Abstract
Seed size is a key determinant of evolutionary fitness in plants and is a trait that often undergoes tremendous changes during crop domestication. Seed size is most often quantitatively inherited, and it has been shown that Sw4.1 is one of the most significant quantitative trait loci (QTLs) underlying the evolution of seed size in the genus Solanum—especially in species related to the cultivated tomato. Using a combination of genetic, developmental, molecular, and transgenic techniques, we have pinpointed the cause of the Sw4.1 QTL to a gene encoding an ABC transporter gene. This gene exerts its control on seed size, not through the maternal plant, but rather via gene expression in the developing zygote. Phenotypic effects of allelic variation at Sw4.1 are manifested early in seed development at stages corresponding to the rapid deposition of starch and lipids into the endospermic cells. Through synteny, we have identified the Arabidopsis Sw4.1 ortholog. Mutagenesis has revealed that this ortholog is associated with seed length variation and fatty acid deposition in seeds, raising the possibility that the ABC transporter may modulate seed size variation in other species. Transcription studies show that the ABC transporter gene is expressed not only in seeds, but also in other tissues (leaves and roots) and, thus, may perform functions in parts of the plants other than developing seeds. Cloning and characterization of the Sw4.1 QTL gives new insight into how plants change seed during evolution and may open future opportunities for modulating seed size in crop plants for human purposes.
Author Summary
Given fixed resources, plants have a choice whether to produce many small seeds or a few large seeds. In terms of reproductive fitness, there are costs and benefits to both strategies. As a result, plant species vary more than 100,000-fold in both seed size and seed output. The current study focuses on understanding the molecular and developmental basis of a single genetic locus (or quantitative trait locus) that determines seed size between the cultivated tomato and its wild relatives. We show that the cause of size variation can be traced to a gene encoding an ABC transporter protein. The gene apparently exercises its control on seed size through expression in the developing seeds and not the mother plant that nurtures those seeds. A comparison with the model plant Arabidopsis thaliana suggests that the ABC transporter identified in tomato may also control seed size in other plants, opening research opportunities for understanding plant adaptation and for potentially modulating seed size in crop plants for human purposes.
Introduction
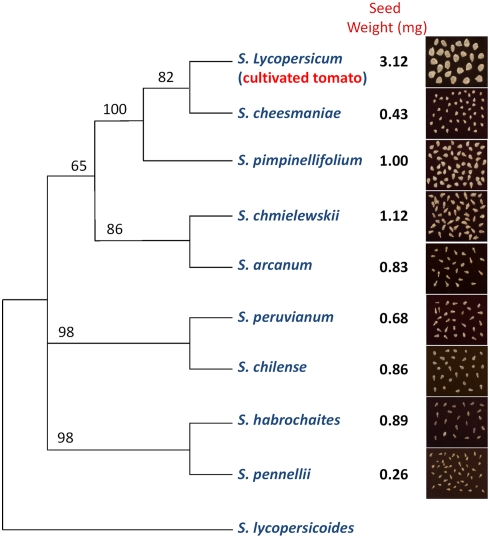
Seeds represent the vehicle by which plants vie for evolutionary success. A key feature of seeds is their size, which in turn is one of the most variable traits in the plant kingdom. Seeds range in weight from less than 1 microgram in the Coral-root orchid (Corallorhiza maculate) to more than 10 kg in the Coco-de-mer palm (Lodoicea maldivica). This large range can be observed not only among taxa, but also within taxa. For example, contained in the genus Solanum are a set of 9 cross compatible species closely related to the tomato. Despite their close taxonomic affinities, these species show a 10-fold range in seed size suggesting a rapid rate of evolutionary change (Figure 1).
Figure 1. Phylogenetic relationships of species in the genus Solanum most closely related to the cultivated tomato.
Numbers at nodes indicates bootstrap values. Modified from Spooner et al. [55] and Peralta et al. [56].
Why plants vary so much in seed size is not known. However, evolutionists and ecologists have long noted this great variation and hypothesized its importance in adaptation. In terms of survival, there are both risks and benefits for a species to increase (or decrease) seed size. Because maternal resources are limited, a species has to “decide” whether to invest energy into a few large seeds or many small seeds [1],[2]. Intra- and interspecific studies of offspring fitness in plant communities have demonstrated that plants producing a small number of large seeds often have higher tolerance to drought [3], herbivory [4], shading [5], and nutrient-deficient soils [6]. However, plants producing a large number of small seeds exhibit superior colonization abilities with the advantage of dispersal due to the abundance of seeds and higher likelihood to escape from predation [7],[8].
Scientific interest in seed size relates not only to its importance in evolution, but also to crop domestication. Crops domesticated for consumption of their seeds (e.g. soybean, wheat, sunflower) often produce seeds significantly larger than their wild ancestors [9]–[12]. It is likely that early humans consciously selected for larger seeds in an effort to increase yield and improve harvest efficiency. However, seed size also increased during domestication in crops not harvested for their edible seed. For example, domesticated tomatoes produce seeds up to several fold larger than their wild ancestors (Figure 1) [13]. Likewise cultivated squash (Cucurbita pepo) produce seed more than two fold larger than their wild counter parts [14]. Why seed size increased during domestication in crops not consumed for their seeds is unclear. However, it has been conjectured that seed size increased in these species due to indirect selection for greater seedling vigor and germination uniformity under field production [15].
Despite the importance of seed size in plant evolution and crop domestication, relatively little is known about the genetic and molecular processes underlying natural variation in seed size. Most of our knowledge comes from quantitative trait mapping studies which have revealed a fairly large number of QTL affecting seed size in a variety of plants – e.g. Arabidopsis [16], rice [17]–[22], soybean [23],[24], sunflower [11], [25]–[28]. However, most of these studies have not gone beyond the mapping stage and hence provide little insight into the developmental and molecular mechanisms underpinning seed size variation. The exception is rice where three seed size QTLs have been recently cloned. These encode a previously unknown RING-type E3 ubiquitin ligase [29], a putative transmembrane protein [30], and deletion of a gene of unknown function [17].
Tomato is one of the few species not domesticated for edible seeds, where extensive QTL mapping for seed size has been conducted. Over the past 25 years, mapping studies involving crosses between the cultivated tomato and related wild species have revealed approximately 20 QTLs which account for most seed size variation [13], [31]–[34]. Different subsets of these QTLs were identified in different studies. However, a common feature of all studies was that a major QTL on chromosome 4 (referred to as Seed weight 4.1 or Sw4.1) invariably accounted for a large portion of the genetic variation for seed size. Sw4.1 is responsible for up to 25% of the total phenotypic variation in segregating populations and up to 54% of the seed weight variation in crosses between nearly isogenic lines [13]. The conservation of Sw4.1 across tomato species, and its potential role in the evolution and domestication of cultivated tomato, makes Sw4.1 a prime candidate for characterization and cloning. Thus the objective of this study was to uncover the genetic, developmental and molecular mechanisms underlying modulation of seed size by the Sw4.1 QTL.
Results/Discussion
Sw4.1 Controls Seed Weight through Zygotic Effects
The size or weight of a seed can potentially be affected by the genotype of three different plant parts/tissues: a) the female plant bearing the fruit which contains the developing seed and contributes the testa; b) the triploid endosperm which nourishes the developing embryo and c) the diploid embryo. A maternal effect would be caused by a substantial contribution of the maternal genotype from the testa (a) and/or endosperm (b) to the seed development, while a zygotic effect would be attributed to the equal contribution of maternal and paternal genotypes from the zygote (c).
