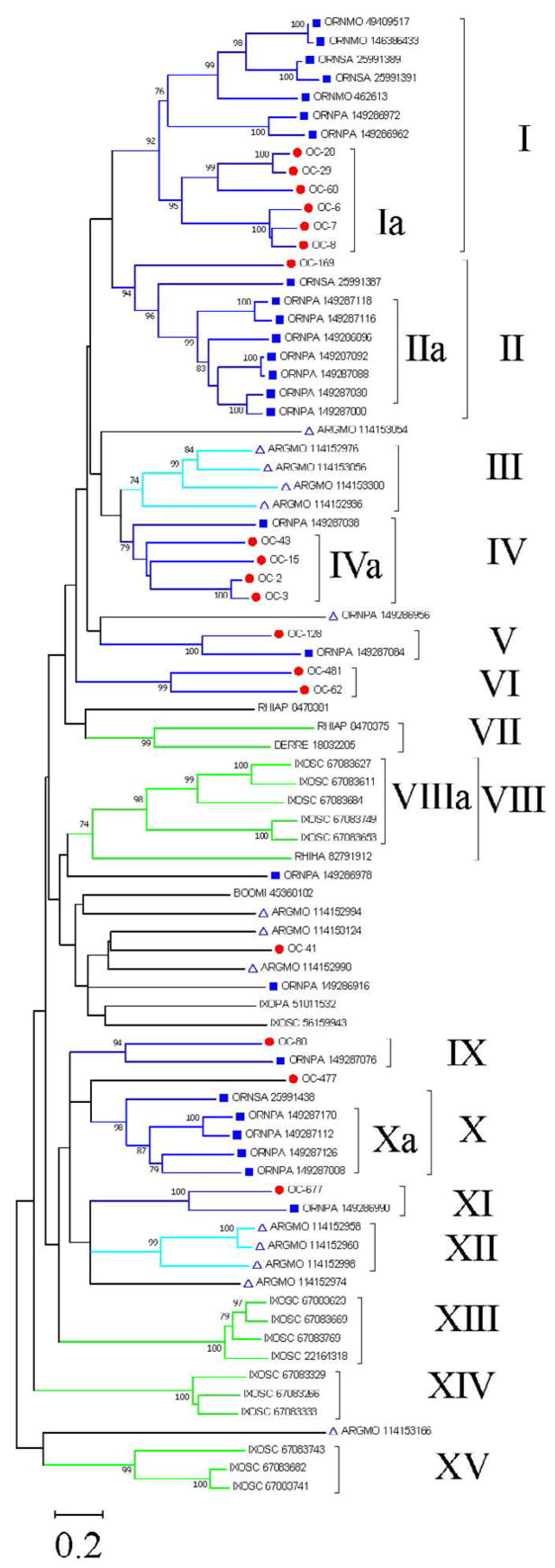
Eighteen sequences of salivary lipocalins (one of which is truncated) were mined from the O. coriaceus sialotranscriptome (Supplemental Table S2). Some of these may be alleles from the same gene, such as the pairs OC-2/OC-3, as they are more than 90% identical, or they may reflect recent or conserved gene duplication events. These sequences were aligned using the program ClustalW [17] with other tick lipocalins existing in the NR database of the NCBI, which gave sequence similarity (using BLASTP) to either of the 18 O. coriaceus lipocalins. The alignment was submitted to the MEGA package [27] to produce the phylogram shown in Figure 1. Bootstrap support (above 75%) was obtained for 15 clades as indicated by the Roman numerals in Figure 1. Inspection of these clades indicates: i) no metastriate sequence grouped with prostriate-derived sequences, as expected from the phylogeny, possibly a reflection of the rapid evolution rate of this protein family to the point of losing recognition of a common ancestor. ii) Monophyletic clades (Ia, IIa, III, IVa, VI, VIIIa, Xa, XII, XIII, XIV, and XV) indicate either recent gene duplication events in different species or the occurrence of concerted evolution or gene conversion that can homogenize tandem arrayed genes within a degree of similarity [45, 46], as previously proposed for the evolution of tick proteins [47, 48]. Clade Ia (Fig. 1) contains the most abundant group of expressed lipocalins in O. coriaceus salivary glands (OC-6, OC-7 and OC-8, OC-28, OC-29, and OC-60) and groups in Clade I with moubatin, OMCI, TSGP2, and TSGP3, proteins previously described for Ornithodoros savigny and Ornithodoros moubata [49, 50], and several other lipocalins from O. parkeri [8]. OC-169 is a single protein sequence from O. coriaceus in Clade II, that has an expanded family of seven O. parkeri sequences. Of interest, clade I abounds with O. coriaceus, O. moubata and O. savigny sequences but has only two O. parkeri sequences. This result may reflect either recent gene expansions or gene conversion mechanisms, as indicated above. Clade IV contains four O. coriaceus lipocalins grouping with a single O. parkeri sequence. Clade V has one protein each of O. coriaceus and O. parkeri, as is the case with clades IX and XI. The function of these proteins can only be speculated upon. The members of clade IVa OC-43, OC-2, and OC-3 have the PFAM motif of histamine-binding lipocalins, and do have the motif C-D-[VIL]-x(7,17)-E-L- [WY]-x(11,30)-C representing the AA fold involved in histamine binding as deduced from the crystal structure of lipocalins co-crystallized with biogenic amines (unpublished observations). As expected, they are abundantly expressed proteins that would bind these biogenic amines, which accumulate to near micromolar levels during inflammation and hemostasis. The lipocalins in clade Ia, however, are also abundantly expressed. They significantly co-localize within clade I with lipocalins from African ticks, such as moubatin, TSGP2, and TSGP3, and O. parkeri proteins, some of which have recently shown to bind leukotriene B4 and thromboxane A2 (unpublished observations). A branch without significant bootstrap support leads to clades IX–XII, which includes the O. savignyi lipocalin (gi|25991438) and the A. monolakensis protein (gi|114152974), both of which were shown to avidly bind cysteinyl leukotrienes such as LTC4 (unpublished observations). No O. coriaceus sequence was found to group with the O. moubata inhibitor of complement activation (gi|49409517) [43], which is a member of clade I, and appears to be exclusive of African Ornithodoros species (unpublished observations). It is possible that in addition to these mediators, lipocalins will be found that bind platelet aggregation factor or adenosine nucleotides, as is the case with Rhodnius [51, 52].
Fig. 1.
Unrooted phylogram of the O. coriaceus salivary lipocalins with other tick salivary lipocalins. The O. coriaceus sequences are indicated by OC-X where X is the number shown in Supplemental Table S2. The remaining sequences derived from the non-redundant (NR) protein database of the National Center for Biotechnology Information (NCBI) and are represented by five letters followed by the NCBI gi| accession number. The five letters derive from the first three letters of the genus and the first two letters from the species name. The protein sequences were aligned by the Clustal program [17], and the phylogram was done with the Mega package [27] after 5,000 bootstraps with the neighbor-joining algorithm. The bar at the bottom represents 20% amino acid substitution; circles indicate O. coriaceus sequences reported in this work; squares represent sequences deriving from other Ornithodoros ticks; triangles represent sequences from A. monolakensis. Remaining sequences are from metastriate ticks. Roman numerals represent groups of two or more sequences that reside on a clade with bootstrap support larger than 75%, as indicated by the numbers on the tree nodes.
3.5.2. Kunitz-domain containing polypeptides
The Kunitz domain characterizes many protease inhibitors. It was first identified in the pancreatic Kunitz inhibitor, also known as aprotinin or bovine pancreatic trypsin inhibitor [53]. Multiple Kunitz domains may occur in a single protein, thus providing for inhibition in multiple protease amplification cascades such as occur with the vertebrate tissue factor pathway inhibitor of the extrinsic blood coagulation cascade [54]. The tick sialomes have been studied to date contain a large number of Kunitz domain-containing polypeptides [2, 6–9, 55]. Some of these tick proteins have been characterized functionally and shown to have both expected and unexpected functions. Expected functions include inhibition of blood clotting at different target proteases, such as the proteins named ixolaris [56, 57] and penthalaris [58]. Unexpected functions are related to inhibition of platelet aggregation, a function that evolved by a unique incorporation of an RGD motif into the Kunitz domain, first described by Mans et. al in 2002 [59]. The RGD peptide competes with fibrinogen binding to platelet receptor αIIbβ3, thus preventing platelet aggregation [59]. So far, this RGD acquisition into a Kunitz domain has been found exclusively in the Argasidae.
