Abstract
In eukaryotic cells the TATA-binding protein (TBP) associates with other proteins known as TBP-associated factors (TAFs) to form multisubunit transcription factors important for gene expression by all three nuclear RNA polymerases. Computer searching of the complete Saccharomyces cerevisiae genome revealed five previously unidentified yeast genes with significant sequence similarity to known human and Drosophila RNA polymerase II TAFs. Each of these genes is essential for viability. A sixth essential gene (FUN81) has previously been noted to be similar to human TAFII18. Coimmunoprecipitation experiments show that all six proteins are associated with TBP, demonstrating that they are true TAFs. Furthermore, these proteins are present in complexes containing the TAFII130 subunit, indicating that they are components of TFIID. Based on their predicted molecular weights, these genes have been designated TAF67, TAF61(68), TAF40, TAF23(25), TAF19(FUN81), and TAF17. Yeast TAF61 is significantly larger than its higher eukaryotic homologues, and deletion analysis demonstrates that the evolutionarily conserved, histone-like domain is sufficient and necessary to support viability.
The TATA-binding protein (TBP) is a key element in transcription by all three eukaryotic RNA polymerases (1). TBP binds a set of TBP-associated factors (TAFs) to form the transcription factor TFIID, the core factor involved in RNA polymerase II transcription (2). Other subsets of TAFs associate with TBP in different complexes (SL1, TFIIIB, SNAPc) involved in production of RNA polymerase I, RNA polymerase III, and snRNA transcripts, respectively (1). In addition, there is evidence that TBP associates with various groups of TAFs to form TBP complexes distinct from TFIID but of undetermined function (3, 4).
Most information about the function of TAFs has been obtained from in vitro studies on partially purified mammalian and Drosophila factors (5). These studies suggest that TAFs are essential for the response to transcriptional regulatory proteins but not for basal transcription in vitro (6). More recently, several TAFs have been identified in yeast extracts (7–9) and studied in vivo (10, 11). These in vivo analyses have shown that TAFs are important for transcription of specific genes but are not universally required for transcriptional response to activator proteins. In particular, studies both in yeast (11, 12) and in a hamster cell line with conditional expression of TAF250 (13) implicate TAFs in the correct expression of cell-cycle genes. Other experiments show TAFs to be important for the expression of some genes whose transcription is directed by weak TATA elements or initiator elements (10, 14). Taken together, the results of these in vitro and in vivo studies suggest that TAFs play important gene-specific roles in transcription and are potential, but not obligate, targets of trans-activating proteins.
Yeast TBP was originally purified as a single protein (15–18). This stands in contrast to its mammalian counterpart, which remains tightly associated with TAFs throughout multiple chromatographic purification steps. However, affinity purification experiments revealed the presence of a constellation of factors associated with TBP in yeast (7, 8). Cloning of the genes for several of these proteins revealed them to be highly similar in sequence to the known mammalian and Drosophila TAFs. Thus far, genes have been identified that encode yeast TAF130/145, TAF90, TAF60 (7, 8), and TAF25 (9). In addition, an open reading frame with unknown function (FUN81; ref. 19) has been noted to resemble human TAFII18 (20). However, this yeast protein has not previously been shown to associate with TBP.
Assuming the composition of the yeast TFIID complex to be similar to that of mammalian and Drosophila TFIID, we expected that there would be yeast homologues of the other known higher eukaryotic TAFs. The completed sequencing of the Saccharomyces cerevisiae genome (21) allowed us to search for potential homologues of the remaining known TAFs by protein sequence similarity. Here we report the identification of several previously unknown yeast proteins as true yeast TAFs associated with the TFIID complex.
MATERIALS AND METHODS
Sequence Analysis.
Protein sequence similarity searching was performed by blast (22) search of the complete yeast genome [Saccharomyces Genome Database (SGD), Stanford University]. Alignments were made using the gcg and seqvu programs.
TAF Gene Cloning.
The putative TAF genes were amplified by PCR using Vent polymerase (New England Biolabs) and cloned into yeast shuttle vectors using standard molecular biology techniques (23).
Gene Disruptions.