To differentiate maternal from zygotic effects, a reciprocal cross experiment was conducted using a pair of nearly isogenic lines (NILs) (see Material and Methods). In Cross 1, an inbred line homozygous for a “large-seed” (L/L) allele from S. lycopersicum was used as the female in a cross with a nearly isogenic line (NIL) homozygous for the “small-seed” (S/S) allele from S. pimpinellifolium. In Cross 2, the reciprocal cross was performed using the S/S as the female parent. For Cross 1, the F1 seed would develop on a maternal plant of the L/L genotype, whereas with Cross 2, the seed would develop on a maternal plant of the S/S genotype. If Sw4.1 exerts its effect on seed weight through the maternal parent, F1 seed from Cross 1 should be significantly larger than F1 seed from Cross 2. The results from these experiments revealed that reciprocal crosses result in seed indistinguishable in weight: 3.07 mg (Cross 1) versus 3.10 mg (Cross 2) (P = 0.99). Whereas self-pollination of the same parents, L/L and S/S, resulted in seeds weighting 3.34 mg and 2.63 mg respectively (P<0.005).
The equivalency in seed weight for the reciprocal F1s suggests that Sw4.1 does not influence seed weight through any significant effect exerted by the genotype of the maternal environment (i.e., fruit). It should be noted that reciprocal crosses would result in triploid endosperm with different parental allelic dosage (L/L/S versus L/S/S). Such differences in allelic dosage might also cause differences in seed weight between seed produced from reciprocal crosses. The fact that no such differences were observed suggests that the genotype of the developing embryo is most likely the major point of control in the differential seed weight associated with Sw4.1 alleles.
Additive Interaction of the L and S Alleles
By comparing the seed weight values of the self-pollinated L/L and S/S NILs, it was estimated that the additive gene effect of Sw4.1 is approximately 0.36 mg [(LL-SS)/2]. As a result, it is estimated that the genetic effect of a single “large-seed” (L) allele is to increase seed weight by 14% – a value very similar to what was reported previously [33]. Using these same parental values, as well as data from the F1 seed lots, it was also possible to calculate the degree of dominance (d/a or k) for interaction of the L and S alleles. It is thus estimated that the alleles interact in a largely additive manner (d/a = −0.21). This result suggests that the change in seed size affected by the two alleles is not due to a loss-of-function (e.g. deletion) at the locus, in which case a dominant-recessive gene action would be observed. This result is consistent with results from the gene expression experiments to be presented later.
Timing of Sw4.1 Effects during Seed Development
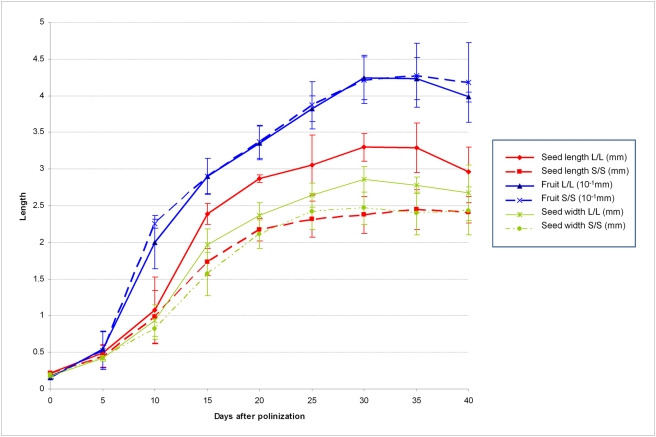
In an effort to identify the time during development at which Sw4.1 alleles modulate seed size, a comparative developmental study was conducted on the large-seeded (L/L) NIL and small-seeded (S/S) NIL. Fruit size was also measured to determine whether Sw4.1 may also affect this character. Developmental plots for the two NILs are shown in Figure 2. No differences were observed for the NILs with respect to fruit size at any time during development. It is therefore concluded that Sw4.1 alleles specifically modulate changes in seed size, and that these changes are not the indirect effect of modulations in fruit size. These results are consistent with the earlier showing that the Sw4.1 effect on seed size is exerted largely through the genotype of the zygote and not the maternal plant.
Figure 2. Plots for developmental changes in fruit length, seed width and seed length for the L/L and S/S NILs.
Bars indicate the standard errors.
Beginning at 10 days after pollination (DAP), seed size (as measured by width and length) was consistently greater for the L/L NIL than for the S/S NIL. For example, seeds from the L/L NIL were on average 23% longer than those of the S/S NIL at 15 DAP (P = 0.008, Figure 2). At maturity, seeds of the L/L NIL were 17% longer than seeds of the S/S NIL (P = 0.001). Overall, L/L NIL showed the greatest change in seed size, relative to the S/S NIL, in the period from 10 DAP to 15 DAP, suggesting that Sw4.1 exerts its largest differentiating effect during this early period of seed development. In an effort to determine the stage of embryo development corresponding to this critical period, paraffin cross sections were prepared from seeds at 10, 15 and 20 DAP. Microscopic examinations of the sections revealed that the embryos are globular at 10 DAP and torpedo-shape to curving at 15 DAP. These stages are similar to those previously reported by Lersten [35] and correspond to the initiation of rapid deposition of starch and lipids into the endospermic cells.
Sw4.1 Exerts Equal Effects on the Size of Both the Embryo and Endosperm but Does Not Affect Seed Viability or Germination Rate
An examination of sections of mature seed from the L/L and S/S NILs revealed that the L/L NIL produces seed that are increased with respect to the size of both the embryo (P = 0.014) and endosperm (P = 0.002) relative to those of the S/S NIL. However, no difference was observed between the two NILs with respect to the ratio of endosperm to embryo (P = 0.956). In both NILs the embryo accounts for the largest portion of the total seed with an embryo∶endosperm ratio of 4∶1. These results suggest that Sw4.1 modulates changes in the amount of both embryo and endosperm tissue during seed development.
Germination tests of the L/L and S/S NILs revealed that both give rise to highly viable seed (germination rates of 97%±3.1 versus 96.3%±2.1, respectively). Therefore, while Sw4.1 affects seed size, it appears to have no detectable affect on seed viability as measured by germination percentage. Another way to characterize seed viability is by germination rate. Germination rate is defined as the speed of germination, which is often associated with seedling vigor [36]. When the L/L and S/S NILs were subjected to germination rate tests, no difference was observed (P = 0.106). However, since these tests were done under laboratory conditions, we cannot rule out the possibility that allelic variation at Sw4.1 does affect seed germination under natural or field conditions.
High-Resolution Mapping of Sw4.1
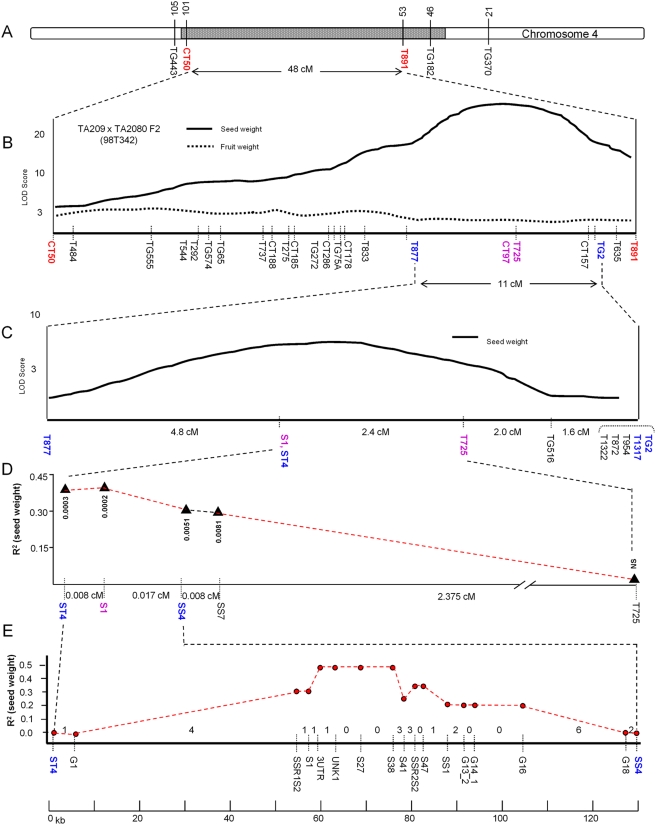
A high-resolution genetic mapping study was conducted in an effort to establish the molecular basis for Sw4.1. This study was facilitated by the availability of the L/L and S/S NILs from which large F2 mapping populations could be derived. The first such F2 population was screened with the markers CT50 and T891 which flank the Sw4.1 QTL (Figure 3A). DNA from individuals showing recombination between these two markers was further screened by an additional 23 markers within the region, allowing the Sw4.1 QTL to be resolved to an 11 cM interval between markers T877 to TG2 (Figure 3B). Within this interval the highest LOD score was observed for marker CT97, 53% of the variation in seed weight was associated with this marker. To further resolve the position of Sw4.1, 140 additional individuals, derived from a F2-heterozygous individual, were then screened for all markers (including 9 additional markers) in the 11 cM interval. From this mapping, Sw4.1 was further resolved to a 7 cM interval between marker T877 and T725 (Figure 3C). Within this interval, seed weight showed the strongest association with marker S1. The position of Sw4.1 was further delimited to a 2.4 cM interval (between markers ST4 and T725) by the screening of an additional 1,000 progeny (Figure 3D). The S1 marker again showed the strongest association with seed weight explaining 41% of the seed weight variation.
