Abstract
A cell's ability to generate different responses to different levels of stimulus is an important component of an adaptive environmental response. Transcriptional responses are frequently controlled by transcription factors regulated by phosphorylation. We demonstrate that differential phosphorylation of the budding yeast transcription factor Pho4 contributes to differential gene expression. When yeast cells are grown in high-phosphate growth medium, Pho4 is phosphorylated on four critical residues by the cyclin–CDK complex Pho80–Pho85 and is inactivated. When yeast cells are starved for phosphate, Pho4 is dephosphorylated and fully active. In intermediate-phosphate conditions, a form of Pho4 preferentially phosphorylated on one of the four sites accumulates and activates transcription of a subset of phosphate-responsive genes. This Pho4 phosphoform binds differentially to phosphate-responsive promoters and helps to trigger differential gene expression. Our results demonstrate that three transcriptional outputs can be generated by a pathway whose regulation is controlled by one kinase, Pho80–Pho85, and one transcription factor, Pho4. Differential phosphorylation of Pho4 by Pho80–Pho85 produces phosphorylated forms of Pho4 that differ in their ability to activate transcription, contributing to multiple outputs.
Differentially phosphorylated forms of the Pho4 transcription factor regulate distinct sets of target genes, thereby mediating specific responses to environmental conditions
Introduction
A cell's survival depends on its ability to respond appropriately to its extracellular environment. An appropriate response often depends not only on the presence or absence of a signal, but also on its magnitude and duration. Many signaling pathways utilize protein kinases, which commonly regulate downstream transcription factors through phosphorylation. Phosphorylation can affect the activity, stability, or subcellular localization of transcription factors (Cohen 2000). Multisite phosphorylation of a protein permits not only the binary combination of the effects of each phosphorylation event, but also the emergence of more complex properties, such as signal integration (Cohen 2000), switch-like behavior (Ferrell and Machleder 1998; Nash et al. 2001), and kinetic proofreading (McKeithan 1995). Although many proteins are phosphorylated on multiple sites, the biological significance of such phosphorylation is in most cases unclear.
When starved for inorganic phosphate, Saccharomyces cerevisiae induces a program of gene expression that includes a phosphate permease, Pho84 (NP_013583); a secreted acid phosphatase, Pho5 (NP_009651); and proteins involved in phosphate storage (Oshima 1982; Ogawa et al. 2000). Induction of the phosphate-responsive gene expression program is controlled by the transcription factor Pho4 (NP_116692) (Oshima 1997). Activity and localization of Pho4 are regulated in response to phosphate availability through phosphorylation by the nuclear cyclin–CDK (cyclin-dependent kinase) complex, Pho80–Pho85 (NP_014642 and NP_015294) (O'Neill et al. 1996; Komeili and O'Shea 1999). Pho4 is unphosphorylated, nuclear-localized, and active when yeast cells are starved for phosphate, but is phosphorylated, cytoplasmic, and inactive when cells are grown in phosphate-rich medium (Kaffman et al. 1994; O'Neill et al. 1996). Pho4 is phosphorylated by Pho80–Pho85 on four functionally important serine residues (O'Neill et al. 1996) (Figure 1A). Together, phosphorylation of sites 2 and 3 results in export of Pho4 from the nucleus by promoting its interaction with the export receptor Msn5 (NP_010622) (Kaffman et al. 1998a). Phosphorylation of site 4 inhibits nuclear import of Pho4 by impairing its binding to the import receptor Pse1 (NP_014039) (Kaffman et al. 1998b). Phosphorylation of Pho4 also regulates its transcriptional activity within the nucleus; phosphorylation of site 6 inhibits the binding of Pho4 to Pho2 (NP_010177), a transcription factor essential for the induction of many phosphate-responsive genes (Komeili and O'Shea 1999).
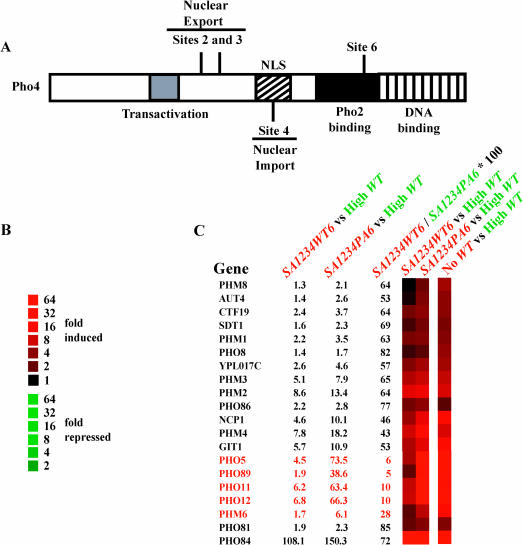
Figure 1. Whole-Genome Expression DNA Microarrays of PHO4SA1234WT6 and PHO4SA1234PA6 Strains.
(A) Schematic showing phosphorylation sites (O'Neill et al. 1996), the transcriptional activation domain (McAndrew et al. 1998), the nuclear localization sequence (Kaffman et al. 1998b), the Pho2-binding domain (Hirst et al. 1994), and the DNA-binding domain of Pho4 (Ogawa and Oshima 1990). Pho4 is phosphorylated on five sites (referred to as sites 1, 2, 3, 4, and 6) by Pho80–Pho85 (O'Neill et al. 1996). Site 1 is phosphorylated inefficiently in vivo and in vitro, and no functional consequence has been attributed to its phosphorylation (O'Neill et al. 1996; Komeili and O'Shea 1999).
(B) Color scheme denoting the fold induction/repression for all of the microarray experiments.
(C) Expression of phosphate-responsive genes in strains containing Pho4SA1234WT6 and Pho4SA1234PA6 grown in high-phosphate medium. Cy5-labeled samples are colored red, and Cy3-labeled samples are colored green. The percent of induction of each gene in the Pho4SA1234WT6 strain compared to its maximal induction in the Pho4SA1234PA6 strain is presented on the right. Several phosphate-responsive genes (PHO5, PHO8, PHO11, and PHO12) share large regions of homology. Cross-hybridization leads to similar response profiles even though the genes probably respond differently.
The kinetics by which Pho80–Pho85 phosphorylates Pho4 could lead to the accumulation of different Pho4 phosphoforms in vivo. In vitro, on average only two of the four functionally relevant phosphorylatable serine residues are phosphorylated on a single molecule of Pho4 during one binding interaction, between Pho80–Pho85 and an unphosphorylated Pho4 dimer (Jeffery et al. 2001). Additionally, in vitro Pho80–Pho85 shows dramatic site preference; site 6 on Pho4 is phosphorylated approximately 50% of the time during any single phosphorylation event (Jeffery et al. 2001). Together, this leads to phosphorylation of site 6 before phosphorylation of both sites 2 and 3 approximately 90% of the time (Jeffery et al. 2001). These in vitro characteristics of Pho4 phosphorylation by Pho80–Pho85 suggest that a partially phosphorylated form of Pho4 predominately phosphorylated on site 6 may accumulate in vivo when Pho80–Pho85 is partially active.