Candidate TAF gene deletions were performed in the diploid yeast strains YSB286 (MATa/MATα, ura3-52/ura3-52, leu2Δ1/leu2-3, 112, his3Δ200/his3Δ200, TRP1/trp1Δ63) or KY320 (MATa/MATα, ade2-101/ade2-101, his3Δ200/his3Δ200, leu2::PET56/leu2::PET56, trp1Δ1/trp1Δ1, ura3-52/ura3-52) by standard one-step gene disruption-deletion using either LEU2 or ADE2 as the disrupting marker (24). Disruption of one copy of the gene of interest was verified by Southern blotting in all cases. The heterozygous diploids were sporulated and tetrads were dissected onto rich growth medium to determine viability of the resulting haploids. Details of the disruption constructs will be provided on request.
Creation of Epitope-Tagged Alleles.
The plasmid ZM253 (HIS3, CEN/ARS), containing the inducible GAL1 promoter upstream of three copies of the influenza hemagglutinin (HA) epitope (25), was constructed to allow immunological detection of the TAF proteins. Each putative TAF protein coding sequence was cloned into this vector in frame with the triple epitope tag. The resulting plasmids were each introduced into a haploid strain containing a chromosomal deletion of the same gene using plasmid shuffling (26) on plates containing 2% galactose and 5-flouro-orotic acid (FOA). In each strain, the chromosomal deletion of the gene was fully complemented by the epitope-tagged copy of the gene. All strains required galactose for growth.
Immunoprecipitations and Western Blotting.
Each strain expressing an epitope-tagged TAF homologue (or epitope-tagged Fus3 as a control) was grown to mid-logarithmic phase in 100 ml of YP medium (1% yeast extract/2% bacto-peptone) containing 2% galactose and 0.2–0.5% dextrose. Cells were harvested and whole-cell extracts were made by glass bead lysis in 450 mM Tris·acetate (pH 7.8), 150 mM potassium acetate, 60% (vol/vol) glycerol, 3 mM EDTA, 3 mM dithiothreitol, and 1 mM phenylmethylsulfonyl fluoride (27). In early experiments, yeast whole-cell extracts were further processed by chromatography on BioRex 70 (Bio-Rad) as described (27), and the BioRex 70 0.6 M eluate was used for immunoprecipitations (see Fig. 3, TAF17 and TAF40). This fractionation was subsequently found to be unnecessary, and all further experiments were performed with unfractionated whole-cell extract. TAF17 and TAF40 immunoprecipitations from whole-cell extract yielded the same result as shown in Fig. 3.
Figure 3.
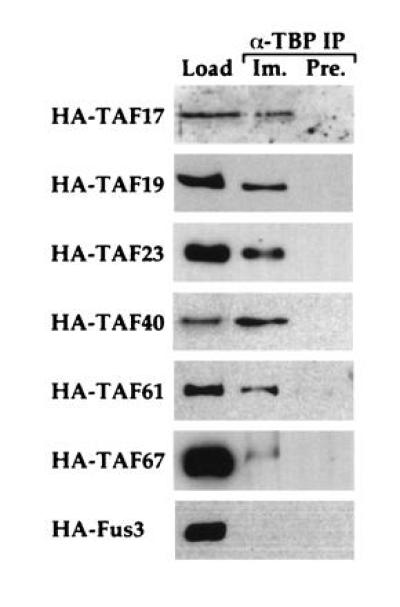
Candidate yeast TAFs are associated with TBP. Extracts were prepared from strains carrying epitope-tagged alleles of the indicated proteins and immunoprecipitated with anti-TBP antibodies. The TBP-associated proteins were blotted and probed with the monoclonal antibody recognizing the epitope tag. Lanes: Load, 30 μg of total extract; Im., immunoprecipitate from 300 μg of total extract with anti-TBP; Pre., immunoprecipitate from 300 μg of total extract with preimmune serum. Fus3 serves as a non-TAF negative control.
Immunoprecipitations were performed according to a modification of a protocol provided by N. Kuldel (personal communication). Reactions contained 100–300 μg of protein extract. The sample volumes were adjusted to 500 μl with buffer A [20 mM Hepes, pH 7.6/20% (vol/vol) glycerol/1 mM dithiothreitol/1 mM EDTA] containing 125 mM potassium acetate, and Nonidet P-40 was added to a final concentration of 1%. The protein extract was precleared by incubation for 1 hr with 50 μl of preswollen 10% (vol/vol) Protein A Sepharose beads (Sigma) in buffer A. Protein A beads were coupled to either rabbit polyclonal α-TBP or to rabbit preimmune serum as described (28). Reactions of extract with 50 μl (10%, vol/vol) of coupled beads were allowed to proceed overnight at 4°C on a rotater apparatus. Antigen–antibody complexes were recovered by centrifugation and were washed five times with 1 ml of buffer A containing 125 mM KOAc and 1% Nonidet P-40. Samples were boiled in SDS/PAGE loading buffer and subjected to SDS/PAGE analysis.