Figure 3. High resolution mapping of Sw4.1 QTL on tomato chromosome 4.
(A) TA2080 S/S NIL showing the introgression region from S. pimpinellifolium containing Sw4.1 QTL (shaded). (B) Sw4.1 mapping of F2-population of 150 individuals (TA209 – L/L NIL×TA2080 – S/S NIL) within a 48 cM region. (C) Sw4.1 mapping within a 11 cM region from 140 individuals from a F2 heterozygous individual (98T342-95). (D) Sw4.1 mapping within a 2.4 cM region from recombinants selected from 1,000 seeds from F2 heterozygous individuals. The S1 marker was then used to isolate and sequence the 130 kb BAC clone LE_HBa0077O05. (E) Sw4.1 mapping within the ∼130 kb BAC LE_HBa0077O05 from recombinants selected from 9,000 seeds from F2 heterozygous individuals. The numbers between markers represent the number of crossover events in each interval.
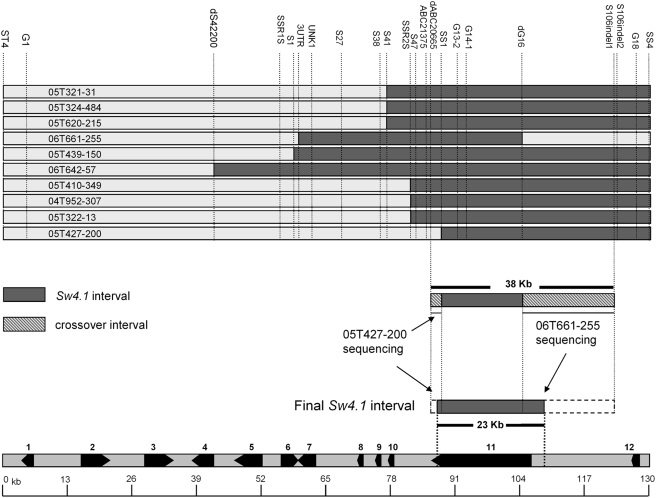
The S1 marker was then used to isolate and sequence a 130 kb BAC (LE_HBa0077O05) from the Sw4.1 region of chromosome 4. The markers SS4 and ST4, derived from the end sequence of this BAC, were subsequently used to screen an additional 9,000 F2 plants and identify 25 recombinants within the BAC interval. However, sufficient seed for analysis was obtained for only 13 of the recombinant individuals. The R2-plot for the seed weight association among the 13 recombinants indicated that the cause of the Sw4.1 QTL resides within the central portion of this BAC (Figure 3E). To gain further precision on the location of Sw4.1 within the BAC, selfed progenies from 10 selected recombinants were examined. These individuals contained crossovers between markers SSR1S2 and G18 (∼73 kb apart) - where the maximum marker association with seed weight was localized by fine mapping (Figure 3E, 4). A comparison of seed weight from homozygous recombinant and non-recombinant progeny allowed positioning of the Sw4.1 QTL relative to the crossover point in each stock (Table 1, Figure 4).
Figure 4. Results from progeny testing of key recombinants to delineate the position of Sw4.1 in BAC.
Shaded area indicates where Sw4.1 QTL is assigned in each recombinant based on progeny tests (see Table 1). Cumulative results pinpointed Sw4.1 to 38 kb interval. Sequencing of two key flanking recombinants (05T427-200 and 06T661-255) further delineated Sw4.1 to a 23 kb interval containing single gene in BAC (gene 11, ABC transporter) (bottom).
Table 1. Progeny analysis of 10 heterozygous individuals with recombination within the BAC.
| F3 recombinant | pedigree | Recombinant/ non-recombinant | A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | n | SWE (mg) | One tail t-test | P value |
| 05T321-31 | 07T542 | R | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 5 | 2.338 | NR>R | 0.105 |
| NR | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 6 | 2.552 | ||||
| 05T324-484 | 07T543 | R | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 6 | 2.419 | R>NR | 0.039 |
| NR | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 6 | 1.992 | ||||
| 05T620-215 | 07T544 | R | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 6 | 3.121 | R>NR | 0.011 |
| NR | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 5 | 2.5 | ||||
| 06T661-255 | 07T540 | R | 1 | 1 | 1 | 1 | 1 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 6 | 2.458 | NR>R | 0.004 |
| NR | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 5 | 3.075 | ||||
| 05T439-150 | 07T541 | R | 3 | 3 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 6 | 2.664 | NR>R | 0.21 |
| NR | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 6 | 2.795 | ||||
| 06T642-57 | 07T539 | R | 1 | 1 | 1 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 1 | 1 | 7 | 2.725 | NR>R | 0.01 |
| NR | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 6 | 3.072 | ||||
| 05T410-349 | 07T545 | R | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 | 2.919 | NR>R | 0.42 |
| NR | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 | 2.956 | ||||
| 04T952-307 | 07T546 | R | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 6 | 2.776 | R>NR | 0.001 |
| NR | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 5 | 1.891 | ||||
| 05T322-13 | 07T547 | R | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | 2.942 | R>NR | 0.003 |
| NR | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 2.497 | ||||
| 05T427-200 | 07T548 | R | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 6 | 2.84 | R>NR | 0.001 |
| NR | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 4 | 2.095 | ||||
| X | X | X | X | X | X | X |
“1” indicates homozygous for corresponding marker from L/L NIL. “3” indicates homozygous for corresponding marker from S/S NIL. SWE = seed weight. In italic and bold is the region indicated by the one-tail t-test of either recombinant versus non-recombinant (R>NR) or non-recombinant versus recombinant (NR>R) for the positioning of the Sw4.1 QTL. Shown at bottom (X) is the consensus region for the location of Sw4.1 based on progeny tests from all 10 recombinants. Marker A = ST4; B = G1; C = dS4220; D = SSR1S2; E = S1; F = 3UTR; G = UNK1; H = S27; I = S38; J = S41; K = SSR2S2; L = S47; M = ABC21375; N = dABC20665; O = SS1; P = G13-2; Q = G14-1; R = G16; S = S106indel1; T = S106indel2; U = G18; V = SS4; W = SS7; X = T725.