We wished to determine whether partially phosphorylated forms of Pho4 accumulate under physiological conditions and whether such forms of Pho4 have activity that is different from Pho4, that is, either completely phosphorylated or unphosphorylated. We demonstrated a mutant form of Pho4 that is partially phosphorylated efficiently activates transcription of a subset of phosphate-responsive genes. We also uncovered a physiological growth condition in which only a subset of the phosphate-responsive genes is induced significantly. The differential gene expression observed under this growth condition correlates with the appearance of a partially phosphorylated form of Pho4 that is phosphorylated predominately on site 6.
Results
Regulation of Either the Activity or Nuclear Localization of Pho4 Is Sufficient to Prevent Induction of PHO5, but Not PHO84, in High-Phosphate Medium
Motivated by our in vitro observations that Pho80–Pho85 preferentially phosphorylates Pho4 on site 6, we wished to systematically explore the transcriptional properties of Pho4 phosphorylated only on site 6. Our previous studies had demonstrated that this form of Pho4 was localized to the nucleus but could not efficiently activate transcription of PHO5 (Komeili and O'Shea 1999). We wished to determine whether Pho4 phosphorylated only on site 6 could activate transcription of any phosphate-responsive genes.
To address this question, we made use of a mutant form of Pho4 that can only be phosphorylated on site 6; this mutant contains serine-to-alanine substitutions at phosphorylation sites 1, 2, 3, and 4 (referred to as SA1234). Pho4SA1234WT6 is localized to the nucleus and phosphorylated on site 6 in high-phosphate conditions, but it cannot efficiently activate transcription of PHO5 (Komeili and O'Shea 1999). We compared the transcriptional properties of Pho4SA1234WT6 to those of a second mutant form of Pho4, which is refractory to phosphorylation because it has serine-to-alanine substitutions at sites 1, 2, 3, and 4 of Pho4 and a proline-to-alanine substitution at site 6 (referred to as SA1234PA6; it is necessary to mutate the proline because the serine at site 6 is required for the full induction of phosphate-responsive genes) (Komeili and O'Shea 1999). Pho4SA1234PA6 is localized to the nucleus and unphosphorylated on site 6 in high-phosphate conditions and efficiently activates transcription of PHO5 (Komeili and O'Shea 1999). Strains expressed both Pho4SA1234PA6 and Pho4SA1234WT6 at levels that were indistinguishable from wild-type Pho4 expression as determined by fluorescence-activated cell sorting (FACS) and fluorescence microscopy (see the Supplementary Microscopy Section below).
Yeast strains expressing Pho4SA1234PA6 and Pho4SA1234WT6 were analyzed by whole-genome expression profiling using cDNA microarrays. The gene expression profile of the PHO4SA1234PA6 strain grown in high-phosphate medium was very similar to the profile observed for a wild-type strain grown in no-phosphate medium, where Pho4 is completely unphosphorylated; all phosphate-responsive genes, including PHO5, are induced (Figure 1C) (Ogawa et al. 2000). In contrast, the PHO4SA1234WT6 strain grown in high-phosphate medium expressed only a subset of genes normally induced in no-phosphate medium (Figure 1C). The 20 phosphate-responsive genes examined largely fell into two classes when we compared the percent induction of each phosphate-responsive gene in the PHO4SA1234WT6 strain to its maximal induction in the PHO4SA1234PA6 strain. Four genes were induced less than 11%, whereas 15 genes were induced more than 40% (Figure 1C). The class induced less than 11% was comprised mainly of phosphate-responsive phosphatases, whereas the class induced more than 40% consisted mainly of genes involved in phosphate storage, mobilization, and transport. From the first category, we chose to analyze PHO5 (encoding a secreted acid phosphatase) further because it was both highly induced in no-phosphate medium and well characterized. From the second category, we chose to analyze PHO84 (encoding a high affinity phosphate transporter) because it was induced over 100-fold in both the PHO4SA1234WT6 and PHO4SA1234PA6strains.
Physiological Conditions That Cause Differential Phosphorylation of Pho4
Our observations with the Pho4 mutants led us to ask whether there exist physiologically relevant conditions in which partially phosphorylated Pho4 accumulates in the nucleus and causes differential gene expression. We predicted that as the extracellular phosphate concentration drops, Pho80–Pho85 would become partially inactivated, and because of the kinetic properties of Pho80–Pho85, a nuclear pool of Pho4 phosphorylated on site 6 (but not on sites 2 and 3) would accumulate. We anticipated that only at much lower concentrations of phosphate would a nuclear pool of completely unphosphorylated Pho4 build up and activate transcription of all phosphate-responsive genes.