Proteins were resolved on 10% or 12% acrylamide gels and then electroblotted to nitrocellulose. Antibody probing and washing was performed according to standard techniques (28), and enhanced chemiluminescent detection was used to visualize bands as recommended by the manufacturers (Amersham; Kirkegaard & Perry Laboratories).
Deletion Analysis of TAF61.
Deletion alleles of TAF61 were constructed using both PCR cloning and existing restriction sites in the TAF61 gene and were inserted into a yeast TRP1, CEN/ARS plasmid. The plasmids were transformed into a haploid strain containing a chromosomal disruption of TAF61 complemented by a URA3 plasmid bearing the wild-type TAF61 gene. The resulting transformants were transferred to FOA-containing medium (26) to determine whether each deletion plasmid could support viability.
RESULTS
Yeast Homologues of Human and Drosophila TAFs.
Protein similarity searches (22) of the complete S. cerevisiae genome were carried out using all published mammalian and Drosophila RNA polymerase II TAFs as query sequences. Each search revealed a unique yeast sequence with significant similarity to the known mammalian or Drosophila TAF. The one exception was Drosophila TAFII110 (29, 30), for which no similar yeast sequence was found. For simplicity, we have designated each gene according to the predicted molecular weight of the encoded protein. Sequence alignments are shown in Fig. 1.
Figure 1.
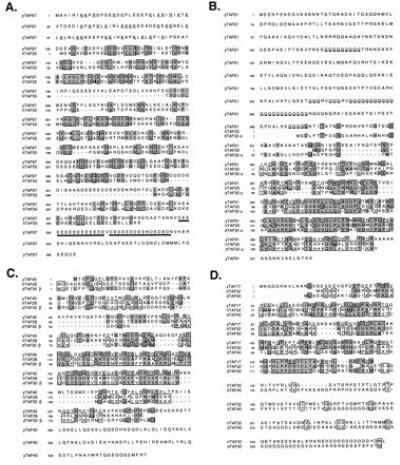
Candidate yeast TAF proteins are similar to known human and Drosophila TAF proteins. (A) Yeast TAF67. Basic residues in the amino-terminal region are underscored; acidic stretches in the carboxyl-terminal region are underlined. (B) Yeast TAF61. Polyglutamine stretches are underscored. (C) Yeast TAF40. (D) Yeast TAF17.
Human TAFII55 (31, 32), for which no Drosophila homologue has yet been described, is similar to a yeast open reading frame encoding a protein of predicted molecular mass 67 kDa (Fig. 1A). Relative to the human protein, the yeast protein (SGD accession no. YMR227C) has additional nonhomologous protein sequences at both the amino and carboxyl termini. Interestingly, the amino-terminal extension is rather basic, while the carboxyl-terminal extension is quite acidic.
Searches with human TAFII28 (20), which has been implicated in the transcriptional response to retinoic acid receptor (33), revealed a predicted yeast protein of 40 kDa (SGD accession no. YML015C). The same yeast protein was also matched with searches using the Drosophila homologue TAFII30β (34). The yeast protein is slightly larger than its counterparts, with the nonoverlapping sequences appearing at the carboxyl terminus of the protein (Fig. 1C).
Human TAFII30 occurs in a subset of TFIID and is thought to be a potential target for the estrogen receptor AF-2 activation domain (35). No Drosophila homologue is known, but a sequence search against the yeast genome uncovered a predicted protein of 23 kDa (SGD accession no. YDR167W) with significant similarity to the human protein. This protein was recently independently shown to be a component of yeast TFIID (9) with an observed molecular mass of 25 kDa and was thereby designated TAF25. The alignment of the human and yeast proteins is presented in that paper.
