Summary
In the fission yeast S. pombe, nuclei are actively positioned at the cell center by microtubules. Here we show that cytoplasmic microtubules are mechanically coupled to the nuclear heterochromatin through proteins embedded in the nuclear envelope. This includes an integral outer nuclear membrane protein of the KASH family (Kms2) and two integral inner nuclear membrane proteins: the SUN-domain protein Sad1 and the novel, conserved protein Ima1. Ima1 specifically binds to heterochromatic regions and promotes the tethering of centromeric DNA to the SUN-KASH complex. In the absence of Ima1, or in cells harboring mutations in the centromeric Ndc80 complex, inefficient coupling of centromeric heterochromatin to Sad1 leads to striking defects in the ability of the nucleus to tolerate microtubule-dependent forces, leading to changes in nuclear shape, loss of spindle pole body components from the nuclear envelope, and partial dissociation of SUN-KASH complexes. This work highlights a novel means of communication between cytoplasmic microtubules and chromatin.
Introduction
To alter nuclear position, the cytoskeleton must exert sufficient force on the nucleus to drive its movement, while, simultaneously, the nucleus must buffer these forces so that its entire mass is efficiently propelled through the cytoplasm without affecting nuclear integrity. How the cytoskeleton and nuclear envelope (NE) interface to achieve this balance is poorly understood. The first molecular details of the proteins that form this interface have come from the identification and characterization of the conserved SUN and KASH domain-containing proteins (Figure 1A). In metazoa, integral outer nuclear membrane (ONM) proteins possessing KASH domains link the NE to all major classes of cytoskeletal elements (Starr and Fischer, 2005; Wilhelmsen et al., 2006), and tether the primary microtubule organizing center (MTOC), the centrosome, to the NE (Malone et al., 2003). In turn, KASH domain proteins bind (in the NE lumen) to integral inner nuclear membrane (INM) proteins containing the conserved SUN domain (Tzur et al., 2006). Thus, integral membrane proteins of the NE link the cytoplasmic cytoskeleton to the nuclear interior. Disruption of SUN-KASH interactions leads to expansion of the NE lumen, illustrating the importance of this complex in maintaining nuclear structure (Crisp et al., 2006). However, given the large forces exerted on SUN-KASH complexes (potentially up to 100 pN during mammalian cell migration (Szabo et al., 2004)), it is likely that additional macromolecules reinforce these NE-cytoskeletal connections. Candidates include additional NE membrane proteins, soluble proteins that reside in the NE lumen, and structural elements within the nucleus. In metazoa, the nuclear lamina, a meshwork of the intermediate filament lamins, provides one important structural scaffold to which SUN domain proteins are coupled (Crisp et al., 2006; Gruenbaum et al., 2005; Haque et al., 2006).
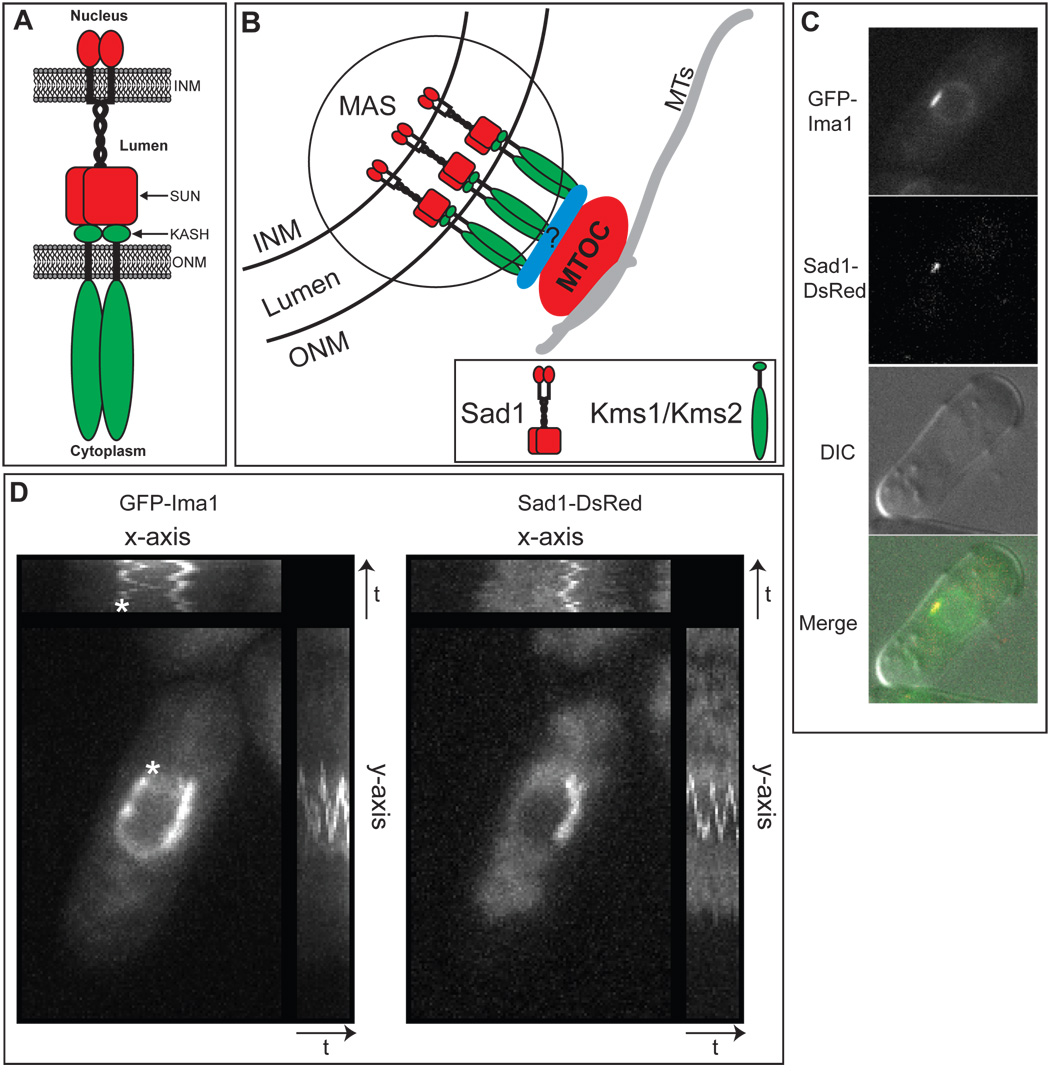
Figure 1. Ima1 is a conserved, integral inner nuclear membrane protein that is enriched at the site of MTOC attachment.
A. Cartoon of SUN-KASH interactions at the nuclear envelope (NE). The SUN domain protein (red) is integrated into the inner nuclear membrane (INM). The protein has an N-terminal nucleoplasmic domain followed by a single transmembrane segment, a lumenal coiled-coil region, and the conserved SUN domain. The KASH domain protein (green) is integrated into the outer nuclear membrane (ONM). The variable N-terminal cytoplasmic domain interacts with cytoskeletal elements and the C-terminus contains the KASH domain, which is composed of the transmembrane segment (black) and a small lumenal tail (labeled KASH). Both proteins are shown as homodimers. B. Diagram of the microtubule organizing center (MTOC) attachment site (MAS) at the NE of S. pombe (circled). SUN domain protein Sad1 (red) and KASH domain proteins Kms1 and/or Kms2 (green) interact within the lumen of the NE to link the MTOC to the NE either directly or through an as yet unidentified adapter protein(s) (blue). MTs = microtubules. C. GFP-Ima1 localizes to the NE and MAS. Fluorescent micrographs, DIC and merged images are shown of a representative single cell of strain MKSP58 expressing GFP-Ima1 and Sad1-DsRed. D. GFP-Ima1 comigrates with Sad1-DsRed as the SPB oscillates along the NE. A composite image of time-lapse frames taken every 15 seconds for 5 minutes of one MKSP58 cell (see above) expressing GFP-Ima1and Sad1-DsRed. The asterisk indicates a second focus of GFP-Ima1 that oscillates along the NE. The time axes are indicated by the arrow and “t”.
We have chosen to further study the molecular linkages between the cytoskeleton and the NE in the fission yeast, Schizosaccharomyces pombe. Several aspects of S. pombe physiology are conserved with that of metazoans and are notably absent in the budding yeast, S. cerevisiae. First, the primary MTOC in S. pombe (termed the spindle pole body, or SPB), is tethered to the ONM during interphase (Figure 1B) (Ding et al., 1997). Second, S. pombe contain clear homologues of both SUN and KASH domain proteins. Tethering of the SPB to the ONM likely involves one or both S. pombe KASH domain proteins, named Kms1 and Kms2 (Miki et al., 2004; Niwa et al., 2000; Shimanuki et al., 1997), although the contribution of these proteins to the SPB-NE interface during interphase has not been examined in detail. Kms1 and Kms2 interact with the S. pombe SUN domain protein, Sad1(Miki et al., 2004), thus providing a means to couple the SPB to the nuclear interior. Sad1 is required for SPB duplication at the onset of mitosis (Hagan and Yanagida, 1995) and oscillates along the NE in a microtubule-dependent fashion, suggesting that it is coupled to the SPB (Tran et al., 2001). Importantly, although Sad1 colocalizes with SPB components at the level of the light microscope, Sad1 is an integral INM protein. Therefore, Sad1 defines a specific region of the NE to which the SPB is attached (Figure 1B). We call this discrete region of the NE the MTOC attachment site, or MAS. As the MAS spans both the INM and ONM, its components include inner MAS proteins (Imas) and outer MAS proteins (Omas). In addition to the SPB, a second type of interface between the NE and microtubules (MTs) exists in S. pombe, called the interphase MTOC (iMTOC (Sawin and Tran, 2006)). Although poorly characterized, iMTOCs play important roles in regulating the interphase MT architecture and also contain the MAS protein Sad1(Tran et al., 2001).