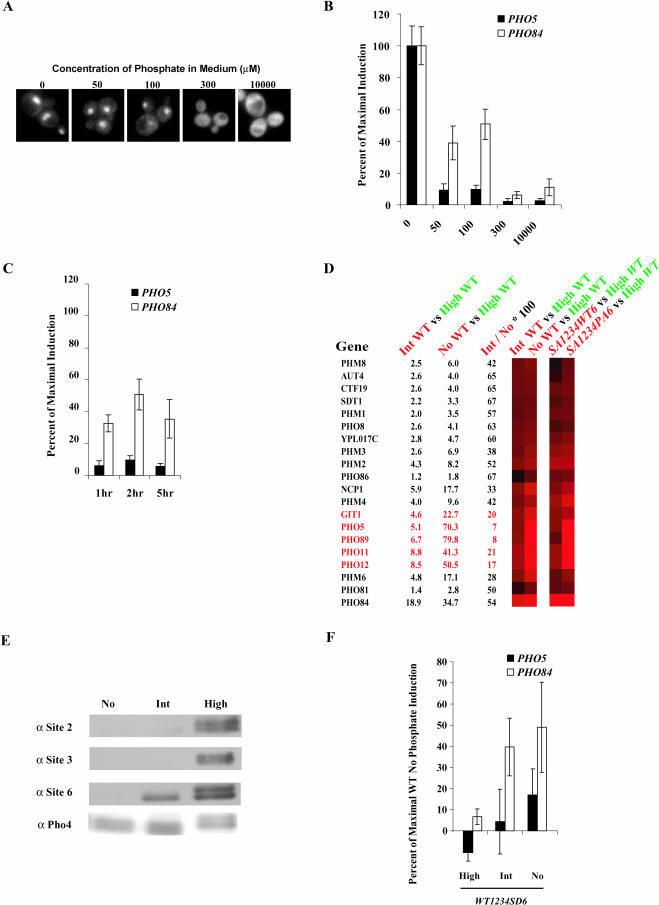
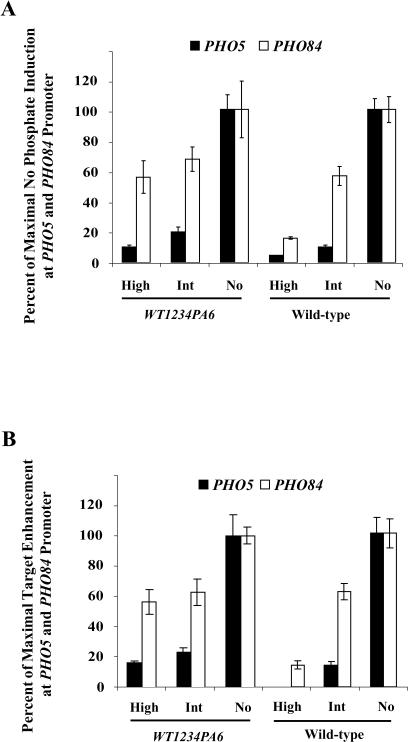
To test these predictions, we grew cells in medium containing different levels of phosphate (0, 50, 100, 300, and 10,000 μM) and monitored localization of a Pho4–GFP (green fluorescent protein) fusion protein and transcription of PHO5 and PHO84. We observed three different responses to different phosphate conditions. First, the high-phosphate response occurs at extracellular phosphate concentrations greater than 300 μM phosphate. Pho4 is cytoplasmic, and PHO5 and PHO84 are expressed at only basal levels under these conditions (Figure 2A and 2B). Second, the no-phosphate response occurs when there is no phosphate in the medium. Pho4 is nuclear, and PHO5 and PHO84 are expressed maximally under these conditions (Figure 2A and 2B). Third, the intermediate-phosphate response occurs at extracellular phosphate concentrations less than 100 μM phosphate and greater than no phosphate. Pho4 is nuclear, but under these conditions there is only approximately 10% of maximal expression of PHO5, while there is approximately 50% of maximal expression of PHO84 (Figure 2A and 2B).
Figure 2. Growth of Yeast Cells in Intermediate-Phosphate Medium Leads to Differential Phosphorylation of Pho4 and Differential Expression of PHO5 and PHO84.
(A) Fluorescence microscopy of yeast cells containing Pho4–GFP grown in no, 50 μM, 100 μM (intermediate [int]), 300 μM, or 10,000 μM (high) phosphate medium.
(B) Quantitation of RNA levels by Northern blot analysis of PHO84, PHO5, and ACT1 in wild-type cells grown in medium containing different concentrations of phosphate.
(C) Quantitation of RNA levels by Northern blot analysis of PHO84, PHO5, and ACT1, in wild-type cells grown for 1, 2, or 5 h in intermediate-phosphate medium.
(D) Expression of genes in the phosphate-responsive cluster (Carroll et al. 2001) for a wild-type strain grown in intermediate- or no-phosphate medium compared to wild-type cells grown in high-phosphate medium. Cy5 and Cy3 samples are colored red and green, respectively. The percent of induction of each gene in intermediate-phosphate medium compared to its maximal induction in no-phosphate medium is presented on the right.
(E) Analysis of Pho4 protein and phosphorylation by Western blotting for wild-type cells grown in no-, intermediate-, and high-phosphate medium. Samples were probed with phosphopeptide antibodies specific to sites 2, 3, and 6 of Pho4 and by a polyclonal antibody that recognizes Pho4.
(F) Quantitation of RNA levels by Northern blot analysis of PHO84, PHO5, and ACT1, in a PHO4WT1234SD6 strain.
To further characterize the properties of the intermediate-phosphate response, we examined the time and concentration dependence of PHO5 and PHO84 expression. Differential gene induction was observed in medium containing less than approximately 150 μM phosphate and more than 1 μM phosphate medium (data not shown). To confirm that the differential transcriptional response of PHO5 and PHO84 observed in intermediate-phosphate medium is not a kinetic artifact, we monitored the amount of PHO5 and PHO84 mRNA over several hours (Figure 2C). The expression pattern of PHO5 and PHO84 was stable over a range of both time and external phosphate concentration, indicating that the intermediate-phosphate response is robust. After 4–6 h, cells adapted to intermediate-phosphate medium and Pho4 was relocalized to the cytoplasm (data not shown).
Whole-genome expression profiling of cells grown in intermediate-phosphate medium was used to assess the number of phosphate-responsive genes that behave like PHO5 or PHO84. Genes fell into two classes when we compared the induction of each phosphate-responsive gene in intermediate-phosphate medium to its induction in no-phosphate medium (Figure 2D). These two classes were similar to the two classes observed when we compared the strain expressing Pho4SA1234WT6 to the strain expressing Pho4SA1234PA6 (Figure 2D). This was anticipated because both a wild-type strain grown in intermediate-phosphate medium and a PHO4SA1234WT6 strain grown in high-phosphate medium have nuclear Pho4, which is predominately phosphorylated on site 6.
To determine whether Pho4 is preferentially phosphorylated on site 6 in intermediate-phosphate medium, we generated phosphopeptide-specific antibodies that recognize phosphorylation on sites 2, 3, and 6 of Pho4. Cells were grown for 2 h in no-, intermediate-, and high-phosphate medium and analyzed by Western blotting, which indicated that there is more unphosphorylated Pho4 present in intermediate-phosphate conditions than in high-phosphate conditions (Figure 2E). In high-phosphate medium, when Pho80–Pho85 kinase activity is high, sites 2, 3, and 6 were phosphorylated. In no-phosphate medium, when Pho80–Pho85 kinase activity is low, there was no detectable phosphorylation on sites 2, 3, and 6. In intermediate-phosphate medium, as predicted by in vitro Pho80–Pho85 kinetics, site 6 was phosphorylated, but there was only minimal phosphorylation on sites 2 and 3. Thus, differential phosphorylation of Pho4 occurs in physiologically relevant conditions in vivo and site 6 phosphorylation in vivo correlates with differential gene expression.