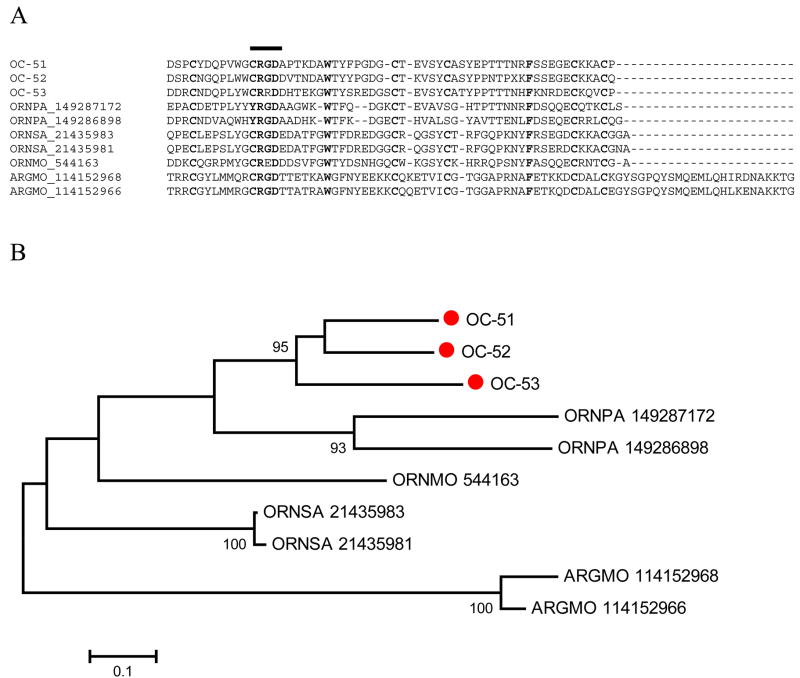
Supplemental Table S2 presents the full-length sequences of six polypeptides containing Kunitz domains with RGD-motifs, three with a single and three with double domains. Alignment of the mono Kunitz proteins of O. coriaceus with the other known Kunitz/RGD peptides of the Argasidae shows that two of the three proteins of O. coriaceus have the canonical RGD domain, and one has instead a RRD domain (Fig. 2). Of note, the O. moubata homologue (gi|544163) has a RED instead of a RGD but is a potent inhibitor of platelet aggregation [60], as are those of O. savigny [59, 61]. The O. coriaceus proteins have six conserved cysteine residues, as do those derived from the African species, O. moubata and O. savigny, but differ from the American tick O. parkeri, which have only four conserved cysteines. A. monolakensis Kunitz/RGD peptides, named monogrins [10] contain the canonical RGD domain and six-cysteine backbone, but additionally contain an extra carboxyterminus domain. The bootstrapped phylogram derived from the alignment (Fig. 2B) shows that the proteins cluster by species, indicating either that gene duplication events occurred after the lineages split from the common ancestor or that they have evolved in a concerted way within each genome. The exclusive finding of this family among Argasidae suggests either that this was an ‘invention’ of soft ticks after the split of the ancestral tick into the two existing lineages or that this gene family was lost in the hard ticks.
Fig. 2.
The single-Kunitz family of Argasidae. A) ClustalW alignment of the known single-Kunitz peptides from the Ornithodoros and Argas genera (excluding the signal peptide region). The line above the alignment marks the RGD domain region. Cysteine residues are marked in black background. Other identical residues are marked in yellow (gray) background. B) Unrooted phylogram of the alignment. OC-labeled sequences are from O. coriaceus. Remaining sequences are from O. parkeri, O. moubata, O. savignyi, and A. monolakensis. For other details, see legend of Fig. 1.
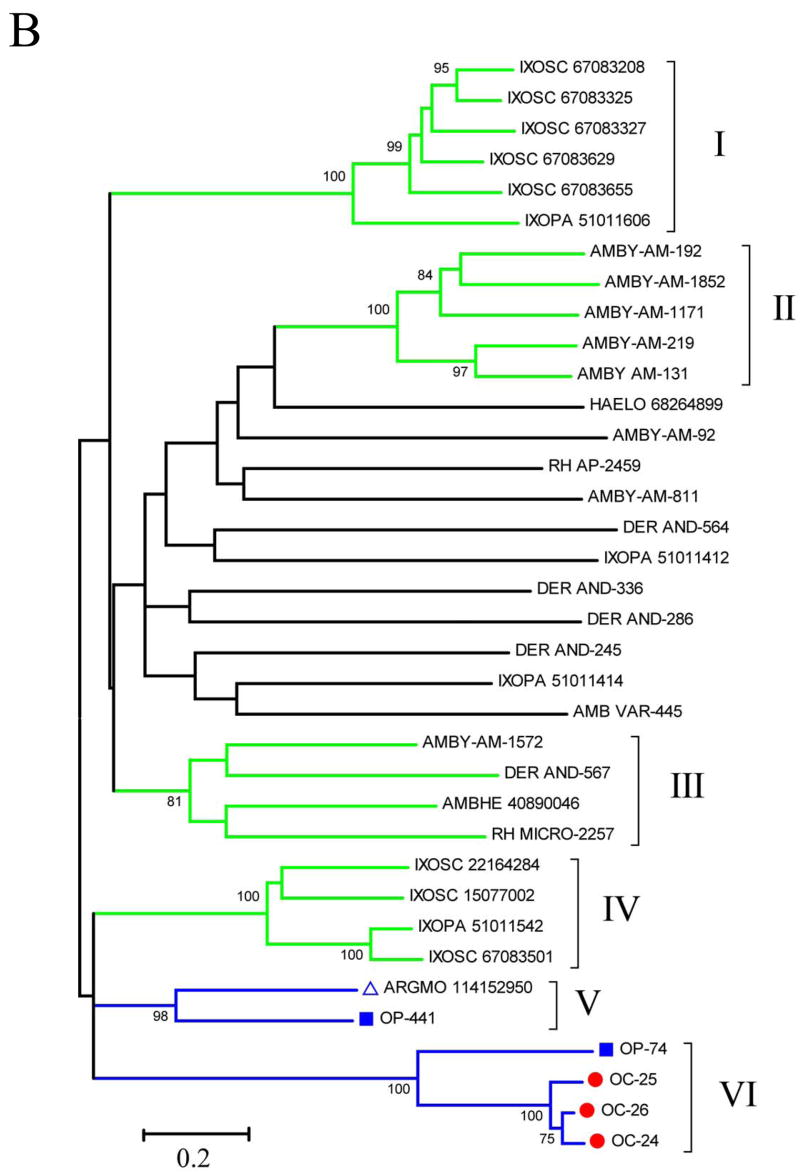
The three double-Kunitz domain-containing proteins from O. coriaceus shown in Supplemental Table 2 were compared by ClustalW with known tick salivary proteins also containing two Kunitz domains with sizes varying from 140 to 200 AA residues (Fig 3A). The resulting alignment reveals a common cysteine backbone and a single additional conserved phenylalanine residue, indicating the vast divergence of this tick superfamily. The bootstrapped phylogram (Fig 3b) indicates monophyletic family expansions, as indicated by clades I, II, IV, and VI, but also families conserved between genera, as in clades III, V, and VI, the latter containing all O. coriaceus sequences and one O. parkeri sequence. To date, the only member of this superfamily shown in Fig 3B to have a known function is Ixolaris, from Ixodes. scapularis (gi|15077002 shown in clade IV), which is a blood-clotting inhibitor acting on the extrinsic Xase complex [56, 62]. The divergence of this superfamily suggests other proteases may be targeted. The C-terminal Kunitz domains of the proteins in clade VI also possess the RGD-motif found in the single-domain RGD proteins (Fig. 3A). This was previously observed for the O. parkeri protein found in clade VI [8] and suggests that this clade forms an orthologous group with a probable conserved function that involves targeting of integrins.
Fig. 3.
Comparison of the double-Kunitz/Bilaris salivary proteins of O. coriaceus with other tick-derived double-Kunitz proteins. A) ClustalW alignment (excluding the signal peptide region). B) Unrooted phylogram of the alignment. OC-labeled sequences are from O. coriaceus. For additional information, see legend of Fig. 1.
Double-Kunitz domain thrombin inhibitors (Monobin, Ornithodorin, and Savignin) that belong to the Kunitz family have been characterized in the salivary glands of A. monolakensis, O. moubata and O. savignyi, respectively, indicating that thrombin inhibitors were conserved in the ancestral soft tick lineage. Single Kunitz-domain fXa inhibitors (fXaI and TAP) have also been described for O. moubata and O. savignyi, but were absent in A. monolakensis [10, 63–65]. It was suggested that the fXa-inhibitors evolved exclusively in the Ornithodorinae lineage [10]. Of interest are the absence of both thrombin and fXa-inhibitors in the sialome of O. coriaceus, similar to O. parkeri [8]. It would thus seem that the thrombin and fXa inhibitors have been lost in related Nearctic Ornithodoros genera. Whether these gene losses were due to ancestral host preference or a chance event that occurred in the ancestral lineage of Nearctic Ornithodorinae remains to be determined. The fXa inhibitors could also have evolved after the divergence of African and Nearctic Ornithodoros species.