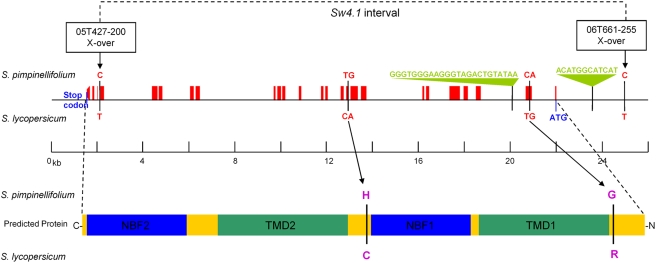
Two independent crossover events (05T427-200 and 06T662-255) delineated the cause of the Sw4.1 QTL to a 38 kb interval extending from markers dABC20665 to S106 indel1 (Table 1, Figure 4). The exact position of the crossover in each of these recombinant stocks was established by PCR sequencing through the crossover boundary region for each stock. As a result, the cause of the Sw4.1 QTL could be further narrowed to a smaller, 23 kb interval (Figure 4). Based on annotation, this 23 kb interval contains a single gene encoding a putative ATP binding cassette (ABC) transporter protein (gene 11 in Figure 4).
Identifying the Arabidopsis Ortholog to the Sw4.1 ABC Transporter Gene
ABC transporters represent a super family of ATP-binding cassette proteins found in a wide range of species [37]. They are used in transmembrane transport of diverse substances – including peptides, sugars, lipids, heavy metal chelates, polysaccharides, alkaloids, steroids, inorganic acids and glutathione conjugates [38]–[42]. The Arabidopsis genome contains at least 129 ABC transporter-like genes, and a number have already been investigated with regards to function [43]. As an aid to annotation of the tomato ABC transporter gene, and in the hope of gaining possible insights into its function, an effort was made to determine which of these Arabidopsis ABC transporter gene(s) might be orthologous to the tomato gene associated with Sw4.1.
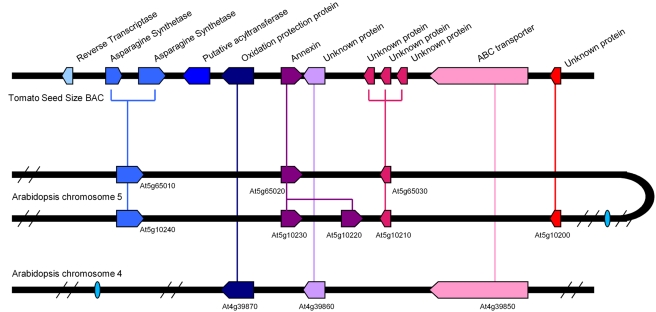
Synteny has proven a very powerful method for establishing orthology [44]. A Reciprocal Best Match (RBC) approach [45] was thus used to identify microsyntenic genomic region(s) between Arabidopsis and tomato for the Sw4.1 region. With this approach, an effort was made to identify the putative orthologs of the 12 genes annotated in the tomato BAC containing the ABC transporter gene (Table S1). As a result, three microsyntenic blocks were identified in Arabidopsis – two blocks located on chromosome 5 and one on chromosome 4 (Figure 5). Only the syntenic block on chromosome 4 contains an ABC transporter (At4g39850) (Figure 5). It is therefore concluded that the Arabidopsis ABC transporter gene At4g39850 is orthologous to the tomato ABC transporter associated with Sw4.1. It is worth noting that At4g39850 is located at the same chromosomal position as a QTL associated with variation in both seed length and width in a cross between Arabidopsis ecotypes, raising the possibility that this gene may also underlie natural variation in seed size in this species [16],[46].
Figure 5. Relationship of genes in tomato BAC containing Sw4.1 and corresponding syntenic regions in Arabidopsis genome.
Sequence alignment of Arabidopsis At4g39850 cDNA with the genomic sequences of both the L allele (S. lycopersicum) and S allele (S. pimpinellifolium) allowed the prediction of intron and exon boundaries in the tomato gene (Figure 6). It also allowed prediction of the full-length tomato ABC transporter protein, which is characterized by 4 functional domains, two of which are ATP-binding cassette (ABC) or nucleotide binding folds (NBFs) and two of which are hydrophobic integral membrane domains (TMDs) (Figure 6) [43]. While both the length of coding region and the intron positions are highly conserved between the tomato and Arabidopsis orthologs, the introns are more variable in length and are generally longer in tomato than Arabidopsis (data not shown).
Figure 6. Annotation of tomato ABC transporter gene associated with Sw4.1 QTL.
Exons are shown in red. Conserved functional domains are shown in blue, ATP-binding cassette (ABC) or nucleotide binding folds (NBFs), and green, hydrophobic integral membrane domains (TMDs).
Several T-DNA insertion mutants have been isolated for the Arabidopsis ortholog At4g39850. Some of these mutants affected seed size [47]. However, the Arabidopsis mutants were associated with an increase in seed size, whereas in tomato the RNAi transgenics produced smaller seeds. Another difference between Arabidopsis and tomato, is that some of the Arabidopsis mutants also affected seed germination, whereas no germination effects were observed for Sw4.1 in tomato. Further, one of the mutants (cts-2) produced seeds with significant higher levels of fatty acids. These results led to the suggestion that the Arabidopsis ABC transporter protein might be involved, not only in lipid metabolism during germination, but also in lipid accumulation during seed development – possibly explaining why the mutants produced larger seeds [47]. In this regard, it is worth noting that in tomato the Sw4.1 QTL produces its largest effects during the stages of seed development associated with lipid deposition (see previous section, Figure 2). Thus, it is possible that the tomato ABC transporter gene modulates seed size by controlling the accumulation of lipids during seed development.
Testing the Effects of the ABC Transporter Gene on Seed Size via RNAi Transformation Experiments
Transformation experiments were used to test whether the ABC transporter gene has the ability to modulate seed size. This gene is quite long (due to many introns) – spanning more than 20 kb from the start to stop codon (Figure 6).
The large size of the gene, absence of efficient enzymatic sites for cloning and lack of a full-length cDNA, precluded complementation analysis with the full-length genomic copy of the gene. Therefore, a gene silencing approach, via RNAi hairpin formation, was employed as an alternative strategy for testing the potential role of the ABC transporter gene in determining seed size.
Efficient post-transcriptional silencing has been reported when 3′UTR regions are targeted for RNAi machinery [48],[49]. Hence, a 278 bp fragment from the 3′UTR of the ABC transporter gene was inserted in the pHELLSGATE2 (Invitrogen) binary vector, the pSP13-1 construct (Figure S1). Sequence specificity was assessed by blasting the 278 bp fragment against the tomato unigene database in the SGN website (www.sgn.cornell.edu). The retrieval of a unique unigene that corresponds to this ABC transporter gene suggests this sequence is specific to this gene and therefore there would present a low risk to silence other genes in the same ABC transporter family. This construct was transformed into both the L/L and S/S NILs. Multiple independent T0 and non-transgenic controls were then analyzed for both NIL sets. L/L transformants were highly fertile, yet produced seeds weighing on average 38% less than those from the non-transgenic controls (P = 0.03, Table 2). S/S transformants also produced smaller seed (11% less heavy) than the non-transgenic controls, however the statistical difference did not quite reach statistical significance (P = 0.09, Table 2). It is worth noting that transformation/complementation with two other genes from the BAC (annexin and a gene of unknown function – genes number 6 and 7, respectively, in Figure 4) did not show statistical difference between transgenic and non-transgenic plants (data not shown). The transformation experiments thus appear to corroborate the results from high-resolution mapping – both pointing to the ABC transporter gene as the cause of the Sw4.1 QTL.