Here we describe the function of a previously uncharacterized integral membrane protein (Ima1) that resides at the inner face of the MAS and supports the buffering of MT forces at the NE. Surprisingly, Ima1 achieves this role by linking the MAS to heterochromatic regions within the nucleus. By examining cells lacking Ima1 we uncover a general role for the centromeric heterochromatin in buffering MT-dependent forces on the NE. This provides a framework for functional cooperation and communication between the cytoplasmic cytoskeleton and chromatin throughout the cell cycle.
Results
Ima1 is a Conserved Inner Nuclear Membrane Protein
A proteomic approach in mammalian cells first suggested that Ima1 is a NE membrane protein (NET5; (Schirmer et al., 2003)). A single gene homologous to Ima1 is present in all metazoa and the fission yeast S. pombe (systematic name SPCC737.03c), but is absent in the budding yeast S. cerevisiae. Across species, an N-terminal cysteine-rich region is the most highly conserved domain of Ima1 (Figure S1 available online).
Consistent with the expectation that Ima1 resides at the NE, a GFP fusion of Ima1 localizes specifically to the nuclear rim in S. pombe (Figure 1C). Using immunoelectron microscopy and antibodies directed against the GFP tag, we found that the majority of gold particles associated with the NE are found along the INM (90%, n=40; Figure S1), suggesting that Ima1 resides at the inner face of the NE. The presence of the GFP antigen within the nucleus combined with glycosylation analysis (Figure S2) suggests that S. pombe Ima1 adopts the topology indicated in Figure S2. In all species, the C-terminal hydrophilic domain contains a nuclear localization signal, which likely promotes trafficking of Ima1 to the INM (Lusk et al., 2007).
At the nuclear rim, GFP-Ima1 is enriched in distinct regions of the NE (Figure 1C). Using time-lapse imaging of live cells, we found that GFP-Ima1-enriched regions of the NE are dynamic and oscillate along the NE largely parallel to the long axis of the cell (Movie S1). On average, Ima1 foci undergo one full oscillation (returning to the same location at the NE) in 189 +/− 50 seconds (n=25), with the average oscillation being 1.7 +/− 0.7 µm in size. Such oscillations are reminiscent of the movement of the SPB as it is pushed by polarized MT bundles (Hagan et al., 1990; Tran et al., 2001), suggesting that GFP-Ima1 may enrich in the MAS. Consistent with this, such GFP-Ima1-rich regions frequently colocalize with the MAS protein, Sad1 (Figure 1C). To better illustrate the amplitude and path of GFP-Ima1 oscillations and investigate their association with the MAS, we created a single, composite image of GFP-Ima1 and Sad1-DsRed localization over five minutes by overlaying an entire time-lapse series (Figure 1D). Each individual oscillation is revealed in the side plots that display signal over time along the x- and y-axes. Here, it is evident that the brightest focus of GFP-Ima1 oscillates along the NE in conjunction with Sad1-DsRed. Oscillation of Ima1 is MT-dependent, as GFP-Ima1 associated with the MAS remains static and colocalized with Sad1-DsRed if cells are treated with the MT- destabilizing agent carbandazim (MBC; Figure S3). Interestingly, GFP-Ima1 also accumulates in a second, discrete region of the NE that oscillates similarly to the SPB (asterisk, Figure 1D), which may represent its accumulation in the iMTOC. Consistent with this, in some cells GFP-Ima1 colocalizes with Sad1-DsRed (a known iMTOC component) at the NE in both the SPB and the iMTOC (Figure S3).
Cells Lacking Ima1 Grow Poorly and have NE and MAS Defects
To investigate the function of Ima1, we generated S. pombe cells lacking Ima1 by gene replacement. ima1Δ cells grow poorly on rich media compared to the WT control at all temperatures tested (Figure S4).
We first investigated whether the absence of Ima1 affects the NE and/or the MAS. To facilitate this, strains were generated expressing a GFP fusion of the transmembrane nucleoporin Cut11 (West et al., 1998) and Sad1-DsRed. WT cells undergo deformations of the NE at the MAS as the plus ends of MT bundles reach the poles of the cell and push back on the nucleus (Figure 2A; Tran et al., 2001). Such deformations are transient and mildly affect the overall spherical shape of the nucleus. The dynamics of the NE in response to MT forces in ima1Δ cells are markedly different. First, deformations of the NE are more pronounced, often leading to large extensions of the NE (Figure 2A). To further visualize these deformations, we imaged ima1Δ cells expressing GFP-tubulin and an mCherry fusion of Heh1 as a NE marker. Heh1 (systematic name SPAC18G6.10) is an S. pombe paralogue of the S. cerevisiae Heh1 and Heh2 proteins, which localize specifically to the INM (King et al., 2006). As shown in Figure 2B, a large extension of the NE takes place at the MT-NE interface as the MT bundles grows to the left and pushes on the right end of the cell. Thus, it appears that ima1Δ cells are poorly equipped to buffer MT forces exerted on the NE. We quantitated the failure of the NE to maintain its shape under MT force by creating overlays of SPB position and determining the range of SPB oscillations in size-matched cells (Figure 2C). In WT cells the average amplitude of maximum SPB oscillation was 1.9 µm, compared to 3.2 µm for ima1Δ cells (Figure 2C). The ability of the SPB to undergo larger oscillations in ima1Δ cells reflects the greater deformation of the NE and its inability to counteract MT-driven forces. In addition, while many NE extensions appear to be led by the MAS and SPB (Figure 2A, arrows), others appear to extend from regions of the NE that are not associated with visible Sad1 (Figure 2A, asterisks), raising the possibility that MT forces are not coupled efficiently or specifically to the MAS. Importantly, these defects are not caused by aberrant formation of intranuclear MT bundles during interphase, nor do ima1Δ cells possess apparent changes in MT architecture (Figure S5).
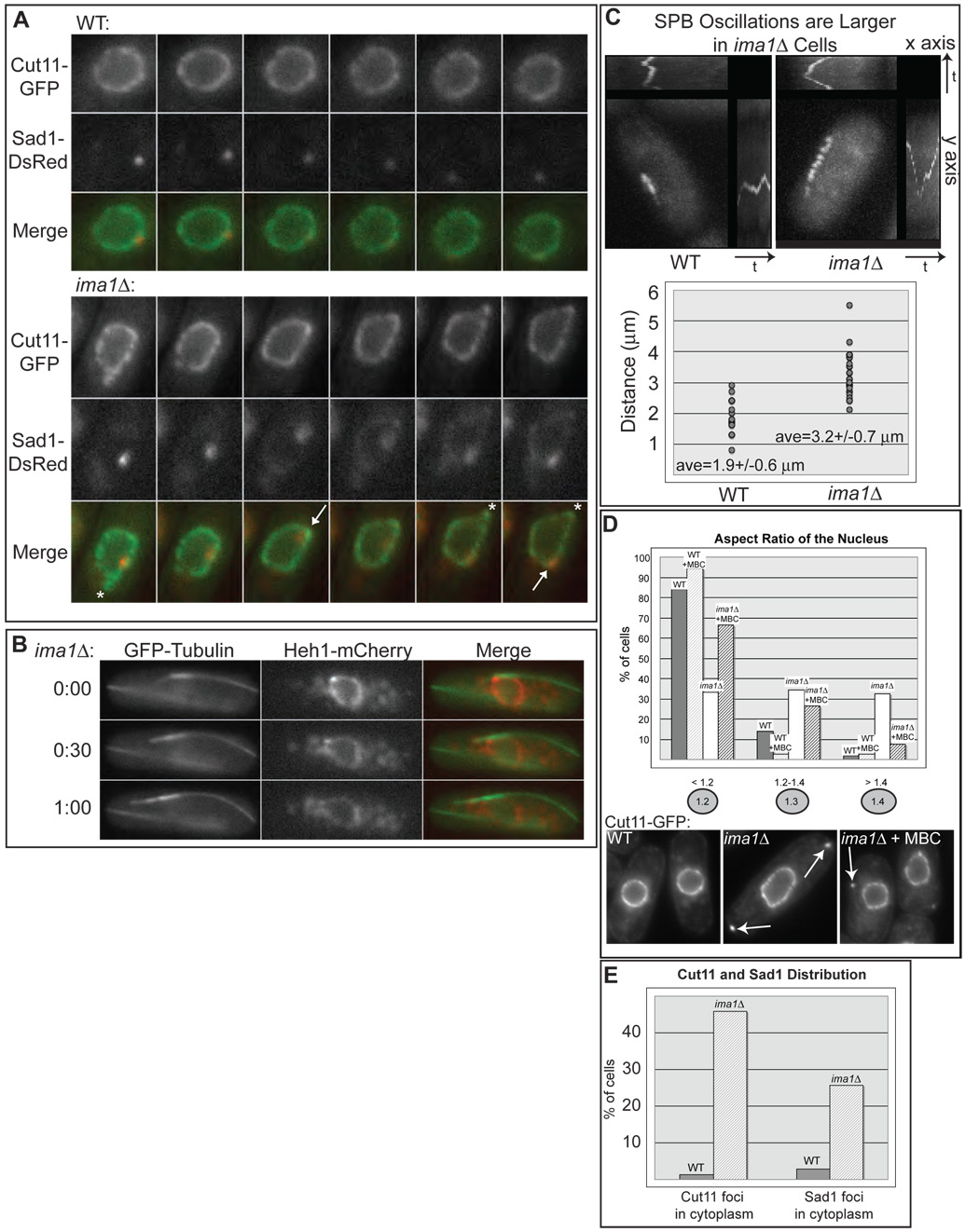
Figure 2. ima1Δ cells have NE and MAS defects.