3.5.3. Cystatin-like molecules
All cystatins studied to date are tight binding inhibitors of papain-like cysteine proteinases (e.g., cathepsins B and L). At the structural level, three distinct loops form the interaction interface of cystatins with their target enzymes. Highly conserved AA can be found in these loops (underlined in the multiple alignment shown in Fig. 4), i.e., a conserved glycine in the N-terminal loop (marked with the number ‘1’ in Fig. 4), a QXVXG in the second loop (also known as the cystatin motif, marked with the numeral ‘2’ in Fig. 4), and a PW dipeptide in the third loop (marked with the numeral ‘3’ in Fig. 4) [66]. Since the first description of chicken egg-white cystatin [67], members of this family have been described in protozoa, invertebrates, vertebrates, and plants. We have previously described two cystatins in the sialotranscriptome of hard ticks [2, 68], while cystatins have been also characterized from other organs of soft and hard ticks [69–71]. Actually, the salivary cystatins from the hard tick I. scapularis (named as sialostatin L and sialostatin L2) are the first to be described as lacking the PW motif in their third loop. Moreover, they show a stringent specificity against mainly cathepsin L and cathepsin S, 1thus displaying anti-inflammatory and immunomodulatory properties [72, 73]. In Supplemental Table S2 and Figure 4, we present two cystatin-like sequences obtained from the sialotranscriptome of O. coriaceus. Given their high similarity on AA sequence level, they may correspond either to products of two different alleles of the same gene or to products of two different genes that resulted from a recent duplication event in the genome of this soft tick. Alignment of the two O. coriaceus salivary cystatin protein sequences with one truncated sequence found in the O. parkeri sialotranscriptome, two extensively characterized sequences from I. scapularis, and other vertebrate and invertebrate cystatins reveals that these soft tick cystatin-like proteins not only lack the PW motif within their third loop but also the cystatin motif within the second loop, making it questionable whether they display any activity against papain-like cysteine proteases. Given the difference in the feeding behavior between hard and soft ticks, the functional characterization of these new cystatins may shed light into further differences between these two families in the inhibitory potential of their salivary gland secretion. Finally, all secreted cystatins display a conserved pair of disulfide bonds [66], which in the O. coriaceus proteins is quite out of phase when compared with the other sequences, including the salivary cystatin of O. parker. This observation further enhances the idea of a unique fold and activity—if any—for these ‘atypical’ soft tick cystatins.
Fig. 4.
Alignment of the O. coriaceus salivary cystatins with cystatins. The O. parkeri sequence is a truncated sequence. Other sequences have their signal peptide excluded. The numbers above the figure indicate the conserved regions of vertebrate sequences. Sequences starting with OC are from O. coriaceus. Other sequences are: ORNPA_Sal: O. parkeri salivary cystatin; IXOSC sequences are for sialostatin L and L2, respectively; IXORI_MG is for I. ricinus midgut cystatin; ORMO_MG refers to O. moubata midgut cystatins, and HAELO_MG the midgut cystatin of Haemaphysalis longicornis. HOMSA_SA and HOMSA_SN are for human cystatins, and GALGA_EW is for chicken egg white cystatin. For other details see Fig. 1 and text.
3.5.4. Calcitonin/adrenomedulin family peptide
In a previous study of the sialotranscriptome of O. parkeri [8], a peptide with similarity to vertebrate peptides of the adrenomedulin family was found. Peptides of this family have never before been described in the sialotranscriptomes of blood-sucking arthropods. We presently describe four peptides in the sialotranscriptome of O. coriaceus having weak similarity to adrenomedulin and stronger similarity to vertebrate vasodilatory peptides of the calcitonin-gene related peptide (CGRP) family. Alignment of the 4 sequences of O. coriaceus with the adrenomedulin-like peptide of O. parkeri reveals 21 sites of identities or closely related AA (Fig. 5), including two conserved cysteines. The O. parkeri sequence has a unique carboxyterminus tail similar to vertebrate pro-hormones, and it was suggested [8] it could be cleaved by a serine protease to produce the mature peptide at exactly the site of the last conserved Lys residue (Fig. 5). The function of this peptide family, which appears to be unique to Onithodoros, is unknown, but may be related to vasodilatory function.
Fig. 5.
Alignment of the adrenomedulin/calcitonin peptide family of Ornithodoros ticks. Conserved cysteines are shown in black background. Signal peptide is shown in gray background. Other marked amino acids are conserved in the five sequences shown.
3.5.5. Possible mucin
The protein sequence named OC-57 has a Gly- and Ser-rich carboxy terminal region and possibly functions as a mucin, a broad category of proteins that often do not share any similarity except for high Ser or Thr content, to which glycosyl residues may be attached. These types of transcripts have been found frequently in blood-sucking arthropod sialotranscriptomes.
3.5.6. Antimicrobial peptides and other immunity-related products
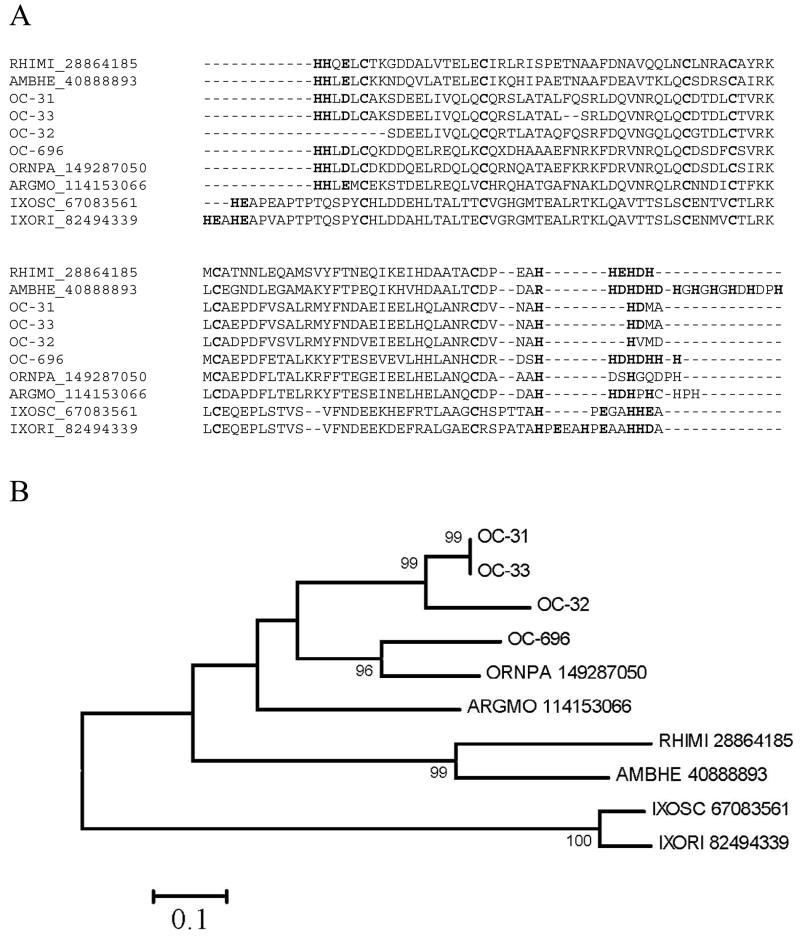
Antimicrobial peptides (AMP) and other immunity-related products have been commonly found in sialotranscriptomes of blood-feeding arthropods. We presently describe three families of such polypeptides in Supplemental Table S2, including members of the hebraein/microplusin, defensin, and TIL domain-containing peptides. These families include: i) Hebraein [74] and microplusin [75] are histidine rich AMP previously described in hard ticks. The alignment of the O. coriaceus proteins with those well characterized proteins of Rhipicephalus microplus and Amblyomma hebraeum, as well as homologues from I. scapularis, Ixodes ricinus, O. parkeri and A. monolakensis (Fig. 6A) shows six conserved Cys and multiple conserved His residues in both the carboxy and amino termini, often followed by acidic residues (Glu or Asp) at positions +1 or +2 following the His repeats. The phylogram (Fig. 6B) shows strong bootstrap support for one O. coriaceus clade, one mixed clade of one O. coriaceus and one O. parkeri clade, one Rhipicephalus and Amblyomma clade, and one Ixodes clade containing the sequences of I. ricinus and I. scapularis. One A. monolakensis sequence roots with the Ornithodoros sequences, but without strong bootstrap support. This family of proteins is apparently a good marker for tick phylogeny. ii) Supplemental Table S2 additionally presents a truncated defensin peptide that shows considerable similarity to other previously described defensins from hard and soft ticks. iii) The trypsin inhibitor-like (TIL) cysteine-rich domain is found in trypsin inhibitors and in other extracellular proteins. Ixodidin, a peptide from hemolymph of R. microplus, has such a domain and exercises chymotrypsin/elastase inhibitory activity as well as antimicrobial activity [76]. Two TIL polypeptides are shown in Supplemental Table S2. OC-100 has a single, while OC-86 has two TIL domains. Their similarity to other tick proteins is revealed mostly in the conserved pattern of Cys residues.