Several of the higher eukaryotic TAFs show some sequence similarity to histone proteins (2). The relevance of these alignments is supported by structural studies showing that the histone-like regions of Drosophila TAF62(60) and TAF42(40) adopt a histone-fold arrangement resembling the histone H3/H4 pair (2). Although a yeast homologue of Drosophila TAF62(60) has been described (7, 8), none have previously been reported for the other histone-like TAFs. Similarity searches with dTAFII42(40) or its human homologue TAFII32 match a yeast open reading frame with a predicted molecular mass of 17 kDa (SGD accession no. YMR236W). Interestingly, the sequence similarity includes the histone H3-like domain and an additional region of strong identity between species (Fig. 1D). However, the yeast protein completely lacks a carboxyl-terminal region that is not conserved between the human and Drosophila proteins.
The putative histone H2B-like TAFs (human TAFII20 (20, 36) and Drosophila TAFII30α or 22/28 (34)) match a predicted yeast protein of 61 kDa (SGD accession no. YDR145W). We note that this protein has been independently cloned as a component of TFIID with an observed molecular mass of 68 kDa (11). The yeast protein is significantly larger than its higher eukaryotic homologues, primarily due to an extra ≈300 aa in the amino-terminal half of the protein. This extra domain is predicted to be very hydrophilic and contains several polyglutamine stretches (Fig. 1B).
Candidate Yeast TAFs Are Essential for Viability.
To determine whether the new genes were essential for cell viability, we performed one-step gene disruptions in diploid yeast. Dissected tetrads from all strains yielded two live and two dead spores each (Fig. 2 and data not shown), with the surviving haploids always lacking the disruption marker. Therefore, each of the five yeast TAF homologue genes is essential for viability.
Figure 2.
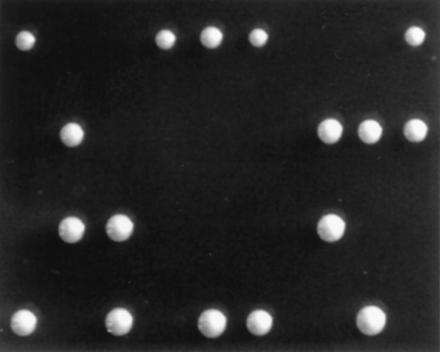
Candidate TAF genes are essential for yeast viability. The four dissected spores from each tetrad of a taf61Δ256::LEU2/TAF61 diploid yeast strain are arranged vertically. Identical results were obtained with deletions of all other candidate TAFs (data not shown).
Candidate Yeast TAFs Are Associated with TBP.
Immunoprecipitations with each of the putative yeast TAFs were performed to determine whether they were indeed associated with TBP in vivo. A HA epitope-tagged allele of each gene was created and introduced into a haploid yeast strain as the sole source of the given protein. Immunoprecipitations were performed by incubating yeast extracts with anti-TBP antibody-coupled beads or with preimmune serum-coupled beads. Bound proteins were subjected to Western blotting using a monoclonal antibody (12CA5) directed against the HA epitope (25). As shown in Fig. 3, each HA-tagged protein was found to be specifically associated with TBP, indicating that these proteins are all truly TAFs. Also tested in this experiment was the essential yeast gene FUN81 (19), which was previously noted to be similar to human TAFII18 (20), but which had not been shown to be associated with TBP. In light of its efficient interaction with TBP, we now refer to this protein as TAF19, in accord with its predicted molecular weight. As a negative control, extract from a strain containing an epitope-tagged Fus3 protein (37) was immunoprecipitated in parallel and failed to bind TBP.
We also performed the converse experiment, in which 12CA5-coupled beads were used for the original immunoprecipitation and then Western blots of the bound proteins were probed with anti-TBP antibody. Again, this experiment shows each of the new proteins to be associated with TBP (data not shown).
Under our experimental conditions, TAF67 appears weakly associated with TBP and requires significantly longer exposures to be detected. Reprobing of the Western blot in Fig. 3 with anti-TAF130 antibody indicates that TAF130 is present in equal amounts in all immunoprecipitations (data not shown), so that the lower affinity of TAF67 for TBP is not a result of failure of this particular immunoprecipitation. We cannot distinguish from this experiment whether there is actually a lower relative amount of TAF67 associated with TBP in vivo, or whether the TAF67 interaction is particularly weak and susceptible to disruption under our experimental conditions.
Candidate Yeast TAFs Are Components of TFIID.
Having determined that the new genes encode true TAFs, we set out to determine whether these TAFs might, like their mammalian and Drosophila counterparts, be part of the TFIID complex. To this end, we probed 12CA5-precipitated proteins from each HA-tagged TAF strain with antibodies directed against TAF130, which has been shown to be an important scaffold for the assembly of TFIID. As shown in Fig. 4, this large TAF is associated with each epitope-tagged TAF, suggesting that the smaller TAFs are part of the TAF130-nucleated TFIID complex.