A. Large deformations of the NE in the absence of Ima1. Fluorescent micrographs of one WT (top panels) and one ima1Δ cell (bottom panels) expressing Cut11-GFP and Sad1-DsRed (MKSP50 and MKSP56, respectively) taken in consecutive 60 second intervals are shown along with the merged images. Deformations led by Sad1-DsRed are labeled with arrows; deformations without visible Sad1-DsRed are labeled with asterisks. B. Large deformations of the NE in ima1Δ cells are driven by MTs. Fluorescent micrographs of one ima1Δ cell expressing GFP-tubulin and Heh1-mCherry (MKSP173) imaged every 30 seconds for one minute (time indicated on the left) and shown with the merged images. C. SPB oscillations are larger in ima1Δ cells. Composite images of time-lapse frames taken every 15 seconds for 5 minutes of one WT cell (MKSP81) and one ima1Δ cell (MKSP152) expressing GFPKms2 (upper panel). Plot of the maximum SPB oscillation amplitude for size-matched WT and ima1Δ cells (n=25, lower panel). The average value (ave) is given with its standard deviation. D. MT forces lead to defects in nuclear shape and cytoplasmic Cut11-GFP foci appear (arrows) in ima1Δ cells. Quantitation of the spherical nature of WT (MKSP10) versus ima1Δ (MKSP22) nuclei. As indicated, WT and ima1Δ cells were also treated for 15 minutes with carbendazim (MBC) prior to imaging. Measurements are presented from at least 100 non-mitotic cells. E. Percentage of WT (MKSP50) and ima1Δ (MKSP56) cells that display Cut11-GFP and Sad1-DsRed cytoplasmic foci. n = >100 interphase cells.
The defects in NE structure observed in ima1Δ cells are reflected by loss of spherical nuclear shape, as well as increases in NEs that appear “ruffled” (Figure 2D). To quantitate this effect, the nuclear aspect ratio of interphase cells was measured by determining the ratio of long to short axes of the nucleus. This ratio is 1 for a sphere, and increases as the nucleus takes on a more ovoid shape. As shown in Figure 2D, over 80% of interphase WT cells have aspect ratios of less than 1.2, while less than 3% have aspect ratios of greater than 1.4. By contrast, only a third of interphase ima1Δ cells had aspect ratios of less than 1.2, while a full third of cells had aspect ratios of greater than 1.4. We anticipated that the changes in NE shape seen in ima1Δ cells require active MT forces. Nearly 95% of WT cells have aspect ratios of less than 1.2 when MT-forces are eliminated by addition of MBC (Figure 2D). Addition of MBC to ima1Δ cells leads to a substantial recovery of the nuclear shape defects apparent in untreated cells, suggesting that the NE distention seen in the absence of Ima1 involves MT-driven forces. Interestingly, this recovery is not complete, suggesting that some irreversible defects in NE structure also arise in ima1Δ cells. To investigate the specificity of this effect, we investigated whether similar defects are seen in cells lacking the INM protein Heh1. Defects in the NE that occur in heh1Δ cells are distinct from those observed in ima1Δ cells and are not manifested in changes in the spherical nature of the nuclei (Figure S6). Further heh1Δ cells do not display any unusual NE deformations (data not shown).
In conjunction with changes in NE dynamics and shape, ima1Δ cells also accumulate cytoplasmic foci containing Cut11-GFP (over 45% of ima1Δ cells, Figure 2D–E) and/or Sad1-DsRed (25% of cells, Figure 2E and Figure S6), often specifically at the poles of the cell (Figure 2D, arrows). The separation of these foci from the NE is not reversible, as they remain in the cytoplasm even after treatment with MBC. Like irrevocable effects on NE shape, we expect that these foci arise from failures of the NE structure under MT force drastic enough to cause fragments of NE to become separated from the nucleus. Since NE-MT interfaces are typically enriched in Sad1, it is not unexpected that we would observe foci containing both Cut11-GFP and Sad1-DsRed (Figure S6).
Disorganization of the MAS in ima1Δ cells
We sometimes observe NE deformations that are not associated with visible Sad1 (Figure 2A, asterisks), but can be reversed by MBC. We hypothesized that the ability of MTs to produce NE extensions would require KASH domain proteins even if Sad1-DsRed is at low levels or is absent from these interfaces. Although Kms1 is dispensable for vegetative growth (Shimanuki et al., 1997), we found that cells lacking Kms2 are almost completely inviable (our unpublished data). GFP-Kms2 localizes specifically to one focus at the MAS, where it colocalizes and comigrates with Sad1 (Figure 3A). We do not detect GFP-Kms2 at iMTOCs.
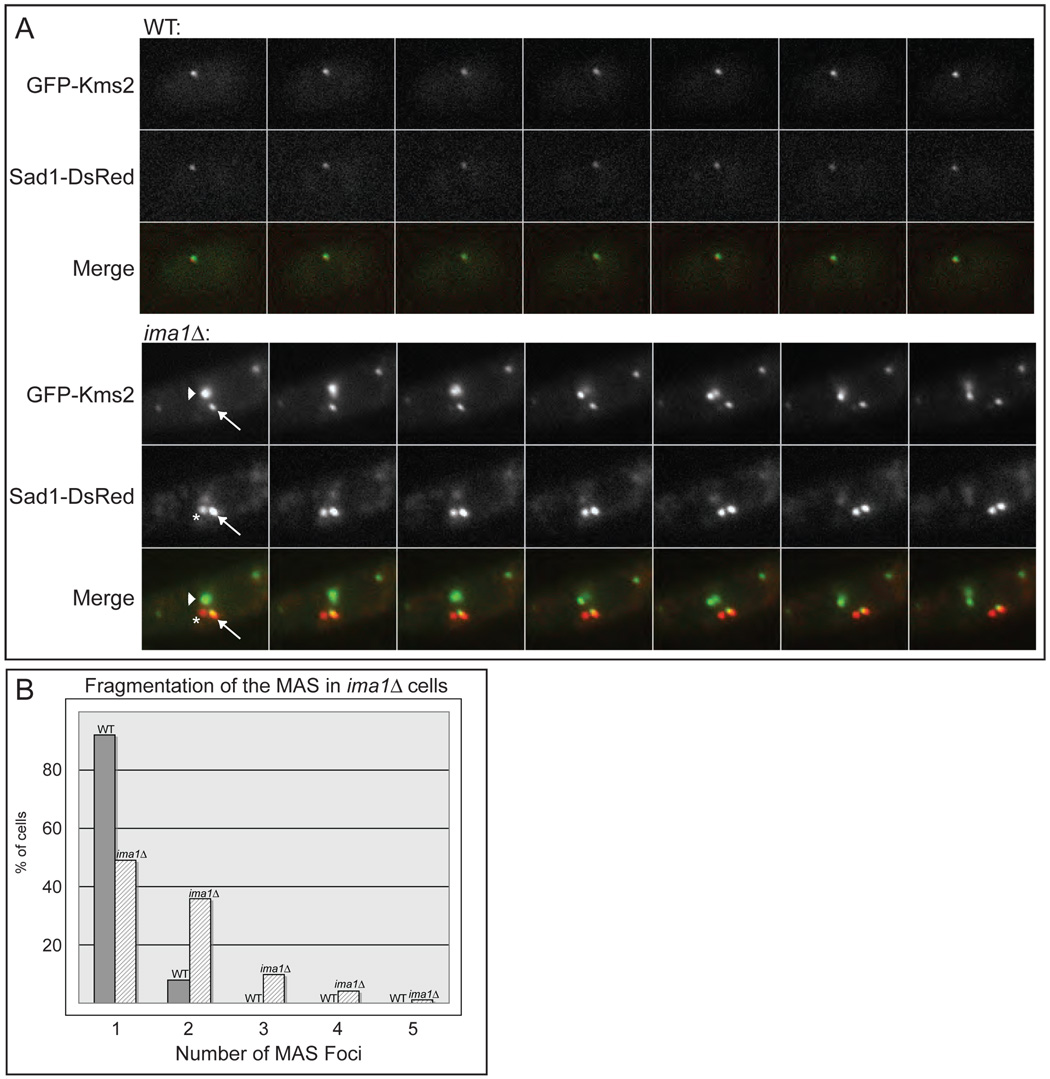
Figure 3. Sad1 and Kms2 become disorganized in the absence of Ima1.