Fig. 6.
The Hebraein/microplusin family of tick antimicrobial peptides. Cysteine residues are indicated in black background. Histidine residues in the amino and carboxy termini are marked in red (gray). Acidic residues following His residues at position +1 or +2 are also indicated. A) Clustal alignment. B) Unrooted phylogram showing bootstrap values >75%. The bar at the bottom indicates 10% amino acid change. Sequences starting with OC- are from O. coriaceus. Remaining sequences are named with the first three letters of the genus and first two letters of the species name, followed by the NCBI accession number.
3.5.7. Enzymes
A fibrinogen-specific metalloprotease activity has been biochemically characterized previously in the saliva of I. scapularis [77, 78], consistent with the sialotranscriptome finding of an expanded gene family coding for this enzyme class [2, 7]. These metalloproteases are also a common finding in snake venoms [79]. Recently, six such enzymes were characterized from the tick Haemaphysalis longicornis [80]. Metalloprotease fragments have been also found in the soft ticks A. monolakensis and O. parkeri [8, 9]. Supplemental Table S2 presents evidence for the presence of such salivary enzyme in O. coriaceus in the form of the 5′ truncated clones OC-372 and OC-676. The longer clone (OC-372) indicates a ORF coding for 217 AA containing the HEXXH zinc-binding motif of the enzyme active site in addition to the CDD domain indicative of salivary arthropod metalloproteases. This enzyme may participate in fibrinolytic reactions in addition to other possible physiologic interventions.
Transcripts coding for secreted phospholipases of the A2 family (PLA2) have been found in sialotranscriptomes of mosquito, sand flies, and both hard and soft ticks; however, only in the hard tick Amblyomma americanum was the activity demonstrated to occur in dopamine-stimulated saliva [81, 82]. It is proposed that the enzyme promotes hemolysis of the ingested red cells and thus facilitates digestion [83]. Three 5′ truncated clones were found in the sialotranscriptome of O. coriaceus having the CDD motif indicative of PLA2 of the bee venom-like class. These enzymes are quite divergent, indicating fast evolution, or selection of different ancestral paralogues to account for a salivary function.
OC-79 matches the C-terminal domain of 5′-nucleotidases, being a candidate for salivary apyrase, which is a common activity in the saliva of blood-sucking arthropods that destroy ADP, an important signal triggering platelet aggregation in vertebrates [84], and also destroys ATP, a pro-inflammatory and pain inducing nucleotide [1]. The salivary apyrase of mosquitoes was shown to derive from an enzyme of the 5′-nucleotidase family [85]. More recently, the salivary apyrase of the mosquito Aedes aegypti was expressed and shown to hydrolyze ADP and ATP, as predicted, but not AMP [86]. 5′-nucleotidases are typically membrane bound, extracellular enzymes. Binding to the membrane is associated with a GPI anchor on the C terminal domain of the enzyme [87, 88]. Notably, hematophagous arthropod salivary apyrases of the 5′-nucleotidase family lack the AA sequence required for a GPI anchor, allowing the enzyme to be secreted. Within this context, OC-79 also lacks the required GPI anchor motif, as predicted by the program FragAnchor [28].
3.6 Polypeptides belonging to unique tick protein families, most having unknown function
3.6.1. Basic or acidic tail proteins found in both Argasidae and Ixodidae
This class of proteins, initially described in hard ticks of the Ixodes genus [2, 7, 68], was also found in the soft ticks A. monolakensis [9]and O. parkeri [8]. They range from 10 to 20 kDa, and have in common the presence of basic (usually lysine) or acidic (usually glutamate) residues on their carboxyterminus, although an Ixodes subfamily lacking the charged tail also exists. The sialotranscriptomes of O. coriaceus are rich in this class of proteins, ten full-length versions of which are shown in Supplemental Table S2. Most of these proteins have the PFAM domain TSGP1, indicative of the tick salivary gland peptide group 1. Only one member of this family, from I. scapularis and named Salp14 (gi|15428308), has been shown to exert anticlotting activity [89].
3.6.2. Cytotoxin family found in Argasids and Ixodids
OC-574 codes for a protein similar to a salivary protein previously described in I. scapularis, and to several deducted protein sequences deriving from tick EST [44]. Homologues from soft ticks have not been previously described. Alignment of these related sequences produce the Prosite block N-R-D-x(7)-G-x(13,16)-C-x(12,13)-C-x(16)-G-x(53,55)-L-N-x(2)-R [90] which is specific to this protein family, as it does not retrieve additional proteins from the NR protein database.
3.6.3. Glycine-rich protein family also found in Ixodidae
OC-130 codes for a Gly-rich protein with similarity to a salivary I. scapularis protein deposited at GenBank, as well as Amblyomma variegatum and Rhipicephalus proteins deducted from publicly available EST [44]. Members of this family have not been found previously in Argasidae. The function of this unique protein family is unknown.
3.6.4. 7DB family found in Argasidae
The sialotranscriptome of A. monolakensis revealed a unique gene family coding for Cys-rich peptides with seven disulphide bonds, named the 7DB family [9]. Members of this protein family were also found in O. parkeri [8]. Supplemental Table S2 presents three additional members of this family found in the O. coriaceus sialotranscriptome. The function of this unique Argasidae protein family is unknown.
3.6.5. 8-kDa family and other peptides unique to Argasidae
OC-98 and OC-197 belong to a protein family also found in the sialome of O. parkeri that includes a conserved framework of six cysteines. The Cys residue pattern corresponds to the 8-kDa cysteine-rich family of soft and hard tick proteins previously described [8, 10]
3.6.6. Peptides unique to the Argasidae
OC-93 and OC-129 belong to a novel family of small (5 kDa) disulphide-rich proteins that is also conserved in O. parkeri. Supplemental Table S2 presents six other peptides of mature molecular masses varying from 4.4–5.6 kDa that have similarities to previously described Ornithodoros peptides, albeit varying from 33% to 47% identity only.
3.6.7. Additional putative secreted peptides of unique protein families
Six additional polypeptide sequences with indication of a signal sequence are presented in Supplemental Table S2. They have no homology to known protein sequences. Some of these peptides may be carboxy terminal regions of transporters, as these proteins tend to have internal domains starting with a Met that may produce a false indication of signal peptide in the SignalP program.
3.7. Housekeeping and transposable element-derived proteins
Supplemental Table S2 describes 31 full-length sequences coding for housekeeping proteins, including 2 proteins with a weak Niemann-Pick type C2 domain (which may be involved in lysosomal metabolism or in immune reactions), 14 ribosomal proteins, and 1 reverse transcriptase fragment.
3.8. Preliminary characterization of the salivary proteome of O. coriaceus
To obtain information on protein expression in the salivary glands of O. coriaceus, we performed a redundant and complementary proteomic approach using i) one-dimensional (1D) polyacrylamide gel electrophoresis (PAGE) protein separation followed by blotting the proteins onto a PVDF membrane and performing Edman degradation of the bands (Fig. 7); ii) two-dimensional (2D) gel electrophoresis separation of the homogenate followed by tryptic digest and MS/MS of the Coomassie blue-stained bands (Fig. 8); and iii) high density 1D gel electrophoresis followed by tryptic digestion and reverse-phase HPLC/MS/MS of the indicated bands (Fig. 9), to attempt increased coverage of lower molecular mass peptides. The data of these three experiments are summarized below, and in Table 4.
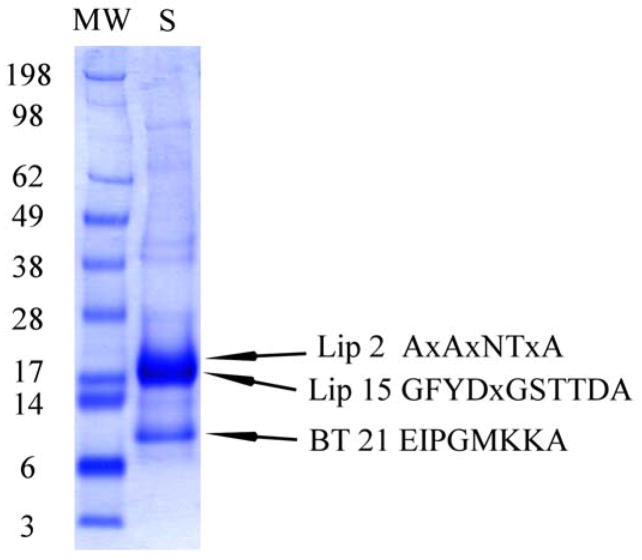
Fig. 7.
1D gel electrophoresis of O. coriaceus salivary gland homogenates. After electrophoresis, the proteins were transferred to a PVDF membrane, stained with Coomassie blue, and bands submitted to Edman degradation. Numbers on the left indicate molecular weight marker positions in the gel. OC-2 and OC-15 are lipocalins; OC-21 is a basic tail protein. For experimental details, see Materials and methods.