Site 6 phosphorylation is lower in intermediate-phosphate medium than high-phosphate medium (Figure 2E). Differential expression of PHO5 and PHO84 could be the result of differential sensitivity of these two promoters to the amount of unphosphorylated Pho4 in the nucleus. To assess this possibility, we created a Pho4 mutant that behaves as if it were phosphorylated on site 6 constitutively. This was achieved by substitution of the serine at site 6 with aspartate, which hinders the interaction between Pho4 and Pho2 (Komeili and O'Shea 1999). Gene expression in a yeast strain expressing Pho4WT1234SD6 was monitored by Northern blotting (Figure 2F) and microarray analysis (data not shown). Even when grown in no-phosphate medium, the strain expressing Pho4WT1234SD6 exhibited differential expression of PHO5 and PHO84. Therefore, differential expression of PHO5 and PHO84 in intermediate-phosphate conditions is not likely to be due to a subset of Pho4 molecules that is unphosphorylated in the nucleus.
Differential Promoter Binding of Pho4
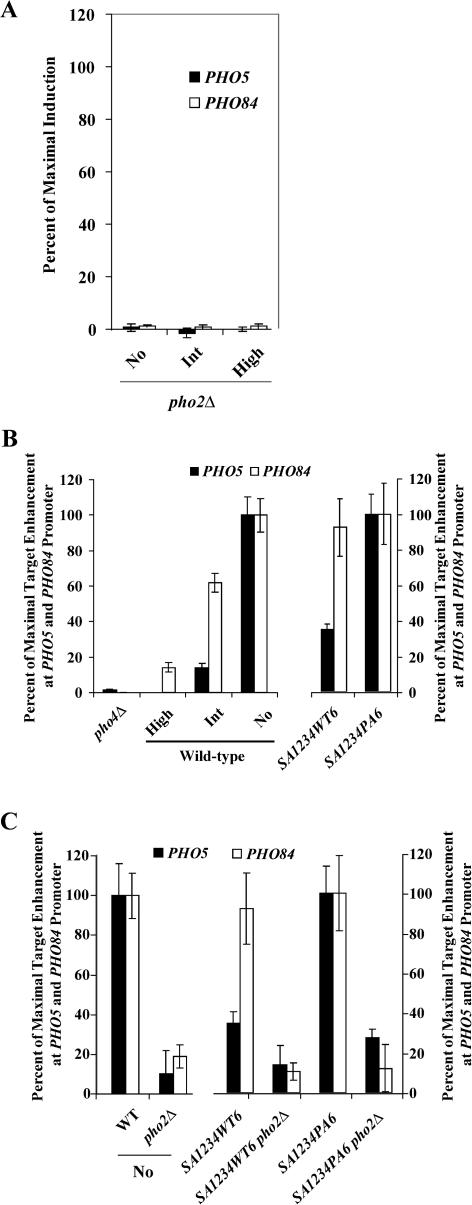
To determine whether differential gene expression results from differential occupancy of Pho4 at PHO5 and PHO84, we examined the binding of Pho4 to these promoters using chromatin immunoprecipitation. There was little enrichment of Pho4 at the PHO5 or PHO84 promoters when cells were grown in high-phosphate medium, whereas in no-phosphate medium, Pho4 was significantly enriched at both the PHO5 and PHO84 promoters (Figure 3B, left). When cells were grown in intermediate-phosphate medium, Pho4 was enriched at the PHO84 promoter, but was not significantly enriched at the PHO5 promoter. Furthermore, chromatin immunoprecipitation experiments carried out with PHO4SA1234WT6 and PHO4SA1234PA6 strains grown in high-phosphate medium resembled immunoprecipitations carried out with wild-type cells grown in intermediate and no-phosphate medium, respectively (Figure 3B, right). Thus, differential expression of PHO5 and PHO84 correlates with differential Pho4 binding to these promoters.
Figure 3. Deletion of PHO2 Abrogates Expression of PHO5 and PHO84 and Binding of Pho4 to These Promoters.
(A) Quantitation of RNA levels by Northern blot analysis in pho2Δ strains grown in no-, intermediate-, or high-phosphate medium.
(B) Chromatin immunoprecipitation analysis of Pho4. Pho4 was immunoprecipitated from extracts of wild-type cells grown in high-, intermediate-, or no-phosphate medium, from a strain lacking Pho4 and from the two mutant Pho4 strains grown in high-phosphate medium. Experiments using the Pho4SA1234WT6- and Pho4SA1234PA6-expressing strains are normalized to the maximal amount of enrichment in a strain expressing Pho4SA1234PA6 in high-phosphate medium. The fold enrichment of PHO5 over ACT1 was 1.03, 0.99, 1.26, 2.84, 2.41, and 5.04 in lanes 1–6, respectively (pho4Δ, wt high, wt int, wt no, PHO4SA1234WT6, and PHO4SA1234PA6). The fold enrichment of PHO84 over ACT1 was 0.99, 1.75, 4.27, 6.28, 10.8, and 11.6 in lanes 1–6, respectively.
(C) Chromatin immunoprecipitation analysis of Pho4. Pho4 was immunoprecipitated from extracts of wild-type cells grown in no-phosphate medium, a mutant lacking Pho2 in no-phosphate medium, Pho4SA1234WT6 and Pho4SA1234PA6 strains grown in high-phosphate medium, and pho2Δ Pho4SA1234WT6 and pho2Δ Pho4SA1234PA6 strains grown in high-phosphate medium. The fold enrichment of PHO5 over ACT1 was 2.84, 1.19, 2.41, 1.49, 5.04, and 2.06 in lanes 1–6, respectively (wt no, pho2Δ no, PHO4SA1234WT6, pho2Δ PHO4SA1234WT6, PHO4SA1234PA6, and pho2ΔPHO4SA1234PA6). The fold enrichment of PHO84 over ACT1 was 6.28, 2.0, 10.8, 2.4, 11.6, and 2.63 in lanes 1–6, respectively.
Because site 6 phosphorylation inhibits the Pho4–Pho2 interaction in vitro (Komeili and O'Shea 1999), the preferential induction of genes in intermediate-phosphate medium could be explained if their transcription does not require Pho2. To determine whether the expression of PHO84 is dependent on Pho2, we deleted PHO2 in a Pho4 wild-type strain and in strains expressing Pho4SA1234WT6 and Pho4SA1234PA6. Northern blotting (Figure 3A; data not shown) and microarray analysis (data not shown) of these pho2Δ strains demonstrated that both PHO5 and PHO84 (and the majority of the phosphate-responsive genes) required Pho2 for induction. To determine whether the transcriptional defect in pho2Δ strains was due to a defect in binding of Pho4, we used chromatin immunoprecipation to examine Pho4 occupancy at the PHO5 and PHO84 promoters in pho2Δ strains. Surprisingly, in a pho2Δ strain, little Pho4 was bound to the PHO5 or PHO84 promoter in either wild-type or mutant Pho4 strain backgrounds (Figure 3C) (Barbaric et al. 1996). Therefore, Pho2 must directly or indirectly facilitate Pho4 binding to the PHO84 promoter, even when Pho4 is phosphorylated on site 6.