A. Top panel. Fluorescence micrographs demonstrate that the KASH domain protein GFP-Kms2 and Sad1-DsRed colocalize at the MAS in WT cells (MKSP81). Time-lapse images taken of one cell every 30 seconds for 3 minutes are shown. Bottom panel. GFP-Kms2 and Sad1-DsRed fragment into multiple foci at the NE and no longer colocalize stoichiometrically in ima1Δ cells. Fluorescence micrographs of one cell of strain MKSP152 taken every 30 seconds for 3 minutes. Arrow indicates a focus with both GFP-Kms2 and Sad1-DsRed; arrowhead indicates a focus with only GFP-Kms2; asterisk indicates a focus with only Sad1-DsRed (see text). B. Quantitation of the number of MAS foci at the NE in WT and ima1Δ cells. Strains (MKSP81 and MKSP152) were analyzed for the total number of foci containing GFP-Kms2 and/or Sad1-DsRed in non-mitotic cells. n = >100 cells.
The distribution of GFP-Kms2 is profoundly altered in ima1Δ cells. Most obviously, Kms2 is found in several foci at the NE (Figure 3A). Further, the relative distribution of GFP-Kms2 and Sad1-DsRed varies greatly within these foci. For example, there are two Sad1-DsRed foci of approximately equal intensity that move together along the NE (Figure 3A, arrow and asterisk). Interestingly, only one of these foci (Figure 3A, arrow) colocalizes with GFP-Kms2. Conversely, the focus of greatest GFP-Kms2 intensity contains a relatively small fraction of Sad1-DsRed (Figure 3A, arrowhead). These observations suggest that the MAS is fragmented in ima1Δ cells, altering the normal stoichiometry of inner MAS proteins and outer MAS proteins. Fragmentation of the MAS in ima1Δ cells is also reflected in the number of MAS foci (the combined number of Sad1-DsRed and GFP-Kms2 foci) that accumulate at the NE (Figure 3B). While over 90% of WT cells have a single MAS focus, over 50% of ima1Δ cells have multiple MAS foci. Further, 15% of ima1Δ cells have three or more MAS foci, a phenotype never observed in WT cells. Although there is an increase in the total fluorescence intensity of GFP-Kms2 and Sad1-DsRed in some ima1Δ cells, we did not detect a substantial increase in Sad1 levels in ima1Δ cells (Figure S7). Further, large NE deformations are often observed in cells that display normal levels of Sad1-DsRed and GFP-Kms2. Thus, it is unlikely that upregulation of Sad1 and Kms2 causes the defects we observe in ima1Δ cells.
Changes in MAS-associated heterochromatin in ima1Δ cells
We next investigated the mechanism by which Ima1 contributes to the organization and/or integrity of the MAS. In order to effectively buffer MT forces, we envision that the SUN-KASH complexes at the NE require additional associated factors to stabilize the MAS. Interestingly, centromeres (and proteins that bind to centromeric DNA) are clustered adjacent to the site of SPB attachment (Funabiki et al., 1993), suggesting a link between chromatin and the cytoskeleton. While little information about the potential importance of centromere localization to the MAS is available, it is clear that telomeres, which are clustered in place of centromeres at the MAS during meiotic prophase, are critical to MAS function during this period. Release of telomeres from the MAS (by deletion of their binding proteins) during meiosis leads to large NE extensions (Chikashige et al., 2006) and accumulation of Sad1 foci in the cytoplasm (Tomita and Cooper, 2007). Because ima1Δ cells display similar phenotypes, we considered whether deletion of Ima1 might affect the interaction of centromeres with the MAS. This model predicts that Ima1 may itself interact with centromeres.
We investigated whether Ima1 interacts with centromeric DNA by chromatin immunoprecipitation (ChIP). When we immunoprecipitated GFP-Ima1, we detected a 5-fold enrichment at the central region of the centromere (cnt1), with less enrichment over the outer centromeric regions, including the innermost (imr) and outer repeats (otr; dg and dh, Figure 4A). This distribution is different from that of heterochromatin binding proteins such as Swi6, the S. pombe homologue of HP1 (Lorentz et al., 1994), which enriches primarily in the repeat regions (Figure S8A). Thus, this finding supports the hypothesis that Ima1 serves to couple centromeric heterochromatin to the NE by interacting with the centromeric core.
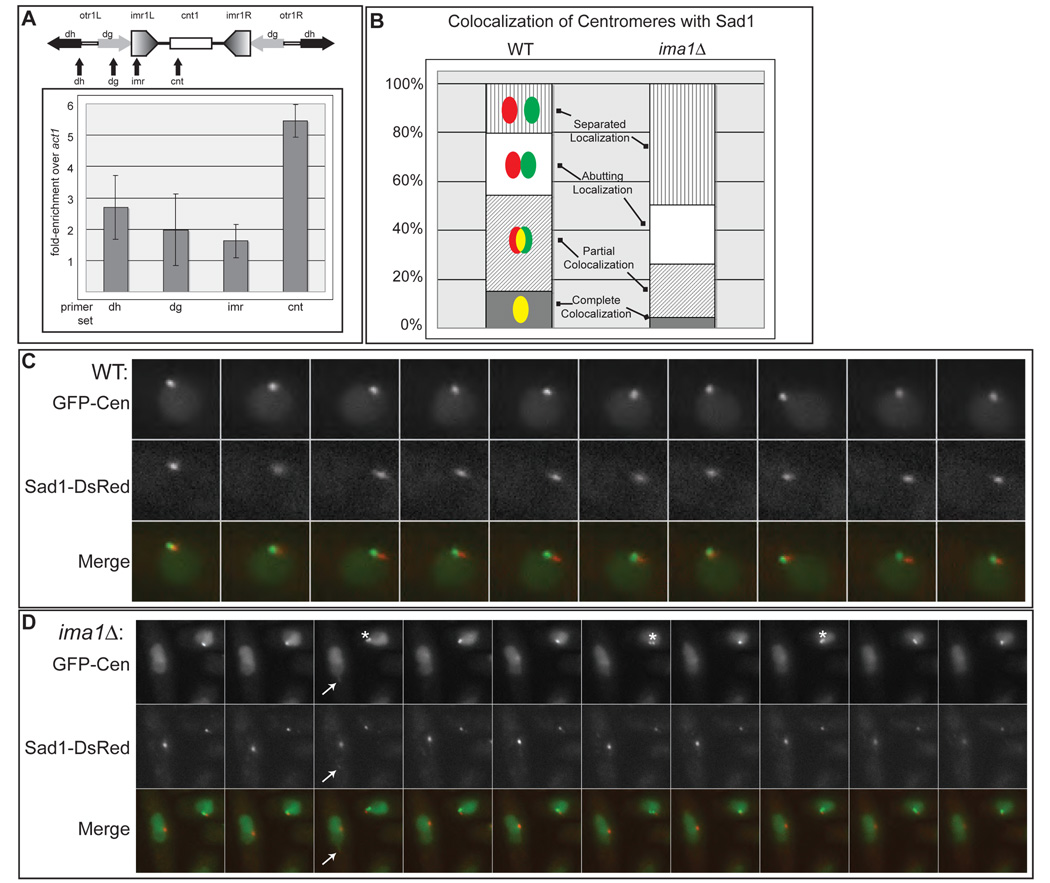
Figure 4. Centromeres becomes uncoupled from the MAS in ima1Δ cells.
A. GFPIma1 associates with centromeres. Schematic representation of the Chromosome I centromere. otr=outer repeat, imr=innermost repeat, cnt1=center region, dg=dg repeats, dh=dh repeats. Arrows indicate positions of primer sets used in real-time PCR analysis. Graph of fold-enrichment of GFP-Ima1 by chromatin immunoprecipitation at the given region over the control actin gene location, act1. The average of three independent immunoprecipitations are presented with their standard deviations. B. Centromeres are separated from Sad1-DsRed in ima1Δ cells. For each cell, the degree of colocalization of LacI-GFP associated with Lac operators integrated into centromere II (cen2:lacOp, green, labeled GFP-Cen) with Sad1-DsRed (red) was categorized as illustrated in the cartoons with yellow representing colocalization. The percentage of cells displaying each localization pattern was plotted. The graphs represent the average of three separate experiments consisting of greater than 100 cells each. C. In WT cells, GFP-Cen remains closely associated with the MAS during SPB oscillation. Time-lapse images of GFP-LacI and Sad1-DsRed in WT cells with integrated cen2:lacOp (MKSP71) taken every 30 seconds for 5 minutes. D. In ima1Δ cells the centromeres of sister chromatids often separate (asterisk) and no longer colocalize with Sad1-DsRed. Time-lapse images of GFP-LacI and Sad1-DsRed in an ima1Δ cell with integrated cen2:lacOp (MKSP158) were taken every 30 seconds for 5 minutes. The arrow indicates the distended nucleus (green channel) led by a weak focus of Sad1-DsRed (red channel), which is not associated with the centromeres.