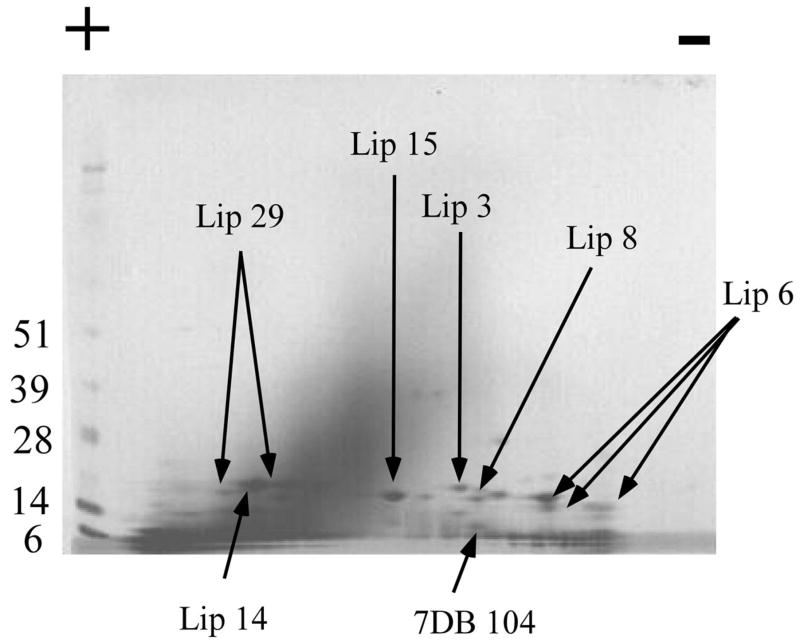
Fig. 8.
2D gel electrophoresis of O. coriaceus salivary gland homogenates. Numbers on the left indicate molecular weight marker positions in the gel. The + and − signs indicate the anode or cathode side of the IEF dimension, which ranged from pH 3–10. Gel bands that were identified to a protein (following tryptic digestion and mass spectrometry) are shown in the gel. In some cases, more than one band accounted for the same protein, possibly due to trailing or multiple isoforms. The bands labeled with Lip OC-X correspond to lipocalins where X is the number of the OC-X protein found in Supplemental Table S2. Similarly, 7DB is for a protein of the 7DB family. For experimental details, see Materials and methods.
3.8.1. 1D-PAGE followed by blotting to PVDF membrane and Edman degradation
Soluble O. coriaceus salivary glands proteins (30 μg) were treated with sodium dodecyl sulfate (SDS) buffer before applying to the gel bed. A strong band is visible above the 17-kDa marker, where the amino terminal sequences for the abundantly transcribed lipocalins OC-2 (predicted mass 15 kDa) and OC-15 (16 kDa) were found. The strong band between the 6- and 14-kDa marker retrieved the predicted mature amino terminus of OC-21, a basic tail protein with predicted mature mass of 7.3 kDa.
3.8.2. 2D-PAGE followed by tryptic digestion and MS/MS
Soluble O. coriaceus salivary gland proteins (90 μg) were separated by 2D SDS-PAGE. The stained protein bands were excised from the gel and submitted to tryptic digestion followed by MS/MS identification. The resulting electropherogram, similar to the 1D gel, is dominated by a cluster of bands indicating a MW ranging from 14–28 kDa and slightly acidic pI (Fig. 8). Many bands are found between the 14- and 6-kDa marker at the limit of resolution of the gel. Several bands were identified as lipocalins by tryptic digestion and MS/MS of the products. Additionally, a member of the 7DB cysteine-rich protein family was found. These results are summarized in Figure 7, in Table 4, and in column AL of Supplemental Table S2.
3.8.3. 1D-PAGE followed by tryptic digestion, RP-HPLC, and MS/MS
To further explore the expression of polypeptides in O. coriaceus salivary glands, we submitted both the soluble (S) and insoluble pellet (P) fractions of salivary gland homogenates to PAGE followed by in-gel tryptic digestion of 30 (S) or 31 (P) gel fragments, and reverse-phase HPLC/MS/MS of the digested products. Results of this experiment lead to a much greater identification of polypeptides, as shown in Figure 9, in Table 4, and in Supplemental Table S2. Twenty of the 21 lipocalins indicated in Supplemental Table S2 were identified by at least two unique peptides derived from the same gel fraction. Peptide coverage greater than 30% of the mature protein was found for 15 lipocalins. Two MS fragments indicating lipocalin OC-80 were identified in a gel position between the 36.5- and 55-kDa marker on the (S) gel, suggest that this lipocalin may be multimeric or associated with other masses. The remaining lipocalins are generally found in close agreement with their estimated molecular masses or slightly higher. Six basic tail peptides were identified, all in their expected gel positions except for OC-517, which shows itself at a location consistent with twice the expected molecular mass. Three double-Kunitz proteins were identified, as well as a protein similar to Ixodid cytotoxins. Cystatins were also identified. The Gly-rich protein (OC-130) was identified at its expected gel location. MS peptide fingerprints matching the 5′-nucleotidase was found at the 66-Kda marker, the expected position for this enzyme. The metalloproteases (OC-372) and phospholipase (OC-92) enzymes were also identified, both near the 36.5-kDa marker. A phospholipase has been characterized previously in A. americanum [81, 82], with an estimated molecular mass (by gel filtration) of 45 kDa.
It is interesting to consider that the solubilised pellet fraction allowed identification of many proteins not found in the soluble fraction, even though the proteins identified are typically secreted proteins. Perhaps because the homogenization was made in water to avoid ions that could interfere with electrophoresis, the proteins lacked the ‘salt in’ effect and precipitated. Alternatively, the high concentration of proteins and membranes may have caused many proteins bind to each other or to the membranes, as might be the case of the basic tail proteins that tend to bind to negatively charged membranes [91]. Increased concentration of the pellet may have also helped to increase the MS signal following tryptic digestion, thus enhancing finding of MS matches to our database.
4. Conclusion
This sialotranscriptome study is the third produced to date from salivary glands of soft ticks, joining those of O. parkeri [8] and A. monolakensis [9]. Taking into consideration the several salivary transcriptomes of hard ticks, including the genera Ixodes, Amblyomma, Dermacentor, and Rhipicephalus, this work serves to consolidate and generalize the profile of common proteins found in the salivary glands of both hard and soft ticks as well as a subset of protein families that appear to be unique to soft ticks. Additionally, transcripts coding for orphan transcripts may indicate the unique pathways taken by blood-sucking arthropods in their fast evolution of salivary pharmaceuticals.
Multigenic gene families common to all ticks include the lipocalin, Kunitz, cystatin, and basic tail superfamilies. As indicated before, the basic tail family appears to descend from a thrombospondin-repeat ancestor [2]. Metalloproteases, phospholipase A2, and 5′-nucleotidase/apyrase were also found in both Argasidae and Ixodidae. Antimicrobial peptides of the hebraein and defensins families are also found in both soft and hard ticks, as well as TIL domain-containing peptides that might act as antimicrobial peptides. The truncated product OC-574 matches a salivary protein from I. scapularis and other ixodid salivary proteins, indicating a possible common ancestry of this unique protein without significant matches to other known proteins.
Protein families that appear unique to soft ticks found in the transcriptome of O. coriaceus include polypeptides weakly similar to the vertebrate protein calcitonin gene-related peptide, which aligns well with the adrenomedulin-like protein of O. parkeri (Fig. 5). Notice also that the RGD motif-containing Kunitz family also appears to be exclusive to soft ticks. Other expanded salivary protein families of the Argasidae include the 7DB family, which might have evolved from the basic tail family [9] and the argasid 7-kDa family. Supplemental Table S2 indicates three other proteins from O. coriaceus having best matches with similar-sized proteins of O. parkeri, albeit at a low degree of sequence conservation, indicating the rapid evolution of these proteins. For example, OC-64 has only 33% identity to its O. parkeri homologue, while OC-194 has 47% identity over 86% of the O. parkeri homologue and much poorer matches to other proteins found in the NR database, numbering over 4 million proteins at present. The peptide OC-64 was found in the 1D gel experiment (Fig. 9). Supplemental Table S2 additionally presents nine peptides ranging in molecular mass from 3.4 to 6.3 that have no similarity to any other protein in the NR database when compared by the blastp program. As in all sialotranscriptome studies done thus far, the vast majority of the identified proteins have no confirmed or known function.
Supplementary Material
Supplemental Table S1 can be retrieved from
http://exon.niaid.nih.gov/transcriptome/O_coriaceus/Sup1/O-cori-Sup1.xls.
Supplemental table S2 can be retrieved from
http://exon.niaid.nih.gov/transcriptome/O_coriaceus/Sup2/O-cori-Sup2.xls.