Figure 4.
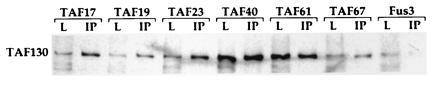
Candidate yeast TAFs are associated with TFIID. Extracts were prepared from strains carrying epitope-tagged alleles of the indicated proteins and immunoprecipitated with the 12CA5 monoclonal antibody. To determine whether the candidate yeast TAFs were associated with TFIID, the precipitated proteins were blotted and probed with anti-TAF130. Lanes: L, 15 μg of the indicated extract; IP, immunoprecipitate from 150 μg of extract. Fus3 serves as a non-TAF negative control.
The Conserved Region of Yeast TAF61 Is Sufficient and Necessary to Support Viability.
As mentioned above, yeast TAF61 is significantly larger than its human or Drosophila homologues, with the homology being entirely confined to the carboxyl terminus of the yeast protein. This domain includes the region similar in sequence to histone H2B. To determine the importance of the nonhomologous region, we created several truncated alleles of this TAF and used plasmid shuffling to analyze their ability to promote normal cell growth. As shown in Fig. 5, the amino-terminal two-thirds of this protein may be deleted without any adverse effects on cell growth, whereas carboxyl-terminally deleted alleles are unable to support life. Strains with amino-terminally deleted alleles as the only source of TAF61 show no phenotypic abnormalities at low (15°C) or high (37°C) temperature or on medium containing galactose, raffinose, or copper sulfate.
Figure 5.
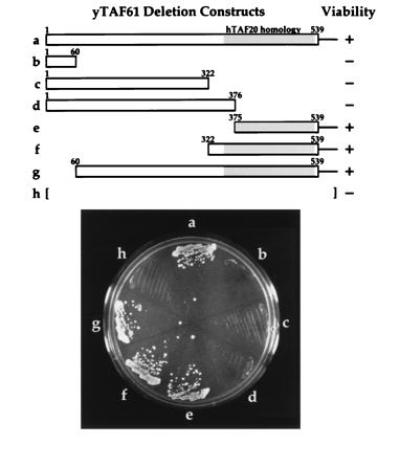
The carboxyl-terminal domain of yeast TAF61 is necessary and sufficient to support viability. Plasmid shuffling was used to introduce constructs containing the indicated amino acids into a yeast strain with a chromosomal deletion of the TAF61 gene. Shading indicates the region of the protein with similarity to human TAFII20 and Drosophila TAF30α.
DISCUSSION
We have identified five essential yeast genes and the previously known yeast gene FUN81 as bona fide yeast TAFs. Two of these (TAF23 and TAF61) have been independently identified in TBP-containing complexes as proteins with apparent molecular masses of 25 and 68 kDa, respectively (9, 11). Previously reported preparations of TBP-associated factors (7, 8) also contain proteins with apparent molecular masses that could be consistent with those of TAF40 and TAF67. Small molecular weight proteins corresponding to TAF17 and TAF19/FUN81 were not reported, but they may not have been visible in the gel systems used.
With one exception (see below), every published higher eukaryotic TAF has a unique, essential yeast homologue (Table 1). Human TAFII18 is similar to two yeast genes: TAF19/FUN81 and SPT3 (20). However, unlike TAF19/FUN81, SPT3 is not essential for yeast viability (39) and is less similar to the human protein. Therefore, it is clear that yeast TAF19 fulfills a role that is not completely redundant with that of Spt3. The one higher eukaryotic TAF for which there is no yeast homologue is Drosophila TAFII110, which has been implicated by in vitro experiments in the response to the glutamine-rich activator SP1 (29). The absence of a yeast homologue to Drosophila TAFII110 is in accord with the inability of glutamine-rich activators to function in yeast (40) and suggests that perhaps a lack of appropriate coactivator(s) prevents this class of activators from functioning in yeast. We note that the yeast TAF61 sequence contains long polyglutamine stretches reminiscent of those found in dTAF110; however, these sequences can be completely deleted with no obvious effects on cell growth (Fig. 5).
Table 1.