We next examined whether clustering of the centromeres at the MAS was disrupted in ima1Δ cells expressing LacI-GFP in which Lac operators were integrated into the centromeric region of chromosome II (GFP-Cen; Yamamoto and Hiraoka, 2003). In WT cells, GFP-Cen is usually found colocalized with or adjacent to the MAS (Sad1-DsRed; Figure 4B and C). In ima1Δ cells, centromeres continue to oscillate with the SPB (Figure 4D), however, we observe several defects in the association of centromeres with the MAS. The centromeres in ima1Δcells often become physically separated from the Sad1-DsRed focus (the MAS, Figure 4D); GFP-Cen and Sad1-DsRed are found colocalized (completely or partially) in only about 25% of cells and are found completely separated in about half of all cells (Figure 4B and Figure S9). Further, we often observe that the two centromeres of the sister chromatids in ima1Δ cells break apart into separated foci (Figure 4D, asterisk) during oscillatory movement. Similar results were obtained when we imaged the total centromeric heterochromatin using a GFP-fusion of Swi6 (Figure S8). Together, these findings suggest that Ima1 supports the coupling of centromeric heterochromatin to the MAS directly by simultaneously binding centromeres and the NE. We suggest that the loss of tight coupling between centromeres and the MAS leads to the increased amplitude of SPB oscillation associated with NE deformations. Further, these results predict that force delivered by MTs to fragmented MAS components lacking associated centromeric heterochromatin may lead to large deformations of the NE. Indeed, in the absence of Ima1 we observe large deformations of the nucleus (visualized by nuclear LacI-GFP) led by an MAS fragment of Sad1-DsRed that is uncoupled from the centromeres (Figure 4D, arrow). Such deformations are MT-dependent, as they are never observed in cells treated with MBC (data not shown). Taken together, these results are consistent with the possibility that heterochromatin supports the MAS – a role potentially carried out by centromeres during vegetative, interphase growth, and by telomeres during meiotic prophase.
Release of centromeres from the MAS causes NE and MAS defects
A model in which centromeric heterochromatin supports the MAS predicts that release of centromeres from the MAS would be expected to cause defects that mimic those associated with loss of Ima1. To test this, we utilized a strain harboring a temperature-sensitive mutation in Nuf2, a component of the Ndc80 complex that is essential for the colocalization of centromeres with Sad1 during interphase (Appelgren et al., 2003). To prevent mitotic defects associated with loss of kinetochore function, all results using this allele, nuf2-1 (Nabetani et al., 2001), were obtained at a permissive temperature of 25°C.
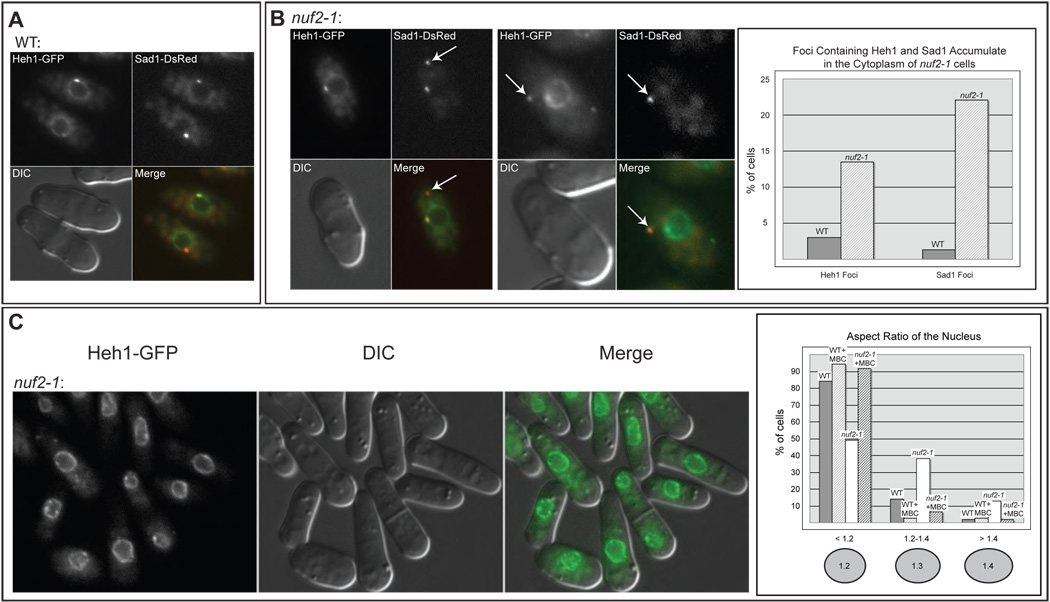
We examined the state of the NE and MAS in nuf2-1 cells expressing Sad1-DsRed and Heh1-GFP. In WT cells, Heh1-GFP localizes to the NE, where it sometimes enriches in the MAS (Figure 5A). In nuf2-1 cells, we observed NE and MAS defects similar to those observed in ima1Δ cells. First, foci of Heh1-GFP and Sad1-DsRed accumulated in the cytoplasm of nuf2-1 cells (Figure 5B). The proportion of nuf2-1 cells with cytoplasmic Sad1-foci (22%, Figure 5B) is similar to that seen in ima1Δ cells (25%, Figure 2E). This suggests that compromising the interaction of centromeres with the MAS is sufficient to cause MAS and SPB dysfunction. Further, like ima1Δ cells, nuf2-1 cells have defects in nuclear shape (Figure 5C). While the defect in nuclear shape is somewhat less pronounced than that seen in ima1Δ cells, nuf2-1 cells clearly deviate from WT cells, having aspect ratios of less than 1.2 in only 50% of cells, and aspect ratios of more than 1.4 in over 10% of cells. Further, the NEs of nuf2-1 cells appear locally perturbed, with ruffles and invaginations (Figure 5C). Like ima1Δ cells, the defects in nuclear shape in nuf2-1 cells can be rescued by addition of MBC (Figure 5C). Thus, weakening the interaction of centromeres with the MAS is sufficient to cause defects in NE and MAS function.
Figure 5. Release of centromeric heterochromatin from the MAS causes NE and MAS defects.
A. Heh1-GFP is an INM protein that is enriched at the MAS. Micrographs of MKSP47 cells expressing Heh1-GFP and Sad1-DsRed shown with the DIC and merged images. B. A temperature-sensitive mutation in Nuf2 leads to NE and MAS defects. Fluorescence micrographs of cells expressing Heh1-GFP and Sad1-DsRed in the nuf2-1 strain (MKSP109) at 25°C. Sad1-DsRed foci and Heh1-GFP foci are seen in the cytoplasm of nuf2-1 cells (arrows). Quantitation of Heh1 and Sad1 foci in WT versus nuf2-1 cells was determined as in Figure 2. C. The nuf2-1 mutation leads to defects in NE shape. Fluorescence micrographs of strain MKSP109 at 25°C. Quantitation of nuclear shape was carried out as in Figure 2.
The heterochromatic state of centromeres affects MAS function
In S. pombe, there are three major regions of heterochromatin: centromeres, telomeres, and the mat locus (Grewal and Jia, 2007). In these heterochromatic regions, histone H3 is methylated by the methyltransferase Clr4 at lysine 9 (H3K9me); Swi6 fails to localize to centromeres and telomeres in clr4Δ cells (Ekwall et al., 1996). Importantly, while deletion of Clr4 causes defects in centromeric silencing and chromosome segregation, it is dispensable for clustering of centromeres adjacent to the SPB (Ekwall et al., 1996). Thus, using a strain lacking Clr4, we can test whether there is a specific role for heterochromatin (rather than just chromatin in general) in supporting MAS function.
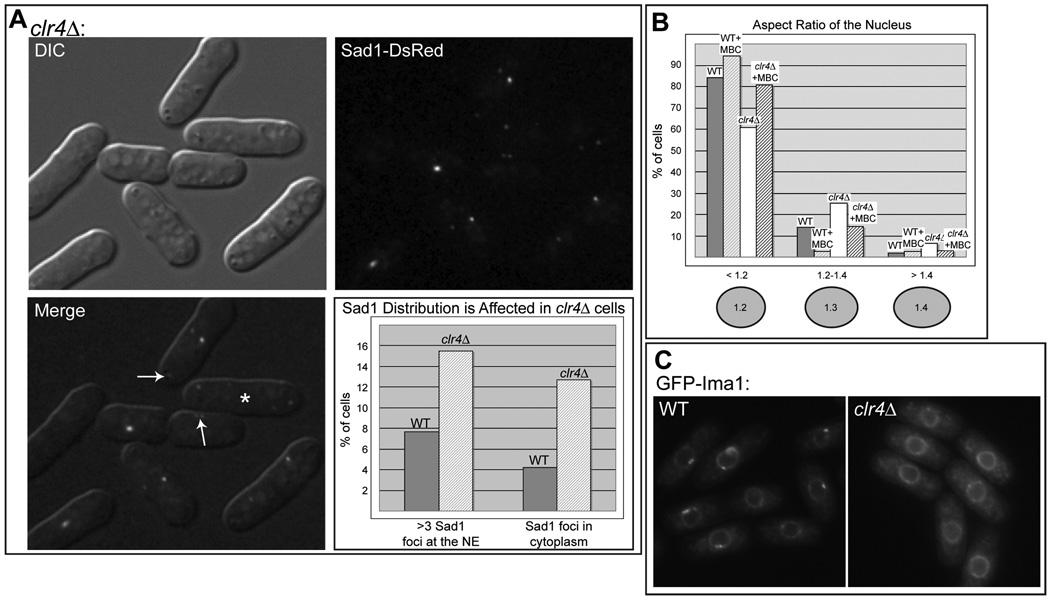
As shown in Figure 6A, Sad1-DsRed localization is perturbed in several ways in clr4Δ cells. First, as seen in ima1Δ cells and nuf2-1 cells, Sad1-DsRed can be observed in cytoplasmic foci (Figure 6A, upper arrow), albeit with less frequency (12% of clr4Δ cells (Figure 6A) compared to 25% of ima1Δ cells (Figure 2E)). Further, the nuclei of clr4Δ cells often harbor greater than three Sad1 foci (15% of non-mitotic cells, Figure 6A). This likely underestimates the percentage of cells in which the MAS is disrupted, as it is difficult to differentiate between the normal localization of Sad1-DsRed to foci associated with the SPB and iMTOC(s) and fragmentation of the MAS. For example, in many cases two Sad1-DsRed foci arise because the MAS is perturbed (see Figure 6A, lower arrow). Finally, clr4Δ cells also have detectable defects in nuclear shape, which can be rescued by addition of MBC (Figure 6B).