Stand-alone versions of the above files can be retrieved from
http://exon.niaid.nih.gov/transcriptome/O_coriaceus/Sup1/O-cori-Sup1-SA.zip
http://exon.niaid.nih.gov/transcriptome/O_coriaceus/Sup2/O-cori-Sup2-SA.zip
Acknowledgments
This work was supported by the Intramural Research Program of the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health. This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract NO1-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organization imply endorsement by the government of the United States of America. We are grateful to the NIAID Core facility led by Dr. Robert Hohman for their support, and to NIAID intramural editor Brenda Rae Marshall for assistance.
Footnotes
Because I.M.B.F., V.M.P., and J.M.C.R. are government employees and this is a government work, the work is in the public domain in the United States. Notwithstanding any other agreements, the NIH reserves the right to provide the work to PubMedCentral for display and use by the public, and PubMedCentral may tag or modify the work consistent with its customary practices. You can establish rights outside of the U.S. subject to a government use license.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ribeiro JM, Francischetti IM. Role of arthropod saliva in blood feeding: sialome and post-sialome perspectives. Annu Rev Entomol. 2003;48:73–88. doi: 10.1146/annurev.ento.48.060402.102812. [DOI] [PubMed] [Google Scholar]
- 2.Ribeiro JM, Alarcon-Chaidez F, Francischetti IM, Mans BJ, Mather TN, Valenzuela JG, et al. An annotated catalog of salivary gland transcripts from Ixodes scapularis ticks. Insect Biochem Mol Biol. 2006;36:111–29. doi: 10.1016/j.ibmb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 3.Santos IK, Valenzuela JG, Ribeiro JM, de Castro M, Costa JN, Costa AM, et al. Gene discovery in Boophilus microplus, the cattle tick: the transcriptomes of ovaries, salivary glands, and hemocytes. Ann N Y Acad Sci. 2004;1026:242–6. doi: 10.1196/annals.1307.037. [DOI] [PubMed] [Google Scholar]
- 4.Nene V, Lee D, Kang’a S, Skilton R, Shah T, de Villiers E, et al. Genes transcribed in the salivary glands of female Rhipicephalus appendiculatus ticks infected with Theileria parva. Insect Biochem Mol Biol. 2004;34:1117–28. doi: 10.1016/j.ibmb.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Lambson B, Nene V, Obura M, Shah T, Pandit P, Ole-Moiyoi O, et al. Identification of candidate sialome components expressed in ixodid tick salivary glands using secretion signal complementation in mammalian cells. Insect Mol Biol. 2005;14:403–14. doi: 10.1111/j.1365-2583.2005.00571.x. [DOI] [PubMed] [Google Scholar]
- 6.Alarcon-Chaidez FJ, Sun J, Wikel SK. Transcriptome analysis of the salivary glands of Dermacentor andersoni Stiles (Acari: Ixodidae) Insect Biochem Mol Biol. 2007;37:48–71. doi: 10.1016/j.ibmb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 7.Francischetti IM, My Pham V, Mans BJ, Andersen JF, Mather TN, Lane RS, et al. The transcriptome of the salivary glands of the female western black-legged tick Ixodes pacificus (Acari: Ixodidae) Insect Biochem Mol Biol. 2005;35:1142–61. doi: 10.1016/j.ibmb.2005.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Francischetti IM, Mans BJ, Meng Z, Gudderra N, Veenstra TD, Pham VM, et al. An insight into the sialome of the soft tick, Ornithodorus parkeri. Insect Biochem Mol Biol. 2008;38:1–21. doi: 10.1016/j.ibmb.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mans BJ, Andersen JF, Francischetti IM, Valenzuela JG, Schwan TG, Pham VM, et al. Comparative sialomics between hard and soft ticks: Implications for the evolution of blood-feeding behavior. Insect Biochem Mol Biol. 2008;38:42–58. doi: 10.1016/j.ibmb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mans BJ, Andersen JF, Schwan TG, Ribeiro JM. Characterization of anti-hemostatic factors in the argasid, Argas monolakensis: Implications for the evolution of blood-feeding in the soft tick family. Insect Biochem Mol Biol. 2008;38:22–41. doi: 10.1016/j.ibmb.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen CI, King DP, Blanchard MT, Hall MR, Aldridge BM, Bowen L, et al. Identification of the etiologic agent of epizootic bovine abortion in field-collected Ornithodoros coriaceus Koch ticks. Vet Microbiol. 2007;120:320–7. doi: 10.1016/j.vetmic.2006.10.036. [DOI] [PubMed] [Google Scholar]
- 12.Lane RS, Burgdorfer W, Hayes SF, Barbour AG. Isolation of a spirochete from the soft tick, Ornithodoros coriaceus: A possible agent of epizootic bovine abortion. Science. 1985;230:85–7. doi: 10.1126/science.3898367. [DOI] [PubMed] [Google Scholar]
- 13.Teglas MB, May B, Crosbie PR, Stephens MR, Boyce WM. Genetic structure of the tick Ornithodoros coriaceus (Acari: Argasidae) in California, Nevada, and Oregon. J Med Entomol. 2005;42:247–53. doi: 10.1093/jmedent/42.3.247. [DOI] [PubMed] [Google Scholar]
- 14.Valenzuela JG, Francischetti IM, Pham VM, Garfield MK, Ribeiro JM. Exploring the salivary gland transcriptome and proteome of the Anopheles stephensi mosquito. Insect Biochem Mol Biol. 2003;33:717–32. doi: 10.1016/s0965-1748(03)00067-5. [DOI] [PubMed] [Google Scholar]
- 15.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 16.Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–77. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–8. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 19.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2000;28:263–6. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schultz J, Copley RR, Doerks T, Ponting CP, Bork P. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000;28:231–4. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30:281–3. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 26.Hansen JE, Lund O, Tolstrup N, Gooley AA, Williams KL, Brunak S. NetOglyc: prediction of mucin type O-glycosylation sites based on sequence context and surface accessibility. Glycoconj J. 1998;15:115–30. doi: 10.1023/a:1006960004440. [DOI] [PubMed] [Google Scholar]
- 27.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–63. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 28.Poisson G, Chauve C, Chen X, Bergeron A. FragAnchor: a large-scale predictor of glycosylphosphatidylinositol anchors in eukaryote protein sequences by qualitative scoring. Genomics Proteomics Bioinformatics. 2007;5:121–30. doi: 10.1016/S1672-0229(07)60022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Francischetti IM, Valenzuela JG, Pham VM, Garfield MK, Ribeiro JM. Toward a catalog for the transcripts and proteins (sialome) from the salivary gland of the malaria vector Anopheles gambiae. J Exp Biol. 2002;205:2429–51. doi: 10.1242/jeb.205.16.2429. [DOI] [PubMed] [Google Scholar]
- 30.Arca B, Lombardo F, Valenzuela JG, Francischetti IM, Marinotti O, Coluzzi M, et al. An updated catalogue of salivary gland transcripts in the adult female mosquito, Anopheles gambiae. J Exp Biol. 2005;208:3971–86. doi: 10.1242/jeb.01849. [DOI] [PubMed] [Google Scholar]
- 31.Ribeiro JM, Andersen J, Silva-Neto MA, Pham VM, Garfield MK, Valenzuela JG. Exploring the sialome of the blood-sucking bug Rhodnius prolixus. Insect Biochem Mol Biol. 2004;34:61–79. doi: 10.1016/j.ibmb.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 32.Ribeiro JM, Charlab R, Pham VM, Garfield M, Valenzuela JG. An insight into the salivary transcriptome and proteome of the adult female mosquito Culex pipiens quinquefasciatus. Insect Biochem Mol Biol. 2004;34:543–63. doi: 10.1016/j.ibmb.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 33.Galperin MY, Koonin EV. Conserved hypothetical proteins: prioritization of targets for experimental study. Nucleic Acids Res. 2004;32:5452–63. doi: 10.1093/nar/gkh885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flower DR, North AC, Sansom CE. The lipocalin protein family: structural and sequence overview. Biochim Biophys Acta. 2000;1482:9–24. doi: 10.1016/s0167-4838(00)00148-5. [DOI] [PubMed] [Google Scholar]