Table 2. Summary of transgenic silencing of ABC transporter gene in L/L and S/S NILs based on RNA interference.
| NIL Plant | Transgenic Status | N | Seed Weight (mean±SE – mg) | P value |
| L/L | Transgenic | 10 | 2.016±0.345 | 0.031 |
| Non-transgenic | 3 | 3.235±0.522 | ||
| S/S | Transgenic | 10 | 2.262±0.495 | 0.093 |
| Non-transgenic | 2 | 2.529±0.132 |
Allelic Polymorphisms in the ABC Transporter Gene
Comparing the sequence of the L and S allele, within the 23 kb Sw4.1 interval, revealed 79 SNPs and single indels located in introns, 10 in the promoter region and 6 in exons (Figure S2). In the exons, 2 non-synonymous changes were observed (Figure 6). Either of these might be causal to the phenotypic effects rendered by the L and S alleles. However, neither substitution is located in a conserved functional domain (e.g. nucleotide binding folds or hydrophobic integral membrane domains) (Figure 6). Among the polymorphisms in non-coding regions, a few are worth mentioning as possible causal candidates for the Sw4.1 QTL. One is a 12 nt indel approximately 1.5 kb upstream in the 5′ promoter and the second, a 24 nt indel in the first intron (Figure 6). Either of these indels might cause a change in expression – as could the many other small nucleotide differences observed in non-coding regions of the two alleles.
Expression of ABC Transporter Gene during Seed Development
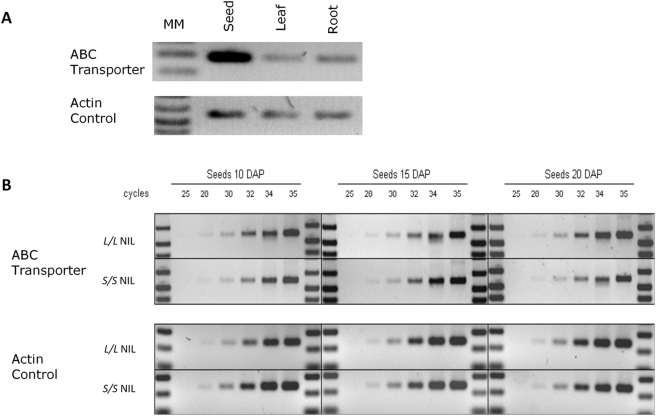
In an effort to determine the expression pattern of the ABC transporter gene, and especially whether the L and S allele differ in regulation/expression during seed development, a set of semi-quantitative RT-PCR experiments were conducted. The first experiment revealed that this gene is expressed at high levels in seeds, and lower levels in shoots and roots (Figure 7A). High expression of the ABC transporter gene in developing seeds is further evidence that this gene is the cause of the Sw4.1 QTL. The subsequent experiment compared expression of the ABC transporter gene in both the L and S NILs at 10, 15 and 20 DAP – the time when the major change in seed size is observed (see previous section) (Figure 7B). Two conclusions can be drawn from this second experiment. First, the ABC gene is expressed in both the L and S NILs – ruling out a loss-of-function as the cause of seed size variation associated with Sw4.1. Second, the failure to detect any major change in expression of the ABC transporter gene between the L and S NILs during seed development would seem to rule out a gross change in the regulation as the cause of the Sw4.1 QTL (Figure 7B). However, we cannot exclude the possibility of a difference in translational/post-translational regulation or small changes in spatial or temporal regulation as the cause of QTL effect. For example, in tomato it was previously shown that a modest change in the timing of allele expression can cause major QTL effects on fruit size [50].
Figure 7. RNA expression studies of tomato ABC transporter gene.
(A) Comparison of expression in seeds, leaves and roots using semi-quantitative RT-PCR. Actin gene used as a comparative control. (B) Semi-quantitative RT-PCR of ABC transporter gene during seed development comparing L/L and S/S NILs.
Conclusions
The results presented herein point to natural variation in an ABC transporter gene as a major cause of the change in seed size that differentiates the cultivated tomato from related wild species. While prior studies in a number of plants have reported the effects of induced mutations on seed size, this example is among the few in which the cause of changes in seed size in nature populations has been pinpointed. Further, the ortholog to this gene in Arabidopsis has been identified through homology/synteny. Results from QTL mapping and mutagenesis studies are consistent with this gene also playing a role in determining seed size in Arabidopsis. However, further studies are required to clearly establish whether this ABC transporter gene operates in a similar manner in both tomato and Arabidopsis. Also, it remains to be established whether variation in ABC transporter genes is a major cause of seed size variation in natural populations of Arabidopsis or other plant species. Having identified the cause of the Sw4.1 QTL in tomato may open he door to addressing these questions in the future.
Materials and Methods
Plant Material and Genetic Markers
Two Solanum lycopersicum nearly isogenic lines (NILs), with alternative alleles at the Sw4.1 locus, were the origin of all genetic stocks used in this study. TA209 carries the large-seeded (S. lycopersicum) allele at Sw4.1 locus. TA2080 is isogenic with TA209, but carries a 55–84 cM segment of chromosome 4 of S. pimpinellifolium LA1589 (Figure 3A) containing the small-seeded allele of Sw4.1 [13],[33]. TA2080 was developed via marker assisted selection during 5 sequential backcrosses of LA1589 into TA209 followed by a single selfing generation (BC5S1). An F2 population, segregating for Sw4.1, was then developed from a cross between TA209 and TA2080. A derived, shorter introgression NIL, derived from TA2080, was used for the reciprocal cross experiments. Likewise, the S/S NIL (TA3820) used for transformation experiments was also a shorter derivative of TA2080. Detailed description of the construction of both derived subNILs can be found in Orsi [46]. The sequence of all markers used in this study can be found in Table S2.
Reciprocal Cross and Gene Action Experiments
Plants were grown in pots in the greenhouse in 12 randomized blocks. Each block was comprised of the following Sw4.1 genotypes and type of pollination: L/L: selfing, S/S: selfing, L/S: selfing; L/L: used as the female in crosses to S/S, S/S: used the female in crosses to L/L. Each plant was either selfed or crossed manually. At maturity, 5 normal fruit (no blossom end rot, non-parthenocarpic) were harvested from each plant and the seed extracted. The fruit were weighed and the average weight per fruit was recorded. From a pool of seeds from 5 fruits, 50 healthy seeds were randomly sampled. From these, an average seed weight was calculated for each plant. A comparison of the least squares means was performed using the adjustment for multiple comparisons Tukey-Kramer (SAS enterprise guide 3.0).
Developmental Analyses
Effects of Sw4.1 on seed development and fruit size: Ten pairs of L/L and S/S NIL plants were grown in the greenhouse. Two to three fruits were harvest from each plant at each of the following stages: anthesis (0 days after pollination –DAP), 5 DAP, 10 DAP, 15 DAP, 20 DAP, 25 DAP, 30 DAP, 35 DAP and 40 DAP. Because of the microscopic size of ovule and developing seeds, it was not possible to collect mass (weight) data. Instead, all traits were recorded as spatial metrics (e.g., length, width). Both fruit and seed were scanned using a HP ScanJet (1200 dpi). Length and width measurements were then extracted from the images using the software Tomato Analyzer Version v.1.2 [51]. A second independent experiment, of identical design, was then conducted. However, based on results from the first experiment, fruit were collected only at 10 DAP, 15 DAP and 20 DAP – periods associated with most rapid changes in seed development. For the verification of seed developmental stages, seeds from 10, 15 and 20 DAP were fixed, dehydrated and embedded in paraplast (Sigma) as described by [52].
Role of Sw4.1 in determining the proportion of embryo to endosperm in mature seed: Five pairs of L/L and S/S individuals were grown in the greenhouse. Five mature fruits were harvested from each individual and the seeds extracted. A random sample of 10 normal seeds was then drawn from each individual. Each individual seed was then dissected longitudinally into halves and the images digitalized under a dissecting scope (ZeissStemi 2000-CS attached to 3CCD camera MTI) using the software Scion Image (www.scioncorp.com). The areas of the entire seed, embryo and endosperm were manually delineated and measured using the software ImageJ 1.31 v (http://rsb.info.nih.gov/ij/).