Figure 6. Disruption of histone H3-K9 methylation leads to NE and MAS defects and mislocalization of GFP-Ima1.
A. Sad1 localizes to multiple foci at the NE and to cytoplasmic foci in clr4Δ cells (MKSP123). Fluorescence micrograph showing Sad1-DsRed in the cytoplasm (top arrow). Sad1 is also found fragmented at the NE (lower arrow), including in cells that have recently completed mitosis, which have two Sad1-DsRed foci per nucleus (asterisk). Quantitation of cytoplasmic Sad1 foci was carried out as in Figure 2. B. clr4Δ cells have a mild defect in nuclear shape. Quantitation of nuclear shape was carried out as in Figure 5, using Heh1-GFP as the NE marker (MKSP119). C. The accumulation of Ima1 to the MAS is reduced in clr4Δ cells. Fluorescence micrographs of GFP-Ima1 in WT (MKSP14) and clr4Δ (MKSP120) cells.
Because Ima1 binds to centromeres and colocalizes with centromeres at the MAS, we also investigated whether the localization of Ima1 is affected by loss of H3K9me in clr4Δ cells. As shown in Figure 6C, deletion of Clr4 reduces the enrichment of GFP-Ima1 at the MAS; GFP-Ima1 instead appears evenly distributed throughout the NE. The failure of clr4Δ cells to recruit normal quantities of Ima1 to the MAS likely plays a role in MAS dysfunction in these cells.
Discussion
Here we characterize a network of integral membrane proteins and heterochromatin that together establish a macromolecular linkage between the nuclear interior and the cytoplasmic cytoskeleton. Ima1, an integral inner nuclear membrane protein, couples centromeres to the NE, thus contributing to their association with the MAS. Without Ima1, or in the absence of functional Ndc80 complex, the release of centromeric heterochromatin from the MAS leads to defects in NE and SPB structure. Thus, chromatin supports the interface between MTs and the NE throughout the cell cycle.
The contribution of chromatin to MAS function
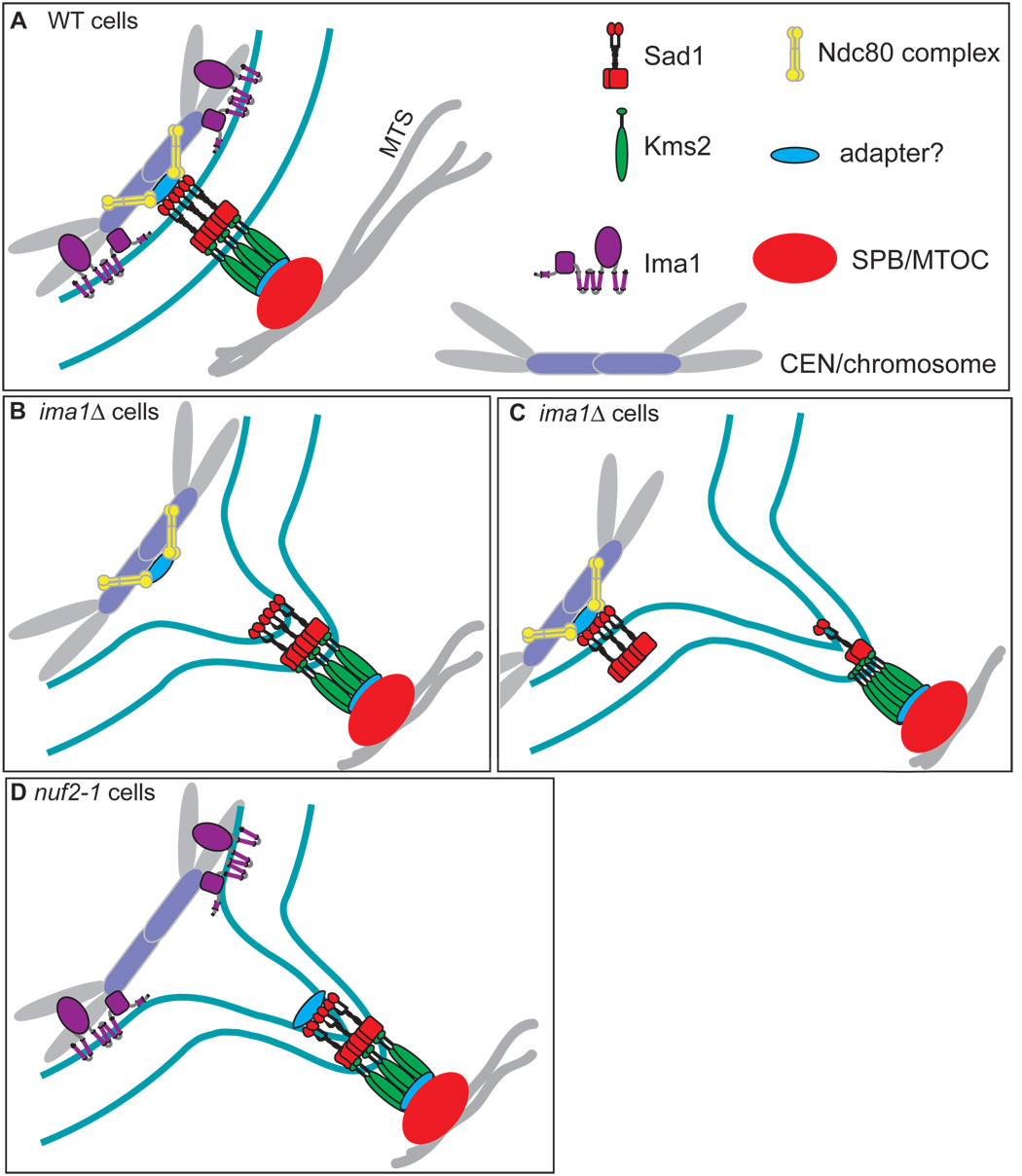
How might heterochromatin stabilize the MAS? When the positive end of a MT bundle pushes on the end of the cell, force is delivered to the SPB. This force would then be transmitted to the MAS, leading to deformation of the NE. At the NE, the integral membrane proteins of the MAS would have to weather these forces. We imagine that these integral MAS proteins serve as a “bolt” to attach the SPB to the NE. To be stable, a bolt must have a “nut” at the end, to counter the forces it bears, as well as to distribute that force more broadly. We suggest that heterochromatin serves as the nut, providing an anchor of tremendous stability to the MAS. Our data suggest that parallel, synergistic contributions from the Ndc80 complex (via an unidentified adapter(s) to Sad1) and Ima1 support coupling of the centromeres to the MAS (Figure 7A). In the absence of Ima1, the ability of the nucleus to withstand MT-mediated forces is compromised, leading to large deformations of the NE at two types of interfaces. First, when the Ndc80 complex (without the support of Ima1) fails to couple centromeric heterochromatin to an intact Sad1-Kms2 complex (Figure 7B), unusually large NE deformations and SPB oscillations occur (see Figure 2A, arrows, and Figure 2C). Second, in many cells we observe further disruption of Sad1-Kms2 complexes, leading to fragmentation of the MAS and large NE protrusions that are no longer coupled to the inner MAS components and heterochromatin (Figure 7C, see Figure 4D, arrows). Finally, Ima1 is not sufficient to stably couple the heterochromatin to the MAS in the absence of Nuf2 function, leading to large NE deformations (Figure 7D).
Figure 7. Model of the MAS in WT, ima1Δ and nuf2-1 cells.
A. In WT cells, the Sad1 (red) / Kms2 (green) bridging complex links the MTOC (SPB, orange) to the centromeric heterochromatin (lavender/gray). Ima1 (purple) resides at the inner nuclear membrane and contributes to the coupling of heterochromatin to the inner MAS. The Ndc80 complex (yellow) also participates in linking the centromeres to the inner MAS, through an unknown adapter (blue) and Sad1. B. Without Ima1, the Ndc80 complex (yellow) is not sufficient to tightly couple centromeres to the MAS, leading to large nuclear envelope (NE) deformations. C. In the absence of Ima1, fragmentation of the MAS occurs and Sad1-Kms2 lumenal interactions are disrupted, decoupling forces delivered on the NE by the SPB from the centromeres and other MAS components. D. In nuf2-1 cells, Ima1 is not sufficient to couple centromeres to the MAS, resulting in large NE deformations.
Recognition of Heterochromatic Regions by Ima1
Ima1 binds specifically to the centromeric core region, but is largely absent from the flanking repeat regions rich in heterochromatic marks (H3K9me) and heterochromatin binding proteins such as Swi6. However, mislocalization of Ima1 by preventing H3K9me (clr4Δ cells) suggests that Ima1-centromere interactions are nonetheless indirectly dependent on heterochromatin. We speculate that localization of Ima1 specifically to the centromere core is either dependent on a component mislocalized in the absence of Clr4 or requires the boundary established by flanking heterochromatin. If the latter was the case, we might predict that Ima1 can associate with other regions of the chromatin that are heterochromatic. Consistent with this possibility, we found that Ima1 is enriched at telomere ends (Figure S10). Further, as at centromeres, enrichment decreases towards the subtelomeric regions (in which Swi6 is prevalent). This suggests that Ima1 plays a general role in tethering heterochromatic regions to the NE, although it does not recognize the heterochromatin per se.