- 35.Andersen JF, Gudderra NP, Francischetti IM, Ribeiro JM. The role of salivary lipocalins in blood feeding by Rhodnius prolixus. Arch Insect Biochem Physiol. 2005;58:97–105. doi: 10.1002/arch.20032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santos A, Ribeiro JM, Lehane MJ, Gontijo NF, Veloso AB, Sant’Anna MR, et al. The sialotranscriptome of the blood-sucking bug Triatoma brasiliensis (Hemiptera, Triatominae) Insect Biochem Mol Biol. 2007;37:702–12. doi: 10.1016/j.ibmb.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Assumpcao TC, Francischetti IM, Andersen JF, Schwarz A, Santana JM, Ribeiro JM. An insight into the sialome of the blood-sucking bug Triatoma infestans, a vector of Chagas’ disease. Insect Biochem Mol Biol. 2008;38:213–32. doi: 10.1016/j.ibmb.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Champagne D, Nussenzveig RH, Ribeiro JMC. Purification, characterization, and cloning of nitric oxide-carrying heme proteins (nitrophorins) from salivary glands of the blood sucking insect Rhodnius prolixus. J Biol Chem. 1995;270:8691–5. doi: 10.1074/jbc.270.15.8691. [DOI] [PubMed] [Google Scholar]
- 39.Andersen JF, Francischetti IM, Valenzuela JG, Schuck P, Ribeiro JM. Inhibition of hemostasis by a high affinity biogenic amine-binding protein from the saliva of a blood-feeding insect. J Biol Chem. 2003;278:4611–7. doi: 10.1074/jbc.M211438200. [DOI] [PubMed] [Google Scholar]
- 40.Sangamnatdej S, Paesen GC, Slovak M, Nuttall PA. A high affinity serotonin- and histamine-binding lipocalin from tick saliva. Insect Mol Biol. 2002;11:79–86. doi: 10.1046/j.0962-1075.2001.00311.x. [DOI] [PubMed] [Google Scholar]
- 41.Ribeiro JMC, Schneider M, Guimaraes JA. Purification and characterization of Prolixin S (Nitrophorin 2), the salivary anticoagulant of the blood sucking bug, Rhodnius prolixus. Biochem J. 1995;308:243–9. doi: 10.1042/bj3080243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Ribeiro JM, Guimaraes JA, Walsh PN. Nitrophorin-2: a novel mixed-type reversible specific inhibitor of the intrinsic factor-X activating complex. Biochemistry. 1998;37:10681–90. doi: 10.1021/bi973050y. [DOI] [PubMed] [Google Scholar]
- 43.Nunn MA, Sharma A, Paesen GC, Adamson S, Lissina O, Willis AC, et al. Complement inhibitor of C5 activation from the soft tick Ornithodoros moubata. J Immunol. 2005;174:2084–91. doi: 10.4049/jimmunol.174.4.2084. [DOI] [PubMed] [Google Scholar]
- 44.Francischetti IMB, Sa-Nunes A, Mans B, Santos IM, Ribeiro JMC. The role of saliva in tick feeding. In: delaFuente J, Kocan KM, Sonenshine DE, editors. Molecular biology of ticks and the tick-pathogen interface. Frontiers in Bioscience; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen JM, Cooper DN, Chuzhanova N, Ferec C, Patrinos GP. Gene conversion: mechanisms, evolution and human disease. Nat Rev Genet. 2007;8:762–75. doi: 10.1038/nrg2193. [DOI] [PubMed] [Google Scholar]
- 46.Palmer GH, Brayton KA. Gene conversion is a convergent strategy for pathogen antigenic variation. Trends Parasitol. 2007;23:408–13. doi: 10.1016/j.pt.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 47.Mans BJ, Louw AI, Neitz AW. Evolution of hematophagy in ticks: common origins for blood coagulation and platelet aggregation inhibitors from soft ticks of the genus Ornithodoros. Mol Biol Evol. 2002;19:1695–705. doi: 10.1093/oxfordjournals.molbev.a003992. [DOI] [PubMed] [Google Scholar]
- 48.Mans BJ, Louw AI, Neitz AW. The influence of tick behavior, biotope and host specificity on concerted evolution of the platelet aggregation inhibitor savignygrin, from the soft tick Ornithodoros savignyi. Insect Biochem Mol Biol. 2003;33:623–9. doi: 10.1016/s0965-1748(03)00047-x. [DOI] [PubMed] [Google Scholar]
- 49.Mans BJ, Venter JD, Vrey PJ, Louw AI, Neitz AW. Identification of putative proteins involved in granule biogenesis of tick salivary glands. Electrophoresis. 2001;22:1739–46. doi: 10.1002/1522-2683(200105)22:9<1739::AID-ELPS1739>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 50.Mans BJ, Louw AI, Neitz AW. The major tick salivary gland proteins and toxins from the soft tick, Ornithodoros savignyi, are part of the tick Lipocalin family: implications for the origins of tick toxicoses. Mol Biol Evol. 2003;20:1158–67. doi: 10.1093/molbev/msg126. [DOI] [PubMed] [Google Scholar]
- 51.Francischetti IM, Andersen JF, Ribeiro JM. Biochemical and functional characterization of recombinant Rhodnius prolixus platelet aggregation inhibitor 1 as a novel lipocalin with high affinity for adenosine diphosphate and other adenine nucleotides. Biochemistry. 2002;41:3810–8. doi: 10.1021/bi011015s. [DOI] [PubMed] [Google Scholar]
- 52.Francischetti IM, Ribeiro JM, Champagne D, Andersen J. Purification, cloning, expression, and mechanism of action of a novel platelet aggregation inhibitor from the salivary gland of the blood-sucking bug, Rhodnius prolixus. J Biol Chem. 2000;275:12639–50. doi: 10.1074/jbc.275.17.12639. [DOI] [PubMed] [Google Scholar]
- 53.Ascenzi P, Bocedi A, Bolognesi M, Spallarossa A, Coletta M, De Cristofaro R, et al. The bovine basic pancreatic trypsin inhibitor (Kunitz inhibitor): a milestone protein. Curr Protein Pept Sci. 2003;4:231–51. doi: 10.2174/1389203033487180. [DOI] [PubMed] [Google Scholar]
- 54.Bajaj MS, Birktoft JJ, Steer SA, Bajaj SP. Structure and biology of tissue factor pathway inhibitor. Thromb Haemost. 2001;86:959–72. [PubMed] [Google Scholar]
- 55.Nene V, Lee D, Kang’a S, Skilton R, Shah T, de Villiers E, et al. Genes transcribed in the salivary glands of female Rhipicephalus appendiculatus ticks infected with Theileria parva. Insect Biochem Mol Biol. 2004;34:1117–28. doi: 10.1016/j.ibmb.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 56.Francischetti IM, Valenzuela JG, Andersen JF, Mather TN, Ribeiro JM. Ixolaris, a novel recombinant tissue factor pathway inhibitor (TFPI) from the salivary gland of the tick, Ixodes scapularis: identification of factor X and factor Xa as scaffolds for the inhibition of factor VIIa/tissue factor complex. Blood. 2002;99:3602–12. doi: 10.1182/blood-2001-12-0237. [DOI] [PubMed] [Google Scholar]
- 57.Monteiro RQ, Rezaie AR, Bae JS, Calvo E, Andersen JF, Francischetti IM. Ixolaris binding to factor X reveals a precursor state of factor Xa heparin-binding exosite. Protein Sci. 2008;17:146–53. doi: 10.1110/ps.073016308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Francischetti IM, Mather TN, Ribeiro JM. Penthalaris, a novel recombinant five-Kunitz tissue factor pathway inhibitor (TFPI) from the salivary gland of the tick vector of Lyme disease, Ixodes scapularis. Thromb Haemost. 2004;91:886–98. doi: 10.1160/TH03-11-0715. [DOI] [PubMed] [Google Scholar]
- 59.Mans BJ, Louw AI, Neitz AW. Savignygrin, a platelet aggregation inhibitor from the soft tick Ornithodoros savignyi, presents the RGD integrin recognition motif on the Kunitz-BPTI fold. J Biol Chem. 2002;277:21371–8. doi: 10.1074/jbc.M112060200. [DOI] [PubMed] [Google Scholar]
- 60.Karczewski J, Endris R, Connolly TM. Disagregin is a fibrinogen receptor antagonist lacking the Arg-Gly-Asp sequence from the tick, Ornithodoros moubata. J Biol Chem. 1994;269:6702–8. [PubMed] [Google Scholar]
- 61.Mans BJ, Neitz AW. The mechanism of alphaIIbbeta3 antagonism by savignygrin and its implications for the evolution of anti-hemostatic strategies in soft ticks. Insect Biochem Mol Biol. 2004;34:573–84. doi: 10.1016/j.ibmb.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 62.Monteiro RQ, Rezaie AR, Ribeiro JM, Francischetti IM. Ixolaris: A factor Xa heparin binding exosite inhibitor. Biochem J. 2004 doi: 10.1042/BJ20041738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Waxman L, Smith DE, Arcuri KE, Vlasuk GP. Tick anticoagulant peptide (TAP) is a novel inhibitor of blood coagulation Factor Xa. Science. 1990;248:593–6. doi: 10.1126/science.2333510. [DOI] [PubMed] [Google Scholar]