Seed Germination and Viability Experiments
Ten pairs of L/L and S/S individuals were grown in the greenhouse. One hundred normal seeds from a pool of 10 fruits of each genotype were germinated in Petri dishes on filter paper saturated with distilled water. The seeds were scanned prior to the germination process for seed length measurement (Tomato Analyzer v.1.2). The number of germinated seed was recorded on a daily basis until no additional seed germination was obtained. For these experiments, a seed was considered germinated once the root tip had emerged. Tests of heterogeneity on the number of germinated seeds at days 3, 4 and 5 (when the majority of seed germination was observed for all the genotypes) were performed for the detection of possible differences in germination rate between NILs (Minitab 15).
Progeny Testing of Selected Recombinants
The genetic stocks used for the progeny analysis were derived from sub-NILs selected from each selected recombinant within the BAC (Figure 4). Ten homozygous recombinant and ten homozygous non-recombinant individuals were selected, via marker analysis, from selfed seed of selected recombinants (Table 1, Figure 4). The selected progeny were grown in a completely randomized design in the greenhouse. From each plant, seeds were extracted from 5 fruits and pooled. From each pool, 50 normal seeds were randomly selected and weighed. For each family, statistical comparisons for seed weight were made between the 10 recombinant and 10 non-recombinant progeny from each family using a one-tailed t-test (Minitab 15) (Table 1).
BAC Annotation and Comparison with the Arabidopsis Genome
BAC LE_HBa0077O05, isolated with marker probe S1, was annotated using the automated annotation tools developed by SGN and refined manually through BLAST searches against EST libraries (Solanaceae, coffee, Arabidopsis). Arabidopsis genes, putatively orthologous to genes in the tomato BAC, were identified using the Best Reciprocal Matches (RBM) in BLAST comparisons [45]. Based on these putative orthologs, it was possible to identify region in the Arabidopsis genome showing conserved microsynteny with the Sw4.1 region of tomato chromosome 4.
Sequencing of Sw4.1 Interval from S. pimpinellifolium and Annotation of Polymorphisms between S. lycopersicum and S. pimpinellifolium
A 38 kb segment, delimited by markers dABC20665 and S106indel1 and known from progeny analyses to encompass the Sw4.1 QTL, was also sequenced via PCR from the genome of S. pimpinellifolium LA1589 – the small-seeded parent of the original mapping population. The objective of the sequencing was to identify polymorphisms that might be causal to the Sw4.1 QTL. Sequence analysis was performed with DNASTAR Lasergene software and alignments with BioEdit version 7.0.9.0 using ClustalW.
Sequencing Tecombinants in the 38 kb Sw4.1 Interval to Pinpoint the Exact Crossover Points
In order to further narrow the Sw4.1 interval, PCR-based sequencing was performed on the two recombinant individuals that define the left (05T427-200) and right (06T661-255) side of the 38 kb interval. By sequencing across the exact crossover point in each recombinant, it was possible to narrow the location of the Sw4.1 QTL to a 23 kb interval (Figure 4).
Transformation Experiments
RNAi transgene construction
A 278 bp fragment from the 3′UTR of the ABC transporter gene orthologous to At4g39850 was inserted in the binary vector pHellsGate 2 (Invitrogen) [53] for the generation of a hairpin construct for the RNAi mechanism induction and gene silencing in L/L and S/S NILs. The schematic representation of plasmid construction is shown in Figure S1. The pHELLSGATE vector was designed such that a single PCR product from primers with the appropriate attB1 and attB2 sites would be recombined into it simultaneously to form the two arms of the hairpin [53]. The recovery of successful recombination (insertion) of both arms of the hairpin was ensured by ccdB genes, which were replaced by the arm sequences (ABC transporter fragment). CcdB gene is lethal in standard E. coli strains such as DH5a, strain that was used for plasmid cloning. The intron retention was ensured by the chloramphenicol-resistance gene (CAM) within the intron. The successful insertion for both arms of the hairpin was confirmed by sequencing analysis (data not shown). Recombinant pHELLSGATE constructs, called pSP13-1, were sent for transformation (Plant Science Initiative, University of Nebraska) for the direct transformation into Agrobacterium for transformation into the L/L and S/S NILs.
Experimental design and statistics
Transgenic analysis of the T0 generation: 10 transgenic L/L NILs, 3 non-transgenic L/L NILs, 10 transgenic S/S NILs and 2 non-transgenic S/S NILs were grown in greenhouse in a random design. Classification of plants as transgenic or non-transgenic was based on the amplification of the insert-vector using the primers AttB1-ABC and XhoI insert-vector for the sense insertion (403 bp) and Att1B1-ABC and XbaI insert-vector for the anti-sense insertion (400 bp). RNA levels for the ABC transporter gene were not measured in seeds from T0 plants since this would have precluded seed size measurements from the same plants. For efficient RNAi mechanism induction, both arms of the hairpin must be present as well as the intronic sequence. Therefore, amplification of both XbaI and XhoI were required for the assignment of transgenic plants. As a positive control for PCR amplification, S85 primers were included in each reaction. S85 amplifies 800 bp from the ABC transporter promoter region, which would be present in transgenic and non-transgenic plants. Five fruits of each plant were harvested and the seeds were extracted. 50–100 seeds were randomly selected from each pool and weighed. One tail t-tests, comparing transgenic and non-transgenic plants were then performed using Minitab 15.
Analysis of ABC Transport Gene Transcript
Experimental design
Ten L/L NIL and ten S/S NIL individuals were paired and grown in greenhouse. Each flower was manually pollinated and the date of pollination noted. Seeds from fruits 10, 15 and 20 DAP were removed and immediately frozen in liquid nitrogen. Seeds at these stages were pooled from 5 healthy plants for the RNA extraction. Frozen seeds were ground to a fine powder in liquid nitrogen and total RNA was isolated using Trizol reagent (Invitrogen). The concentration of total RNA from each sample was determined from 100× diluted solution using spectrophotometry. One microgram of total RNA from each sample was treated with RNase-free DNaseI (amplification grade, Invitrogen). First-strand cDNA was synthesized by reverse transcription with oligo(dT)16 primer following manufacturer's protocol (Invitrogen). Another set of plants was grown and the procedure repeated as an independent experiment for replicate.
Semi-quantitative RT-PCR
ABC transporter transcript levels were detected by using semi-quantitative RT-PCR. The primers used for the amplification are S53R (5′ GGGAAGACGAACCAAATGAA 3′) and ABCp35R (5′ CGGGAACTAGGCGCTATACA 3′). These primers were designed based on the 3′end and 3′UTR of the gene. BLAST search against A. thaliana non-redundant database in NCBI and tomato EST database in SGN (www.sgn.cornell.edu) confirmed the uniqueness of this sequence and specificity to this ABC transporter gene. The internal control was actin TOM52 [54]. For the semi-quantitative approach, from a total reaction of 100 ul, 10 ul were collected at the end of each one of these cycles: 25, 28, 30, 32, 34 and 35.
Supporting Information
Diagram of pSP13-1 construct used for RNAi based gene silencing of ABC transporter gene in transgenic experiments.
(0.75 MB TIF)
Detailed alignment of L and S alleles showing all polymorphisms in the 23 kb region encompassing the Sw4.1 QTL.
(7.55 MB TIF)
Annotation of the 12 genes contained in the tomato BAC LE_HBa0077O05.
(0.04 MB DOC)
Markers for S. lycopersicum and S. pimpinellifolium: marker type, primer sequence, restriction enzyme and fragment size.