Physiological Role of Site 6 Phosphorylation
Site 6 phosphorylation correlates with differential gene expression in wild-type cells and is sufficient to cause differential gene expression in strains expressing Pho4 mutants, but this does not mean site 6 phosphorylation is necessary for differential gene induction. If site 6 phosphorylation is the sole mechanism to inhibit PHO5 expression in intermediate-phosphate conditions, a strain grown in intermediate-phosphate medium (where Pho4 is nuclear) expressing a mutant Pho4 that cannot be phosphorylated on site 6 should significantly induce PHO5. We therefore created a strain expressing a Pho4 mutant, Pho4WT1234PA6, that cannot be phosphorylated on site 6 but is able to bind Pho2. Fluorescence microscopy was used to confirm proper localization of this mutant (data not shown). In contrast to expectations, the mutant that prevents site 6 phosphorylation did not strongly influence the expression of PHO5 or PHO84 in intermediate-phosphate medium (Figure 5A). This suggests that a second mechanism that contributes to differential expression of PHO5 and PHO84 expression exists in intermediate-phosphate conditions. Because a Pho4 mutant that is refractory to phosphorylation, PHO4SA1234PA6 (see Figure 1C), induces all phosphate-responsive genes, we conclude that this second mechanism is likely to work through Pho4 and Pho80–Pho85.
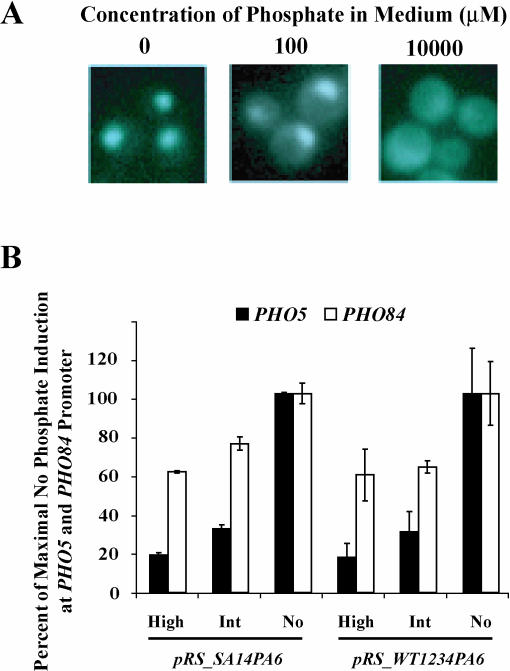
Figure 5. A Strain Expressing Pho4SA14PA6 from a Low Copy Plasmid Differentially Expresses PHO5 and PHO84 in Intermediate-Phosphate Medium.
(A) Fluorescence microscopy of yeast cells containing Pho4SA14PA6–GFP grown in no, 100 μM, or 10 mM phosphate medium.
(B) Quantitation of RNA levels by Northern blot analysis of PHO84, PHO5, and ACT1 from strains expressing Pho4SA14PA6 or Pho4WT1234PA6 from a low copy plasmid grown in no, 100 μM, or 10,000 μM phosphate medium.
Although site 6 phosphorylation is not necessary to inhibit PHO5 expression, it is necessary to keep cells from inducing transcription of PHO84 in high-phosphate medium. A strain expressing Pho4WT1234PA6 grown in high-phosphate medium had levels of PHO5 and PHO84 expression that were similar to those of a wild-type strain grown in intermediate-phosphate medium (see Figure 4A). Site 6 phosphorylation was necessary to inhibit binding of Pho4 to the PHO84 promoter in high-phosphate medium, but was not required to inhibit binding of Pho4 to the PHO5 promoter in intermediate-phosphate medium (see Figure 4B).
Figure 4. A Strain Expressing Pho4 That Cannot Be Phosphorylated on Site 6 Does Not Induce PHO5 in Intermediate-Phosphate Medium.
A strain expressing Pho4WT1234PA6 grown for 2 h in no-, intermediate-, and high-phosphate medium was analyzed by Northern blot analysis (A) and chromatin immunoprecipitation (B). The fold enrichment of PHO5 over ACT1 was 1.71, 2.42, 10.49, 0.99, 1.26, and 2.84 in lanes 1–6, respectively (PHO4WT1234WT6 high, PHO4WT1234WT6 int, PHO4 WT1234WT6 no, wt high, wt int, wt no). The fold enrichment of PHO84 over ACT1 was 24.64, 27.44, 43.74, 1.75, 4.27, and 6.28 in lanes 1–6, respectively.
It is possible that phosphorylation on site 1 or 4 could be contributing to differential expression of PHO5 and PHO84. To assess this possibility, we made a plasmid-expressed version of Pho4, Pho4SA14PA6, which can only be phosphorylated on sites 2 and 3. We used fluorescence microscopy to confirm that, in intermediate-phosphate medium, this mutant is localized to the nucleus (Figure 5A) (see Supplementary Microscopy Section below). Because this mutant is localized to the cytoplasm in high-phosphate conditions, we infer that this mutant can be phosphorylated on sites 2 and 3 (Figure 5A). Pho4SA14PA6-expressing strains grown in intermediate-phosphate medium still induce PHO5 and PHO84 differentially (Figure 5B). This suggests that the second mechanism of inhibition at the PHO5 promoter does not involve phosphorylation of sites 1 or 4. Because a PHO4SA1234PA6 strain in high-phosphate medium mimics a wild-type strain grown in no-phosphate medium (see Figure 2F), it is likely that the second mechanism for differential regulation of PHO5 and PHO84 works through sites 2 and 3.
Supplementary Microscopy Section
To assess whether variations in the nuclear concentration of Pho4 in different mutant backgrounds might affect the expression of PHO5 and PHO84, the levels of Pho4–GFP were quantified by fluorescence microscopy. Nuclear Pho4SA1234WT6 was present at 242 ± 5 arbitrary units, and nuclear Pho4SA1234PA6 was present at 242 ± 6 arbitrary units. Strains expressing either of these two mutants had FACS profiles indistinguishable from that of a wild-type PHO4–GFP strain.
Pho4SA14PA6 expressed from a low copy plasmid had a nuclear concentration of 359 ± 13 arbitrary units when grown in intermediate-phosphate medium. Pho4–GFP had a nuclear concentration of 293 ± 7 arbitrary units when grown in no-phosphate medium.
All strains that were directly compared by FACS or microscopy were grown on the same day, to the same cell density, in the same batch of medium (with only phosphate levels varied when appropriate).