This finding raises the possibility that the same proteins might contribute to both the centromere-NE and telomere-NE interfaces during interphase. Binding of centromeres to the NE by Ima1 supports the integrity of the MAS when under MT force. Might there be a similar role for Ima1 at the telomere-NE interface? In support of this notion, we find that telomeres often undergo periods of highly direct motion at the NE that are MT-dependent (Figure S10). Although little is known about the membrane proteins that tether telomeres to the NE in yeasts (Akhtar and Gasser, 2007), the S. cerevisiae Sad1 homologue, Mps3, was shown to be required for NE-association of telomeres during interphase (Bupp et al., 2007). Colocalization of Sad1 with Ima1 at additional foci at the NE beyond the SPB (Figure 1D, asterisk and Figure S3B) raises the possibility that these proteins might together tether telomeres to the NE during interphase. Further, as these foci are also believed to represent iMTOCs, they may potentially be associated with MTs. Thus, while Ima1 and Sad1 are primarily localized to the MAS and promote coupling of the centromeres to the SPB, these same proteins may serve a similar purpose in a telomeric-iMTOC interface. Further research will be required to validate this idea.
Supporting the MAS in interphase and meiotic prophase
During meiotic prophase, vast changes in chromosome organization take place, highlighted by the clustering of telomeres into a “bouquet” at the MAS. This reorganization is coordinated in part by induction of the meiotic-specific proteins Bqt1 and Bqt2, which together bridge the telomeres (via the telomeric-binding protein Rap1) to Sad1 at the MAS (Chikashige et al., 2006; Tang et al., 2006). Soon thereafter, release of centromeres by dissociation of the Ndc80 complex takes place (Asakawa et al., 2005). Thus, a mechanism exists to ensure that the MAS is continually supported by heterochromatin even during this period of massive genomic rearrangement. In the future, it will be interesting to determine whether Ima1 also functions to support the association of telomeres (via Bqt1/Bqt2) with the MAS during meiosis. Such a notion is consistent with our finding that Ima1 can bind telomere ends (Figure S10), likely promoting their association with the NE. Importantly, disrupting association of centromeres with the MAS (by deletion of Ima1 or mutation of Nuf2) during interphase or abrogating telomere clustering at the MAS during meiotic prophase (by deletion of Bqt1 or Bqt2) causes similar defects, including large NE extensions (Chikashige et al., 2006), SPB dysfunction and accumulation of Sad1 foci in the cytoplasm (Tomita and Cooper, 2007). Thus, association of heterochromatin at the MAS appears to be required not just during meiosis, but rather throughout the S. pombe life cycle.
Specificity for heterochromatin
We observe that abrogation of heterochromatin formation by deletion of Clr4 causes milder defects in NE and MAS function than either Ima1 deletion or mutation of Nuf2. In part, this is likely because heterochromatic marks are inherited, and therefore not completely disrupted in clr4Δ cells without extensive culturing (Hall et al., 2002). In addition, the attenuated defects we observe in clr4Δ cells may reflect the fact that, unlike in ima1Δ cells and nuf2-1 cells, centromeres remain coupled to the MAS (although their heterochromatic state is comprised; Ekwall et al., 1996). Determining what aspects of heterochromatin are necessary to support MAS function will be an important question to address in the future, although loss of Ima1 from the MAS is likely one factor (Figure 6C).
Defects in NE shape and formation of NE foci
In addition to NE protrusions, we observe more general defects in nuclear shape concomitant with the decoupling of heterochromatin from the MAS. Although treatment with the MT-depolymerizing drug MBC can largely rescue nuclear shape in ima1Δ cells (Figure 2D), some irreversible defects persist. This is consistent with the finding that distortions of NE structure lasting longer than ten seconds lead to irreversible changes in chromatin organization and cement the altered NE conformation in mammalian cells (Pajerowski et al., 2007). Taken to the extreme, we suspect that large NE extensions can lead to disruption of NE structure sufficient to cause fragments of NE/MAS to become separated from the nucleus, thus creating cytoplasmic foci. Although we have been unable to visualize this process directly, the frequent deposition of NE foci at the poles of the cell (Figure 2D and Figure S6C) is consistent with the possibility that the NE fragments travel with the MT bundle to the end of the cell, where they are left following MT depolymerization.
Roles for chromatin-MT interfaces across eukaryotes
The general use of SUN-KASH domain complexes to link the nucleus to the cytoskeleton is conserved in all metazoa. Although chromosome organization varies widely across these species, it has long been recognized that the nuclear periphery is rich in heterochromatin. In mammals, we expect that Ima1 acts with a subset of mammalian SUN-KASH domain proteins to link the cytoplasmic cytoskeleton to heterochromatin (Haque et al., 2006; Zhang et al., 2005). However, in metazoa, Ima1 need not be coupled specifically to centromeres and/or telomeres. Rather, Ima1 might associate with additional heterochromatic regions that are widely prevalent throughout metazoan genomes. In addition to a role for heterochromatin, there is ample evidence that SUN domain proteins also help to buffer cytoskeletal forces by coupling them to the nuclear lamina, which would provide further support (Tzur et al., 2006).
The connectivity of chromatin and the cytoskeleton described here poises cytoplasmic MTs to potentially affect chromatin organization in interphase cells. Consistent with this possibility, Franco et al. previously described that deletion of Mto1, which is required for normal cytoplasmic MT organization in S. pombe, leads to a defect in centromere clustering (Franco et al., 2007). As a result, unclustered kinetochores must be retrieved during mitosis, leading to an increased probability of chromosome missegregation. Thus, while heterochromatin appears to buffer the forces exerted by the cytoskeleton on the nucleus, such MT-mediated forces simultaneously ensure the maintenance of centromeres at the MAS. Whether similar chromatin-cytoskeletal connections contribute to the maintenance of the substantial heterochromatin at the nuclear periphery in metazoa remains an open question. Further, cycles of tension and relaxation exerted on the chromatin by the cytoskeleton (either directly or through mass movements of the nucleus) could potentially facilitate undirected chromatin motions that might influence transcription, replication and/or DNA repair. The functions of Ima1, Sad1 and Kms2 described here establishes a novel means of communication between the cytoplasm and nucleus that is independent of the nuclear pore complex, and opens up new paradigms for considering how the cytoskeleton can affect chromatin dynamics.
Experimental Procedures
Cell culture and strain generation
S. pombe strains are described in Supplemental Table 1. Standard manipulations and cell culturing were carried out as described (Moreno et al., 1991). Strains were grown at 30°C except for nuf2-1-containing strains, which were grown at 25°C. For gene replacement, ORFs were replaced with the KanMX cassette; C-terminal GFP tagging was carried out using the GFP-KanMX cassette (Longtine et al., 1998); C-terminal mCherry tagging was carried out using pKS390 as a template (Snaith et al., 2005). N-terminal tagging (KanMX nmt41GFP) was carried out as described (Bahler et al., 1998). All integrations were confirmed by PCR. After genetic crosses, progeny were tested by segregation of markers, PCR, or presence of the relevant fluorescent protein fusion, as appropriate.
Microscopy
S. pombe strains were grown in YE5S media to an OD600 of 0.4 – 0.8, mounted on agarose pads (1.5% agarose in EMM), covered with coverslips and sealed with VALAP (equal parts Vaseline, lanolin and paraffin). All images were acquired on a Carl Zeiss AxioImagerZ.1 (Carl Zeiss Inc.) equipped with an AxioCamMRm camera. Time-lapse images were obtained in a single optical plane. The image of Sad1-DsRed in clr4Δ cells (Figure 6) is a maximum intensity projection of 10 z-sections obtained with 0.2 µM spacing.
Quantitation of cellular phenotypes
Still images of random fields were taken in 5 z-sections with 0.4 µM spacing for quantitation. For determination of aspect ratios, the plane that bisects the nucleus was chosen. The longest dimension of the nuclear envelope and the orthogonal dimension were measured using AxioVision software. For counting of cytoplasmic foci and determining colocalization, the entire 5-section stack was analyzed. Composite time-lapse images were acquired as single plane images over the described time course, and the maximum intensity image was computed from the entire overlayed data set using the AxioVision software. To determine the amplitude of SPB oscillations, the distance between the most distant SPB positions over time was measured.
Chromatin Immunoprecipitation
ChIP was carried out as described by the manufacturer (Agilent Technologies) with the following modifications. All immunoprecipitations were carried out using a polyclonal rabbit anti-GFP antibody (gift of M.P. Rout). DNA extraction was reduced to one phenol extraction, one chloroform/isoamyl alcohol extraction, followed by column PCR purification (Qiagen). Primer set sequences were designed according to Nakagawa et al., and include cnt-340, imr-195, dh-383, and dg-223 (Nakagawa et al., 2002). Real-time PCR was carried out using the Platinum SYBR Green qPCR SuperMix-UDG kit (Invitrogen) and an ABI7900 instrument (Applied Biosystems). Analysis was carried out using the SDS2.0 software. Enrichment was determined by comparing products from each primer pair compared to the actin (act1) products in GFP-Ima1 immunoprecipitates (MKSP14) and mock GFP immunoprecipitates from WT cells (MKSP5).