(0.15 MB DOC)
Acknowledgments
We thank Dr. Sami Doganlar and Nancy Eannetta for assistance in population development and data collection; Nick Van Eck for greenhouse support; Yimin Xu, Geoffrey Vernon and Theresa Eannetta for technical support; Dr. Bin Cong for providing root and leaf RNA material and for comments and discussion; TGRC, UC Davis for providing wild species seeds and Dr. Amy Frary for review and editing of the manuscript.
Footnotes
The authors have declared that no competing interests exist.
Support for this research provided by grants from the USDA-NRI and the National Science Foundation DBI-0421634 to SDT and from CNPq, a Brazilian Government entity for scientific and technological development, to CHO.
References
- 1.Smith CC, Fretwell SD. The Optimal Balance between Size and Number of Offspring. The American Naturalist. 1974;108:499. [Google Scholar]
- 2.Geritz SA, van der Meijden E, Metz JA. Evolutionary dynamics of seed size and seedling competitive ability. Theor Popul Biol. 1999;55:324–343. doi: 10.1006/tpbi.1998.1409. [DOI] [PubMed] [Google Scholar]
- 3.Leishman MR, Westoby M. The Role of Seed Size in Seedling Establishment in Dry Soil Conditions – Experimental Evidence from Semi-Arid Species. The Journal of Ecology. 1994;82:249–258. [Google Scholar]
- 4.Bonfil C. The Effects of Seed Size, Cotyledon Reserves, and Herbivory on Seedling Survival and Growth in Quercus rugosa and Q. laurina (Fagaceae). American Journal of Botany. 1998;85:79–87. [PubMed] [Google Scholar]
- 5.Hewitt N. Seed size and shade-tolerance: a comparative analysis of North American temperate trees. Oecologia. 1998;114:432–440. doi: 10.1007/s004420050467. [DOI] [PubMed] [Google Scholar]
- 6.Jurado E, Westoby M. Seedling Growth in Relation to Seed Size Among Species of Arid Australia. The Journal of Ecology. 1992;80:407–416. [Google Scholar]
- 7.Gomez JM. Bigger is not always better: conflicting selective pressures on seed size in Quercus ilex. Evolution Int J Org Evolution. 2004;58:71–80. doi: 10.1111/j.0014-3820.2004.tb01574.x. [DOI] [PubMed] [Google Scholar]
- 8.Coomes DA, Grubb PJ. Colonization, tolerance, competition and seed-size variation within functional groups. Trends in Ecology & Evolution. 2003;18:283–291. [Google Scholar]
- 9.Isemura T, Kaga A, Konishi S, Ando T, Tomooka N, et al. Genome dissection of traits related to domestication in azuki bean (Vigna angularis) and comparison with other warm-season legumes. Ann Bot (Lond) 2007;100:1053–1071. doi: 10.1093/aob/mcm155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Broich SL, Palmer RG. A cluster analysis of wild and domesticated soybean phenotypes. Euphytica. 1980;29:23–32. [Google Scholar]
- 11.Burke JM, Tang S, Knapp SJ, Rieseberg LH. Genetic analysis of sunflower domestication. Genetics. 2002;161:1257–1267. doi: 10.1093/genetics/161.3.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuller DQ. Contrasting patterns in crop domestication and domestication rates: recent archaeobotanical insights from the old world. Ann Bot (Lond) 2007;100:903–924. doi: 10.1093/aob/mcm048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doganlar S, Frary A, Tanksley SD. The genetic basis of seed-weight variation: tomato as a model system. Theoretical and Applied Genetics. 2000;100:1267–1273. [Google Scholar]
- 14.Smith BD. The Initial Domestication of Cucurbita pepo in the Americas 10,000?Years Ago. Science. 1997;276:932–934. [Google Scholar]
- 15.Harlan JR, de Wet JMJ, Price EG. Comparative Evolution of Cereals. Evolution. 1973;27:311–325. doi: 10.1111/j.1558-5646.1973.tb00676.x. [DOI] [PubMed] [Google Scholar]
- 16.Alonso-Blanco C, Blankestijn-de Vries H, Hanhart CJ, Koornneef M. Natural allelic variation at seed size loci in relation to other life history traits of Arabidopsis thaliana. Proc Natl Acad Sci U S A. 1999;96:4710–4717. doi: 10.1073/pnas.96.8.4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shomura A, Izawa T, Ebana K, Ebitani T, Kanegae H, et al. Deletion in a gene associated with grain size increased yields during rice domestication. Nat Genet. 2008;40:1023–1028. doi: 10.1038/ng.169. [DOI] [PubMed] [Google Scholar]
- 18.Lin HX, Qian HR, Zhuang JY, Lu J, Min SK, et al. RFLP mapping of QTLs for yield and related characters in rice (Oryza sativa L.). TAG Theoretical and Applied Genetics. 1996;92:920–927. doi: 10.1007/BF00224031. [DOI] [PubMed] [Google Scholar]
- 19.Huang N, Parco A, Mew T, Magpantay G, McCouch S, et al. RFLP mapping of isozymes, RAPD and QTLs for grain shape, brown planthopper resistance in a doubled haploid rice population. Molecular Breeding. 1997;3:105–113. [Google Scholar]
- 20.Redoña ED, Mackill DJ. Quantitative trait locus analysis for rice panicle and grain characteristics. TAG Theoretical and Applied Genetics. 1998;96:957–963. [Google Scholar]
- 21.Thomson MJ, Tai TH, McClung AM, Lai XH, Hinga ME, et al. Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. TAG Theoretical and Applied Genetics. 2003;107:479–493. doi: 10.1007/s00122-003-1270-8. [DOI] [PubMed] [Google Scholar]
- 22.Yoon DB, Kang KH, Kim HJ, Ju HG, Kwon SJ, et al. Mapping quantitative trait loci for yield components and morphological traits in an advanced backcross population between Oryza grandiglumis and the O. sativa japonica cultivar Hwaseongbyeo. TAG Theoretical and Applied Genetics. 2006;112:1052–1062. doi: 10.1007/s00122-006-0207-4. [DOI] [PubMed] [Google Scholar]
- 23.Hyten DL, Pantalone VR, Sams CE, Saxton AM, Landau-Ellis D, et al. Seed quality QTL in a prominent soybean population. Theor Appl Genet. 2004;109:552–561. doi: 10.1007/s00122-004-1661-5. [DOI] [PubMed] [Google Scholar]
- 24.Liu B, Fujita T, Yan ZH, Sakamoto S, Xu D, et al. QTL mapping of domestication-related traits in soybean (Glycine max). Ann Bot (Lond) 2007;100:1027–1038. doi: 10.1093/aob/mcm149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al-Chaarani GR, Gentzbittel L, Huang XQ, Sarrafi A. Genotypic variation and identification of QTLs for agronomic traits, using AFLP and SSR markers in RILs of sunflower (Helianthus annuus L.). Theor Appl Genet. 2004;109:1353–1360. doi: 10.1007/s00122-004-1770-1. [DOI] [PubMed] [Google Scholar]
- 26.Bert PF, Jouan I, Tourvieille de Labrouhe D, Serre F, Philippon J, et al. Comparative genetic analysis of quantitative traits in sunflower (Helianthus annuus L.). 2. Characterisation of QTL involved in developmental and agronomic traits. Theor Appl Genet. 2003;107:181–189. doi: 10.1007/s00122-003-1237-9. [DOI] [PubMed] [Google Scholar]
- 27.Mokrani L, Gentzbittel L, Azanza F, Fitamant L, Al-Chaarani G, et al. Mapping and analysis of quantitative trait loci for grain oil content and agronomic traits using AFLP and SSR in sunflower ( Helianthus annuus L.). Theor Appl Genet. 2002;106:149–156. doi: 10.1007/s00122-002-1011-4. [DOI] [PubMed] [Google Scholar]