Discussion
Our results demonstrate that three transcriptional outputs can be generated by a pathway whose regulation is controlled by one kinase, Pho80–Pho85, and one transcription factor, Pho4. Whereas fully phosphorylated Pho4 cannot activate transcription of any of the phosphate-responsive genes and unphosphorylated Pho4 can efficiently activate transcription of all phosphate-responsive genes, partially phosphorylated Pho4 contributes to the activation of some phosphate-responsive genes, but not others. When Pho4 is localized to the nucleus and phosphorylated on site 6, it can efficiently bind to the promoter of PHO84 and activate its transcription, but it does not efficiently bind to or activate PHO5. Our in vitro studies of Pho4 phosphorylation suggested that when Pho80–Pho85 was partially active, Pho4 would accumulate in the nucleus phosphorylated on site 6. In vivo, Pho4 accumulates in the nucleus phosphorylated on site 6 in growth medium containing a range of intermediate-phosphate concentrations, conditions that we anticipate result in partial activity of Pho80–Pho85. Nuclear accumulation of this Pho4 phosphoform correlates with both differential expression of PHO5 and PHO84, as well as with differential binding to the PHO5 and PHO84 promoters.
Although nuclear Pho4 phosphorylated on site 6 differentially binds to the PHO84 and PHO5 promoters and differentially activates transcription, site 6 phosphorylation is not the only mechanism contributing to differential expression of phosphate-responsive genes in intermediate-phosphate conditions. When we prevent site 6 phosphorylation, we still observe differential expression of PHO84 and PHO5 in intermediate-phosphate conditions. We have not yet identified the second mechanism contributing to differential expression, but this mechanism works through Pho80–Pho85-dependent phosphorylation of site 2 and 3 of Pho4. It is possible that this additional mechanism functions redundantly with site 6 phosphorylation to cause differential expression of phosphate-responsive genes.
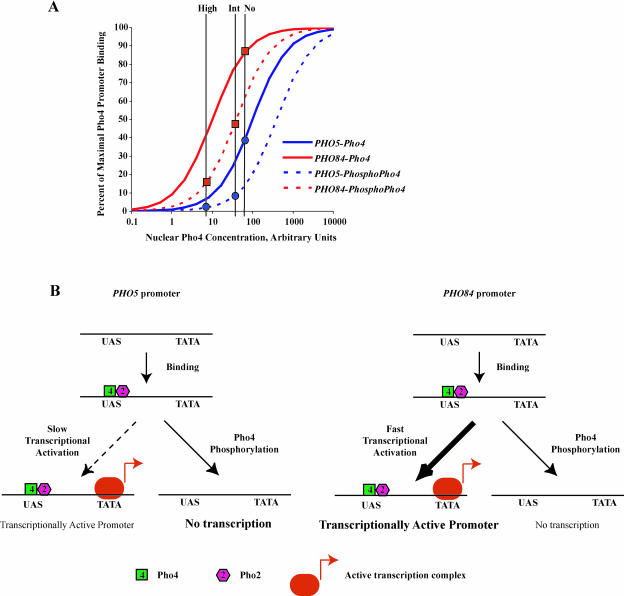
We propose that differences in the regulation of PHO5 and PHO84 are caused by affinity differences and kinetic mechanisms. If the PHO84 promoter has higher affinity for Pho4 than does the PHO5 promoter, differential occupancy of these promoters in intermediate-phosphate conditions may lead to differences in transcription (Figure 6A). For example, the concentration of Pho4 and its affinity for DNA may be such that in no-phosphate conditions Pho4 binding at the PHO84 promoter is saturated, but binding at the PHO5 promoter is not. When Pho4 is phosphorylated on site 6, as in intermediate-phosphate conditions, we expect its affinity for DNA is reduced. This reduced affinity would have a more significant impact on the PHO5 promoter than on the PHO84 promoter because the PHO5 promoter was not saturated for binding. This differential occupancy could account for differences in transcription of these genes in intermediate-phosphate conditions.
Figure 6. Models for Differential Gene Regulation.
(A) Differential affinity of Pho4 for the PHO84 and PHO5 promoters can cause differential gene expression. Simulated curves of the percent occupancy of Pho4 at the PHO84 and PHO5 promoters, assuming Michaelian binding. Pho4 was modeled as having a Kd of 10 at the PHO84 promoter and of 100 at the PHO5 promoter. Phosphorylation of Pho4 was simulated as raising the Kd of Pho4 to 40 at the PHO84 promoter and to 400 at the PHO5 promoter. The nuclear concentration of Pho4 was assumed to be 7.5-fold higher in intermediate-phosphate medium than in high-phosphate medium and 10-fold higher in no-phosphate medium than in high-phosphate medium.
(B) Kinetic diagram of the steps leading to active transcription at the PHO84 and PHO5 promoters. Differences in the kinetic mechanisms of activation of PHO84 and PHO5 can lead to differential gene expression. Even if promoter occupancy is high at the PHO5 promoter, if the transcriptional activation step is slow compared to the rate of Pho4 phosphorylation and inactivation, PHO5 will not be induced.
It is also possible that differences in the kinetic mechanisms leading to transcription of these two genes contribute to differential expression (Figure 6B). For example, if Pho4 can be phosphorylated while bound to DNA and this phosphorylation can compete with formation of an active transcription complex, the amount of transcription of a gene will be determined by the ratio of the rate of phosphorylation of Pho4 to the rate of formation of an active transcription complex. If formation of an active transcription complex is slower for PHO5 than for PHO84, in intermediate-phosphate conditions phosphorylation of Pho4 may compete with formation of an active transcription complex at the PHO5 gene, but not at PHO84. This model is plausible, given what is known about the PHO5 promoter; the promoter is regulated by positioned nucleosomes that need to be “remodeled” in order for the gene to be transcribed (Almer et al. 1986). Chromatin remodeling may represent a slow step in the pathway to transcription that could compete with Pho4 inactivation by phosphorylation on DNA. This mechanism is conceptually similar to kinetic proofreading mechanisms proposed for DNA replication and translation (Hopfield 1974).