- 64.Gaspar AR, Joubert AM, Crause JC, Neitz AW. Isolation and characterization of an anticoagulant from the salivary glands of the tick, Ornithodoros savignyi (Acari: Argasidae) Exp Appl Acarol. 1996;20:583–98. doi: 10.1007/BF00052809. [DOI] [PubMed] [Google Scholar]
- 65.Joubert AM, Louw AI, Joubert F, Neitz AW. Cloning, nucleotide sequence and expression of the gene encoding factor Xa inhibitor from the salivary glands of the tick, Ornithodoros savignyi. Exp Appl Acarol. 1998;22:603–19. doi: 10.1023/a:1006198713791. [DOI] [PubMed] [Google Scholar]
- 66.Vray B, Hartmann S, Hoebeke J. Immunomodulatory properties of cystatins. Cell Mol Life Sci. 2002;59:1503–12. doi: 10.1007/s00018-002-8525-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fossum K, Whitaker JR. Ficin and papain inhibitor from chicken egg white. Arch Biochem Biophys. 1968;125:367–75. doi: 10.1016/0003-9861(68)90672-3. [DOI] [PubMed] [Google Scholar]
- 68.Valenzuela JG, Francischetti IMB, Pham VM, Garfield MK, Mather TN, Ribeiro JMC. Exploring the sialome of the tick, Ixodes scapularis. J ExpBiol. 2002;205:2843–64. doi: 10.1242/jeb.205.18.2843. [DOI] [PubMed] [Google Scholar]
- 69.Grunclova L, Horn M, Vancova M, Sojka D, Franta Z, Mares M, et al. Two secreted cystatins of the soft tick Ornithodoros moubata: differential expression pattern and inhibitory specificity. Biol Chem. 2006;387:1635–44. doi: 10.1515/BC.2006.204. [DOI] [PubMed] [Google Scholar]
- 70.Lima CA, Sasaki SD, Tanaka AS. Bmcystatin, a cysteine proteinase inhibitor characterized from the tick Boophilus microplus. Biochem Biophys Res Commun. 2006;347:44–50. doi: 10.1016/j.bbrc.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 71.Zhou J, Ueda M, Umemiya R, Battsetseg B, Boldbaatar D, Xuan X, et al. A secreted cystatin from the tick Haemaphysalis longicornis and its distinct expression patterns in relation to innate immunity. Insect Biochem Mol Biol. 2006;36:527–35. doi: 10.1016/j.ibmb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 72.Kotsyfakis M, Karim S, Andersen JF, Mather TN, Ribeiro JM. Selective cysteine protease inhibition contributes to blood-feeding success of the tick Ixodes scapularis. J Biol Chem. 2007;282:29256–63. doi: 10.1074/jbc.M703143200. [DOI] [PubMed] [Google Scholar]
- 73.Kotsyfakis M, Sa-Nunes A, Francischetti IM, Mather TN, Andersen JF, Ribeiro JM. Antiinflammatory and immunosuppressive activity of sialostatin L, a salivary cystatin from the tick Ixodes scapularis. J Biol Chem. 2006;281:26298–307. doi: 10.1074/jbc.M513010200. [DOI] [PubMed] [Google Scholar]
- 74.Lai R, Takeuchi H, Lomas LO, Jonczy J, Rigden DJ, Rees HH, et al. A new type of antimicrobial protein with multiple histidines from the hard tick, Amblyomma hebraeum. Faseb J. 2004;18:1447–9. doi: 10.1096/fj.03-1154fje. [DOI] [PubMed] [Google Scholar]
- 75.Fogaca AC, Lorenzini DM, Kaku LM, Esteves E, Bulet P, Daffre S. Cysteine-rich antimicrobial peptides of the cattle tick Boophilus microplus: isolation, structural characterization and tissue expression profile. Dev Comp Immunol. 2004;28:191–200. doi: 10.1016/j.dci.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 76.Fogaca AC, Almeida IC, Eberlin MN, Tanaka AS, Bulet P, Daffre S. Ixodidin, a novel antimicrobial peptide from the hemocytes of the cattle tick Boophilus microplus with inhibitory activity against serine proteinases. Peptides. 2005;27:667–74. doi: 10.1016/j.peptides.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 77.Francischetti IM, Mather TN, Ribeiro JM. Cloning of a salivary gland metalloprotease and characterization of gelatinase and fibrin(ogen)lytic activities in the saliva of the Lyme disease tick vector Ixodes scapularis. Biochem Biophys Res Commun. 2003;305:869–75. doi: 10.1016/s0006-291x(03)00857-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Francischetti IM, Mather TN, Ribeiro JM. Tick saliva is a potent inhibitor of endothelial cell proliferation and angiogenesis. Thromb Haemost. 2005;94:167–74. doi: 10.1267/THRO05010167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Takeya H, Miyata T, Nishino N, Omori-Satoh T, Iwanaga S. Snake venom hemorrhagic and nonhemorrhagic metalloendopeptidases. Meth Enzymol. 1993;223:365–78. doi: 10.1016/0076-6879(93)23058-u. [DOI] [PubMed] [Google Scholar]
- 80.Harnnoi T, Sakaguchi T, Nishikawa Y, Xuan X, Fujisaki K. Molecular characterization and comparative study of 6 salivary gland metalloproteases from the hard tick, Haemaphysalis longicornis. Comp Biochem Physiol B Biochem Mol Biol. 2007;147:93–101. doi: 10.1016/j.cbpb.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 81.Zhu K, Bowman AS, Dillwith JW, Sauer JR. Phospholipase A2 activity in salivary glands and saliva of the lone star tick (Acari: Ixodidae) during tick feeding. J Med Entomol. 1998;35:500–4. doi: 10.1093/jmedent/35.4.500. [DOI] [PubMed] [Google Scholar]
- 82.Bowman AS, Gengler CL, Surdick MR, Zhu K, Essenberg RC, Sauer JR, et al. A novel phospholipase A2 activity in saliva of the lone star tick, Amblyomma americanum (L) Exp Parasitol. 1997;87:121–32. doi: 10.1006/expr.1997.4201. [DOI] [PubMed] [Google Scholar]
- 83.Zhu K, Dillwith JW, Bowman AS, Sauer JR. Identification of hemolytic activity in saliva of the lone star tick (Acari:Ixodidae) J Med Entomol. 1997;34:160–6. doi: 10.1093/jmedent/34.2.160. [DOI] [PubMed] [Google Scholar]
- 84.Ribeiro JMC. Blood-feeding arthropods: Live syringes or invertebrate pharmacologists? Infect Agents Dis. 1995;4:143–52. [PubMed] [Google Scholar]
- 85.Champagne DE, Smartt CT, Ribeiro JM, James AA. The salivary gland-specific apyrase of the mosquito Aedes aegypti is a member of the 5′-nucleotidase family. Proc Natl Acad Sci U S A. 1995;92:694–8. doi: 10.1073/pnas.92.3.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sun D, McNicol A, James AA, Peng Z. Expression of functional recombinant mosquito salivary apyrase: A potential therapeutic platelet aggregation inhibitor. Platelets. 2006;17:178–84. doi: 10.1080/09537100500460234. [DOI] [PubMed] [Google Scholar]
- 87.Ogata S, Hayashi Y, Misumi Y, Ikehara Y. Membrane-anchoring domain of rat liver 5′-nucleotidase: identification of the COOH-terminal serine-523 covalently attached with a glycolipid. Biochemistry. 1990;29:7923–7. doi: 10.1021/bi00486a021. [DOI] [PubMed] [Google Scholar]
- 88.Misumi Y, Ogata S, Ohkubo K, Hirose S, Ikehara Y. Primary structure of human placental 5′-nucleotidase and identification of the glycolipid anchor in the mature form. Eur J Biochem. 1990;191:563–9. doi: 10.1111/j.1432-1033.1990.tb19158.x. [DOI] [PubMed] [Google Scholar]
- 89.Narasimhan S, Koski RA, Beaulieu B, Anderson JF, Ramamoorthi N, Kantor F, et al. A novel family of anticoagulants from the saliva of Ixodes scapularis. Insect Mol Biol. 2002;11:641–50. doi: 10.1046/j.1365-2583.2002.00375.x. [DOI] [PubMed] [Google Scholar]
- 90.Sibbald PR, Sommerfeldt H, Argos P. Automated protein sequence pattern handling and PROSITE searching. Comput Appl Biosci. 1991;7:535–6. doi: 10.1093/bioinformatics/7.4.535. [DOI] [PubMed] [Google Scholar]
- 91.Murray D, Arbuzova A, Hangyas-Mihalyne G, Gambhir A, Ben-Tal N, Honig B, et al. Electrostatic properties of membranes containing acidic lipids and adsorbed basic peptides: theory and experiment. Biophys J. 1999;77:3176–88. doi: 10.1016/S0006-3495(99)77148-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table S1 can be retrieved from
http://exon.niaid.nih.gov/transcriptome/O_coriaceus/Sup1/O-cori-Sup1.xls.
Supplemental table S2 can be retrieved from
http://exon.niaid.nih.gov/transcriptome/O_coriaceus/Sup2/O-cori-Sup2.xls.
Stand-alone versions of the above files can be retrieved from
http://exon.niaid.nih.gov/transcriptome/O_coriaceus/Sup1/O-cori-Sup1-SA.zip
http://exon.niaid.nih.gov/transcriptome/O_coriaceus/Sup2/O-cori-Sup2-SA.zip