- 28.Baack EJ, Sapir Y, Chapman MA, Burke JM, Rieseberg LH. Selection on domestication traits and quantitative trait loci in crop-wild sunflower hybrids. Mol Ecol. 2008;17:666–677. doi: 10.1111/j.1365-294X.2007.03596.x. [DOI] [PubMed] [Google Scholar]
- 29.Song XJ, Huang W, Shi M, Zhu MZ, Lin HX. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet. 2007;39:623–630. doi: 10.1038/ng2014. [DOI] [PubMed] [Google Scholar]
- 30.Fan C, Xing Y, Mao H, Lu T, Han B, et al. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. TAG Theoretical and Applied Genetics. 2006;112:1164–1171. doi: 10.1007/s00122-006-0218-1. [DOI] [PubMed] [Google Scholar]
- 31.Weller JI, Soller M, Brody T. Linkage Analysis of Quantitative Traits in an Interspecific Cross of Tomato (Lycopersicon Esculentum X Lycopersicon Pimpinellifolium) by Means of Genetic Markers. Genetics. 1988;118:329–339. doi: 10.1093/genetics/118.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goldman IL, Paran I, Zamir D. Quantitative trait locus analysis of a recombinant inbred line population derived from a Lycopersicon esculentum×Lycopersicon cheesmanii cross. TAG Theoretical and Applied Genetics. 1995;90:925–932. doi: 10.1007/BF00222905. [DOI] [PubMed] [Google Scholar]
- 33.Grandillo S, Tanksley SD. QTL analysis of horticultural traits differentiating the cultivated tomato from the closely related species Lycopersicon pimpinellifolium. TAG Theoretical and Applied Genetics. 1996;92:935–951. doi: 10.1007/BF00224033. [DOI] [PubMed] [Google Scholar]
- 34.Tanksley SD, Medina-Filho H, Rick CM. Use of naturally occuring enzyme variation to detect and map genes controlling quantitative traits in an interspecific backcross of tomato. Heredity. 1982;49:11–25. [Google Scholar]
- 35.Lersten NR. Flowering Plant Embryology: Blackwell Publishing Ltd; 2004. 212 [Google Scholar]
- 36.Heydecker W. Vigour. In: Roberts EH, editor. Viability of seeds: Chapman and Hall Ltd; 1972. [Google Scholar]
- 37.Theodoulou FL. Plant ABC transporters. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2000;1465:79–103. doi: 10.1016/s0005-2736(00)00132-2. [DOI] [PubMed] [Google Scholar]
- 38.Higgins CF. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 39.Dean M, Allikmets R. Evolution of ATP-binding cassette transporter genes. Curr Opin Genet Dev. 1995;5:779–785. doi: 10.1016/0959-437x(95)80011-s. [DOI] [PubMed] [Google Scholar]
- 40.Decottignies A, Goffeau A. Complete inventory of the yeast ABC proteins. Nat Genet. 1997;15:137–145. doi: 10.1038/ng0297-137. [DOI] [PubMed] [Google Scholar]
- 41.Rea PA, Li ZS, Lu YP, Drozdowicz YM, Martinoia E. From Vacuolar Gs-X Pumps to Multispecific Abc Transporters. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:727–760. doi: 10.1146/annurev.arplant.49.1.727. [DOI] [PubMed] [Google Scholar]
- 42.Gaur M, Choudhury D, Prasad R. Complete inventory of ABC proteins in human pathogenic yeast, Candida albicans. J Mol Microbiol Biotechnol. 2005;9:3–15. doi: 10.1159/000088141. [DOI] [PubMed] [Google Scholar]
- 43.Sanchez-Fernandez R, Davies TG, Coleman JO, Rea PA. The Arabidopsis thaliana ABC protein superfamily, a complete inventory. J Biol Chem. 2001;276:30231–30244. doi: 10.1074/jbc.M103104200. [DOI] [PubMed] [Google Scholar]
- 44.Zheng XH, Lu F, Wang Z-Y, Zhong F, Hoover J, et al. Using shared genomic synteny and shared protein functions to enhance the identification of orthologous gene pairs. Bioinformatics. 2005;21:703–710. doi: 10.1093/bioinformatics/bti045. [DOI] [PubMed] [Google Scholar]
- 45.Wu F, Mueller LA, Crouzillat D, Petiard V, Tanksley SD. Combining bioinformatics and phylogenetics to identify large sets of single-copy orthologous genes (COSII) for comparative, evolutionary and systematic studies: a test case in the euasterid plant clade. Genetics. 2006;174:1407–1420. doi: 10.1534/genetics.106.062455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orsi CH. Genetic, developmental, and molecular dissection of Sw4.1 - the major QTL underlying the evolution of increased seed size that accompanied the domestication of tomato. Ithaca: Cornell University; 2008. 117 [Google Scholar]
- 47.Footitt S, Slocombe SP, Larner V, Kurup S, Wu Y, et al. Control of germination and lipid mobilization by COMATOSE, the Arabidopsis homologue of human ALDP. EMBO J. 2002;21:2912–2922. doi: 10.1093/emboj/cdf300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, et al. 3[prime] UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Meth. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- 49.Yoo MH, Xu XM, Turanov AA, Carlson BA, Gladyshev VN, et al. A new strategy for assessing selenoprotein function: siRNA knockdown/knock-in targeting the 3′-UTR. Rna-a Publication of the Rna Society. 2007;13:921–929. doi: 10.1261/rna.533007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cong B, Liu J, Tanksley SD. Natural alleles at a tomato fruit size quantitative trait locus differ by heterochronic regulatory mutations. Proc Natl Acad Sci U S A. 2002;99:13606–13611. doi: 10.1073/pnas.172520999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brewer MT, Lang L, Fujimura K, Dujimovic N, Gray S, et al. Development of a controlled vocabulary and software application to analyze fruit shape variation in tomato and other plant species. Plant Physiol. 2006;141:15–25. doi: 10.1104/pp.106.077867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jackson D. In situ hybridisation in plants. In: Bowles DJ, Gurr SJ, McPhereson M, editors. Molecular Plant Pathology: A Practical Approach. U.K.: Oxford University Press; 1991. [Google Scholar]
- 53.Wesley SV, Helliwell CA, Smith NA, Wang M, Rouse DT, et al. Construct design for efficient, effective and high-throughput gene silencing in plants. The Plant Journal. 2001;27:581–590. doi: 10.1046/j.1365-313x.2001.01105.x. [DOI] [PubMed] [Google Scholar]
- 54.Barrero LS, Cong B, Wu F, Tanksley SD. Developmental characterization of the fasciated locus and mapping of Arabidopsis candidate genes involved in the control of floral meristem size and carpel number in tomato. Genome. 2006;49:991–1006. doi: 10.1139/g06-059. [DOI] [PubMed] [Google Scholar]
- 55.Spooner DM, Peralta IE, Knapp S. Comparison of AFLPs with other markers for phylogenetic inference in wild tomatoes Solanum L. section Lycopersicon (Mill.) Wettst. Taxon. 2005;54:43–61. [Google Scholar]
- 56.Peralta IE, Spooner DM. Granule-bound starch synthase (GBSSI) gene phylogeny of wild tomatoes (Solanum L. section Lycopersicon [Mill.] Wettst. subsection Lycopersicon). Am J Bot. 2001;88:1888–1902. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Diagram of pSP13-1 construct used for RNAi based gene silencing of ABC transporter gene in transgenic experiments.
(0.75 MB TIF)
Detailed alignment of L and S alleles showing all polymorphisms in the 23 kb region encompassing the Sw4.1 QTL.
(7.55 MB TIF)
Annotation of the 12 genes contained in the tomato BAC LE_HBa0077O05.
(0.04 MB DOC)
Markers for S. lycopersicum and S. pimpinellifolium: marker type, primer sequence, restriction enzyme and fragment size.
(0.15 MB DOC)