Differential gene induction should allow a more finely tuned response to changes in external phosphate concentrations. In high-phosphate conditions, cells can uptake ample phosphate from their environment without inducing any phosphate-dependent genes, so the pathway is not induced. In no-phosphate conditions, regardless of the amount of phosphate transporter at the plasma membrane, it is essential to also scavenge phosphate from the environment. Extracellular phosphatases can liberate inorganic phosphate, which the phosphate transporter can uptake with high affinity. In no-phosphate conditions, it is therefore necessary to induce all the phosphate-responsive genes. In intermediate-phosphate conditions, the concentration of phosphate is near the Km of the Pho84 permease (Bun-Ya et al. 1991; Wykoff and O'Shea 2001). Therefore, it is likely the amount of phosphate that a cell can take up is proportional to the level of permease at the plasma membrane. Induction of PHO84 transcription could help to maintain levels of intracellular phosphate as external levels drop. It is not necessary to secrete extracellular phosphatases at high levels because there is ample inorganic phosphate in the growth medium. An intermediate-phosphate response may thereby allow cells to take up appropriate amounts of phosphate over a wide extracellular concentration range at a minimal energetic cost. Induction of genes involved in phosphate storage and mobilization may also help to buffer the cells from rapid changes in extracellular phosphate and allow the cells time to adjust the level of phosphate permeases at the plasma membrane.
Even though the phosphate regulon involves a single kinase and a single regulated transcription factor, yeast cells can generate multiple programs of gene expression in response to different phosphate levels. Many transcription factors contain multiple phosphorylation sites and may also use differential phosphorylation to respond to different levels of extracellular stimuli with different programs of gene expression. Differential phosphorylation of transcriptional regulators may help generate complex programs of gene expression with only a small set of proteins.
Materials and Methods
Strains and media
The following pRS306-based plasmids were used in this paper: PHO4WT1234PA6–GFP (EB1377), PHO4SA1234WT6–GFP (EB1264), and PHO4SA1234PA6–GFP (EB1265). These plasmids were integrated into the PHO4 locus to create EY1471, EY1022, and EY1023, respectively. EY1022 and EY1023 were also crossed to an isogenic pho2Δ (EY0337) to make EY0778 and EY0779. The following pRS316-based plasmids were used in this paper: PHO4SA14PA6–GFP (EB1487), PHO4WT1234SD6–GFP (EB842), and PHO4WT1234PA6–GFP (EB0843). These plasmids were transformed into a pho4Δ strain (EY0130). PHO4–GFP EY0693 was used as the wild-type strain. The concentration of phosphate in the growth medium was controlled by adding phosphate in the form of KH2PO4 to synthetic medium with dextrose (SD) but lacking phosphate (Huang et al. 2001).
cDNA microarrays
PHO4SA1234WT6, PHO4SA1234PA6, and wild-type (K699) strains were grown in high-phosphate medium (SD with 10 mM phosphate), intermediate-phosphate medium (SD with 100 μM phosphate), or no-phosphate medium (SD with no-phosphate) and were harvested in mid-logarithmic growth. Microarray analysis was performed as described (Carroll et al. 2001). Monofunctional reactive Cy5 (Amersham Biosciences, Little Chalfont, United Kingdom) was used to label reverse-transcribed RNA from the mutant cells, and monofunctional reactive Cy3 was used to label reverse-transcribed RNA from the wild-type cells.
Microarray analysis
Phosphate-responsive genes were analyzed as described previously (Carroll et al. 2001). The percent of maximal induction for each transcript was determined from at least two independent microarray datasets. All error bars are standard error. Genes that were reported met the following criteria: (1) induction was at least 2-fold in an experiment, and (2) there was less than a 2-fold difference between the values calculated from two independent microarray experiments. Microarrays can be accessed at Gene Expression Omnibus (GEO) at the National Center for Biotechnology Information (NCBI) database as platform number GPL423–425 and sample numbers GSM9157–GSM9162.
Fluorescence microscopy
Images were captured with a TE300 inverted Nikon microscope using Metamorph software (Universal Imaging, Downingtown, Pennsylvania, United States) and a Roper Q57 CCD camera. Cells were washed quickly in no-phosphate medium and then grown for 2 h in media of different phosphate concentrations before visualization.
Northern blot analysis
RNA was extracted by standard phenol–chloroform techniques (Steger et al. 2003). Probes to PHO84, PHO5, and ACT1 (NP_116614) (the gene encoding actin) were made by random priming from a PCR product of each full-length gene. PHO84 and PHO5 signal was normalized to signal from ACT1. All experiments were performed at least in triplicate. The reported error is the standard error of the mean. Several phosphate-responsive genes (PHO5, PHO8 [NP_010769], PHO11 [NP_009434], and PHO12 [NP_012087]) share large regions of homology. Cross-hybridization leads to similar response profiles, even though the genes probably respond differently.
Quantitation of extracellular phosphate
Phosphate in the medium was measured with an acidified ammonium molybdate/malachite green G solution (Sigma, St. Louis, Missouri, United States). No more than 50% of the phosphate was depleted in any experiment.
Phosphopeptide generation
Rabbit polyclonal phosphopeptide antibodies were generated (Bethyl Laboratories, Montgomery, Texas, United States) that recognize the following phosphorylated peptides derived from the Pho4 sequence (phosphorylated residues are denoted by square brackets): (1) the peptide containing site 2 is CPRLLY[S]PLIHT; (2) site 3 is VPVTI[S]PNLVACG; and (3) site 6 is VVASE[S]PVIAPCG (Bethyl Laboratories). Antibodies were affinity-purified using the synthetic phosphopeptides. In all cases, there was minimal cross-reactivity with the unphosphorylated peptide.
Western blot analysis
Western blotting was performed with standard techniques using enhanced chemiluminescence (Pierce Biotechnology, Rockford, Illinois, United States) for detection.
Quantitation of Pho4 levels
The level of expression of different versions of Pho4 was quantitated by FACS (Becton Dickinson LSR II; Becton, Dickinson, and Company, Franklin Lakes, New Jersey, United States). The percent nuclear fluorescence of Pho4 grown in different phosphate conditions was determined by quantitating fluorescent pictures of over 100 cells grown in each phosphate condition.
Chromatin immunoprecipitation
The chromatin immunoprecipitation protocol was similar to that previously described (Strahl-Bolsinger et al. 1997). After 2 h of growth, cells were fixed with 1% formaldehyde for 15 min at room temperature and harvested. To obtain DNA, the cell pellet was processed and immunoprecipitated with a polyclonal anti-Pho4 antibody, and copurifying DNA was purified by phenol–chloroform and chloroform extraction. The DNA was then quantified using an Opticon real-time PCR machine (MJ Research, Waltham, Massachusetts, United States) as described previously (Steger et al. 2003). At least three independent extracts were analyzed for each strain and growth condition.
Supporting Information
Accession Numbers
The GenBank accession numbers of the sequences discussed in this paper are NP_009434, NP_009651, NP_010177, NP_010622, NP_010769, NP_012087, NP_013583, NP_014039, NP_014642, NP_015294, NP_116614, and NP_116692.