Supplementary Material
Acknowledgements
We thank the Nurse lab and the Yeast Genomic Resource Center for strains, Ken Sawin for the mCherry tagging template pKS390, Roger Tsien for permission to use the mCherry fusion, Helen Shio for technical support, and M.P. Rout for the anti-GFP antibody. We also thank F. Neumann for technical help and suggesting the “nut and bolt” analogy, and C.P. Lusk for critical reading of the manuscript. This work was supported by a Kirschstein NRSA Fellowship and a Charles H. Revson Senior Fellowship in Biomedical Science (to M.C.K.) and the Howard Hughes Medical Institute (to G.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat. Rev. Genet. 2007;8:507–517. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- Appelgren H, Kniola B, Ekwall K. Distinct centromere domain structures with separate functions demonstrated in live fission yeast cells. J. Cell Sci. 2003;116:4035–4042. doi: 10.1242/jcs.00707. [DOI] [PubMed] [Google Scholar]
- Asakawa H, Hayashi A, Haraguchi T, Hiraoka Y. Dissociation of the Nuf2-Ndc80 complex releases centromeres from the spindle-pole body during meiotic prophase in fission yeast. Mol. Biol. Cell. 2005;16:2325–2338. doi: 10.1091/mbc.E04-11-0996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Bupp JM, Martin AE, Stensrud ES, Jaspersen SL. Telomere anchoring at the nuclear periphery requires the budding yeast Sad1-UNC-84 domain protein Mps3. J. Cell Biol. 2007;179:845–854. doi: 10.1083/jcb.200706040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige Y, Tsutsumi C, Yamane M, Okamasa K, Haraguchi T, Hiraoka Y. Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell. 2006;125:59–69. doi: 10.1016/j.cell.2006.01.048. [DOI] [PubMed] [Google Scholar]
- Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. Coupling of the nucleus and cytoplasm: role of the LINC complex. J. Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R, West RR, Morphew DM, Oakley BR, McIntosh JR. The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol. Biol. Cell. 1997;8:1461–1479. doi: 10.1091/mbc.8.8.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K, Nimmo ER, Javerzat JP, Borgstrom B, Egel R, Cranston G, Allshire R. Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J. Cell Sci. 1996;109:2637–2648. doi: 10.1242/jcs.109.11.2637. [DOI] [PubMed] [Google Scholar]
- Franco A, Meadows JC, Millar JB. The Dam1/DASH complex is required for the retrieval of unclustered kinetochores in fission yeast. J. Cell Sci. 2007;120:3345–3351. doi: 10.1242/jcs.013698. [DOI] [PubMed] [Google Scholar]
- Funabiki H, Hagan I, Uzawa S, Yanagida M. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. The J. Cell Biol. 1993;121:961–976. doi: 10.1083/jcb.121.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. The nuclear lamina comes of age. Nat. Rev. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J. Cell Biol. 1995;129:1033–1047. doi: 10.1083/jcb.129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan IM, Riddle PN, Hyams JS. Intramitotic controls in the fission yeast Schizosaccharomyces pombe: the effect of cell size on spindle length and the timing of mitotic events. J. Cell Biol. 1990;110:1617–1621. doi: 10.1083/jcb.110.5.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SI. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- Haque F, Lloyd DJ, Smallwood DT, Dent CL, Shanahan CM, Fry AM, Trembath RC, Shackleton S. SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol. Cell. Biol. 2006;26:3738–3751. doi: 10.1128/MCB.26.10.3738-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Lusk CP, Blobel G. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature. 2006;442:1003–1007. doi: 10.1038/nature05075. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lorentz A, Ostermann K, Fleck O, Schmidt H. Switching gene swi6, involved in repression of silent mating-type loci in fission yeast, encodes a homologue of chromatin-associated proteins from Drosophila and mammals. Gene. 1994;143:139–143. doi: 10.1016/0378-1119(94)90619-x. [DOI] [PubMed] [Google Scholar]
- Lusk CP, Blobel G, King MC. Highway to the inner nuclear membrane: rules for the road. Nat. Rev. Mol. Cell Biol. 2007;8:414–420. doi: 10.1038/nrm2165. [DOI] [PubMed] [Google Scholar]
- Malone CJ, Misner L, Le Bot N, Tsai MC, Campbell JM, Ahringer J, White JG. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 2003;115:825–836. doi: 10.1016/s0092-8674(03)00985-1. [DOI] [PubMed] [Google Scholar]
- Miki F, Kurabayashi A, Tange Y, Okazaki K, Shimanuki M, Niwa O. Two-hybrid search for proteins that interact with Sad1 and Kms1, two membrane-bound components of the spindle pole body in fission yeast. Mol. Genet. Gen. 2004;270:449–461. doi: 10.1007/s00438-003-0938-8. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enz. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Nabetani A, Koujin T, Tsutsumi C, Haraguchi T, Hiraoka Y. A conserved protein, Nuf2, is implicated in connecting the centromere to the spindle during chromosome segregation: a link between the kinetochore function and the spindle checkpoint. Chromosoma. 2001;110:322–334. doi: 10.1007/s004120100153. [DOI] [PubMed] [Google Scholar]
- Nakagawa H, Lee JK, Hurwitz J, Allshire RC, Nakayama J, Grewal SI, Tanaka K, Murakami Y. Fission yeast CENP-B homologs nucleate centromeric heterochromatin by promoting heterochromatin-specific histone tail modifications. Genes Dev. 2002;16:1766–1778. doi: 10.1101/gad.997702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa O, Shimanuki M, Miki F. Telomere-led bouquet formation facilitates homologous chromosome pairing and restricts ectopic interaction in fission yeast meiosis. EMBO J. 2000;19:3831–3840. doi: 10.1093/emboj/19.14.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE. Physical plasticity of the nucleus in stem cell differentiation. Proc. Natl. Acad. Sci. USA. 2007;104:15619–15624. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin KE, Tran PT. Cytoplasmic microtubule organization in fission yeast. Yeast. 2006;23:1001–1014. doi: 10.1002/yea.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer EC, Florens L, Guan T, Yates JR, 3rd, Gerace L. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science. 2003;301:1380–1382. doi: 10.1126/science.1088176. [DOI] [PubMed] [Google Scholar]
- Shimanuki M, Miki F, Ding DQ, Chikashige Y, Hiraoka Y, Horio T, Niwa O. A novel fission yeast gene, kms1+, is required for the formation of meiotic prophase-specific nuclear architecture. Mol. Gen. Genet. 1997;254:238–249. doi: 10.1007/s004380050412. [DOI] [PubMed] [Google Scholar]
- Snaith HA, Samejima I, Sawin KE. Multistep and multimode cortical anchoring of tea1p at cell tips in fission yeast. EMBO J. 2005;24:3690–3699. doi: 10.1038/sj.emboj.7600838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA, Fischer JA. KASH 'n Karry: the KASH domain family of cargo-specific cytoskeletal adaptor proteins. Bioessays. 2005;27:1136–1146. doi: 10.1002/bies.20312. [DOI] [PubMed] [Google Scholar]
- Szabo B, Kornyei Z, Zach J, Selmeczi D, Csucs G, Czirok A, Vicsek T. Auto-reverse nuclear migration in bipolar mammalian cells on micropatterned surfaces. Cell Mot. Cytoskel. 2004;59:38–49. doi: 10.1002/cm.20022. [DOI] [PubMed] [Google Scholar]
- Tang X, Jin Y, Cande WZ. Bqt2p is essential for initiating telomere clustering upon pheromone sensing in fission yeast. J. Cell Biol. 2006;173:845–851. doi: 10.1083/jcb.200602152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Cooper JP. The telomere bouquet controls the meiotic spindle. Cell. 2007;130:113–126. doi: 10.1016/j.cell.2007.05.024. [DOI] [PubMed] [Google Scholar]
- Tran PT, Marsh L, Doye V, Inoue S, Chang F. A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J. Cell Biol. 2001;153:397–411. doi: 10.1083/jcb.153.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzur YB, Wilson KL, Gruenbaum Y. SUN-domain proteins: 'Velcro' that links the nucleoskeleton to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 2006;7:782–788. doi: 10.1038/nrm2003. [DOI] [PubMed] [Google Scholar]
- West RR, Vaisberg EV, Ding R, Nurse P, McIntosh JR. cut11(+): A gene required for cell cycle-dependent spindle pole body anchoring in the nuclear envelope and bipolar spindle formation in Schizosaccharomyces pombe. Mol. Biol. Cell. 1998;9:2839–2855. doi: 10.1091/mbc.9.10.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmsen K, Ketema M, Truong H, Sonnenberg A. KASH-domain proteins in nuclear migration, anchorage and other processes. J. Cell Sci. 2006;119:5021–5029. doi: 10.1242/jcs.03295. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Hiraoka Y. Monopolar spindle attachment of sister chromatids is ensured by two distinct mechanisms at the first meiotic division in fission yeast. EMBO J. 2003;22:2284–2296. doi: 10.1093/emboj/cdg222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Ragnauth CD, Skepper JN, Worth NF, Warren DT, Roberts RG, Weissberg PL, Ellis JA, Shanahan CM. Nesprin-2 is a multi-isomeric protein that binds lamin and emerin at the nuclear envelope and forms a subcellular network in skeletal muscle. J. Cell Sci. 2005;118:673–687. doi: 10.1242/jcs.01642. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.