Microarrays can be accessed at the Gene Expression Omnibus (GEO) at the National Center for Biotechnlogy Information (NCBI) database as platform number GPL423–425 and sample numbers GSM9157–GSM9162.
Acknowledgments
We thank Hiten Madhani, Barbara Panning, Dave Wang, Andrew Uhl, Adam Carroll, Charles Holst, and the O'Shea laboratory members for careful reading of the manuscript and David Steger for advice with the chromatin immunoprecipitation. This work was supported by the National Institutes of Health (NIH) (GM51377) and the Howard Hughes Medical Institute (to EKO). MS was supported by the National Science Foundation and by a fellowship from the Burroughs–Wellcome Fund, and DDW and NM were supported by postdoctoral fellowships from the NIH (GM20762 and GM19734, respectively).
Abbreviations
- CDK
cyclin-dependent kinase
- FACS
fluorescence-activated cell sorting
- GFP
green fluorescent protein
- SD
synthetic medium with dextrose
- wt
wild-type
Conflicts of Interest. The authors have declared that no conflicts of interest exist.
Author Contributions. MS, DDW, NM, and EKO conceived and designed the experiments. MS, DDW, and NM performed the experiments. MS, DDW, and NM analyzed the data. MS, DDW, and EKO wrote the paper.
Academic Editor: Michael Levine, University of California, Berkeley
References
- Almer A, Rudolph H, Hinnen A, Horz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986;5:2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbaric S, Munsterkotter M, Svaren J, Horz W. The homeodomain protein Pho2 and the basic-helix–loop–helix protein Pho4 bind DNA cooperatively at the yeast PHO5 promoter. Nucleic Acids Res. 1996;24:4479–4486. doi: 10.1093/nar/24.22.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bun-Ya M, Nishimura M, Harashima S, Oshima Y. The PHO84 gene of Saccharomyces cerevisiae encodes an inorganic phosphate transporter. Mol Cell Biol. 1991;11:3229–3238. doi: 10.1128/mcb.11.6.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll AS, Bishop AC, DeRisi JL, Shokat KM, O'Shea EK. Chemical inhibition of the Pho85 cyclin-dependent kinase reveals a role in the environmental stress response. Proc Natl Acad Sci U S A. 2001;98:12578–12583. doi: 10.1073/pnas.211195798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P. The regulation of protein function by multisite phosphorylation: A 25-year update. Trends Biochem Sci. 2000;25:596–601. doi: 10.1016/s0968-0004(00)01712-6. [DOI] [PubMed] [Google Scholar]
- Ferrell JE, Machleder EM. The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science. 1998;280:895–898. doi: 10.1126/science.280.5365.895. [DOI] [PubMed] [Google Scholar]
- Hirst K, Fisher F, McAndrew PC, Goding CR. The transcription factor, the Cdk, its cyclin and their regulator: Directing the transcriptional response to a nutritional signal. EMBO J. 1994;13:5410–5420. doi: 10.1002/j.1460-2075.1994.tb06876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfield JJ. Kinetic proofreading: A new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci U S A. 1974;71:4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Jeffery DA, Anthony MD, O'Shea EK. Functional analysis of the cyclin-dependent kinase inhibitor Pho81 identifies a novel inhibitory domain. Mol Cell Biol. 2001;21:6695–6705. doi: 10.1128/MCB.21.19.6695-6705.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery DA, Springer M, King DS, O'Shea EK. Multi-site phosphorylation of Pho4 by the cyclin–CDK Pho80–Pho85 is semi-processive with site preference. J Mol Biol. 2001;306:997–1010. doi: 10.1006/jmbi.2000.4417. [DOI] [PubMed] [Google Scholar]
- Kaffman A, Herskowitz I, Tjian R, O'Shea EK. Phosphorylation of the transcription factor PHO4 by a cyclin–CDK complex, PHO80–PHO85. Science. 1994;263:1153–1156. doi: 10.1126/science.8108735. [DOI] [PubMed] [Google Scholar]
- Kaffman A, Rank NM, O'Neill EM, Huang LS, O'Shea EK. (1998a) The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature. 396:482–486. doi: 10.1038/24898. [DOI] [PubMed] [Google Scholar]
- Kaffman A, Rank NM, O'Shea EK. (1998b) Phosphorylation regulates association of the transcription factor Pho4 with its import receptor Pse1/Kap121. Genes Dev. 12:2673–2683. doi: 10.1101/gad.12.17.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeili A, O'Shea EK. Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science. 1999;284:977–980. doi: 10.1126/science.284.5416.977. [DOI] [PubMed] [Google Scholar]
- McAndrew PC, Svaren J, Martin SR, Horz W, Goding CR. Requirements for chromatin modulation and transcription activation by the Pho4 acidic activation domain. Mol Cell Biol. 1998;18:5818–5827. doi: 10.1128/mcb.18.10.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeithan TW. Kinetic proofreading in T-cell receptor signal transduction. Proc Natl Acad Sci U S A. 1995;92:5042–5046. doi: 10.1073/pnas.92.11.5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, et al. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- Ogawa N, Oshima Y. Functional domains of a positive regulatory protein, PHO4, for transcriptional control of the phosphatase regulon in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:2224–2236. doi: 10.1128/mcb.10.5.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa N, DeRisi J, Brown PO. New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae revealed by genomic expression analysis. Mol Biol Cell. 2000;11:4309–4321. doi: 10.1091/mbc.11.12.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill EM, Kaffman A, Jolly ER, O'Shea EK. Regulation of PHO4 nuclear localization by the PHO80–PHO85 cyclin–CDK complex. Science. 1996;271:209–212. doi: 10.1126/science.271.5246.209. [DOI] [PubMed] [Google Scholar]
- Oshima Y. Saccharomyces: Metabolism and gene expression. In: Oshima Y, Strathern JN, Jones EW, Broach JR, editors. The molecular biology of the yeast Saccharomyces: Metabolism and gene expression. Plainview, New York: Cold Spring Harbor Laboratory; 1982. pp. 159–180. [Google Scholar]
- Oshima Y. The phosphatase system in Saccharomyces cerevisiae. Genes Genet Syst. 1997;72:323–334. doi: 10.1266/ggs.72.323. [DOI] [PubMed] [Google Scholar]
- Steger DJ, Haswell ES, Miller AL, Wente SR, O'Shea EK. Regulation of chromatin remodeling by inositol polyphosphates. Science. 2003;299:114–116. doi: 10.1126/science.1078062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- Wykoff DD, O'Shea EK. Phosphate transport and sensing in Saccharomyces cerevisiae. Genetics. 2001;159:1491–1499. doi: 10.1093/genetics/159.4.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]