TFIID subunit homologies across species
| Yeast | Human | Drosophila |
|---|---|---|
| TSM1 | 150 | |
| TAF130/145 | 250 | 230 |
| None | 135 | 110 |
| TAF90 | 100 | 80 |
| TAF67 | 55 | |
| TAF61 (68) | 20/15 | 30α (p28/p22) |
| TAF60 | 70 | 60 (62) |
| TAF40 | 28 | 30β |
| TAF30 (TFG3/ANC1) | AF-9/ENL | |
| TAF23 (25) | 30 | |
| TAF19 (FUN81) | 18 | |
| TAF17 | 32 | 40 (42) |
In fact, the only region of TAF61 necessary for cell growth is the carboxyl-terminal region, which is homologous to the much smaller higher eukaryotic TAFs and to histone H2B. The histone-like structure of some TAFs has been proposed to be functionally important, perhaps in TFIID assembly (36, 41) or in DNA wrapping around TFIID (2). Our observation that this is the essential domain of the yeast TAF61 protein supports the idea of a critical role for this histone-like motif in TAF function.
The TFIID TAF subunits are widely believed to mediate transcriptional activation. The ability of yeast cells to activate transcription after TAF depletion (10, 11) might suggest that other proteins have functional redundancy with TAFs in vivo. However, the finding that each higher eukaryotic TFIID subunit has a single homologue in the yeast genome implies that any functional redundancy in response to activators is not due to multiple copies of TAF genes. Because the TAF genes are essential for viability, TAFs must also have other unique critical functions in the cell. These functions remain to be determined.
Acknowledgments
We thank Elaine Elion for the epitope-tagged Fus3 strain, Joseph Reese and Michael Green for the TAF130 antibody, Natalie Kuldell and Joseph Geisberg for advice on immunoprecipitations, and Marie Keaveney, Laurie Stargell, and Bertha Michel for discussion. This work was supported by grants to K.S. (GM30186) and to S.B. (GM46498) from the National Institutes of Health and also by the American Cancer Society and the Pew Scholars Program.
Footnotes
Abbreviations: TBP, TATA-binding protein; TAF, TBP-associated factor; SGD, Saccharomyces Genome Database; HA, hemagglutinin.
References
- 1.Hernandez N. Genes Dev. 1993;7:1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- 2.Burley S K, Roeder R G. Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 3.Timmers H T, Meyers R E, Sharp P A. Proc Natl Acad Sci USA. 1992;89:8140–8144. doi: 10.1073/pnas.89.17.8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poon D, Campbell A M, Bai Y B, Weil P A. J Biol Chem. 1994;269:23135–23140. [PubMed] [Google Scholar]
- 5.Goodrich J A, Tjian R. Curr Opin Cell Biol. 1994;6:403–409. doi: 10.1016/0955-0674(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 6.Chen J L, Attardi L D, Verrijzer C P, Yokomori K, Tjian R. Cell. 1994;79:93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- 7.Reese J C, Apone L, Walker S S, Griffin L A, Green M R. Nature (London) 1994;371:523–527. doi: 10.1038/371523a0. [DOI] [PubMed] [Google Scholar]
- 8.Poon D, Bai Y, Campbell A M, Bjorklund S, Kim Y J, Zhou S, Kornberg R D, Weil P A. Proc Natl Acad Sci USA. 1995;92:82224–82228. doi: 10.1073/pnas.92.18.8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klebanow E R, Poon D, Zhou S, Weil P A. J Biol Chem. 1996;271:13706–13715. doi: 10.1074/jbc.271.23.13706. [DOI] [PubMed] [Google Scholar]
- 10.Moqtaderi Z, Bai Y, Poon D, Weil P A, Struhl K. Nature (London) 1996;383:188–191. doi: 10.1038/383188a0. [DOI] [PubMed] [Google Scholar]
- 11.Walker S S, Reese J C, Apone L M, Green M R. Nature (London) 1996;383:185–188. doi: 10.1038/383185a0. [DOI] [PubMed] [Google Scholar]
- 12.Apone L M, Virbasius C A, Reese J C, Green M R. Genes Dev. 1996;10:2368–2380. doi: 10.1101/gad.10.18.2368. [DOI] [PubMed] [Google Scholar]
- 13.Wang E H, Tjian R. Science. 1994;263:811–814. doi: 10.1126/science.8303298. [DOI] [PubMed] [Google Scholar]
- 14.Pugh B F, Tjian R. Genes Dev. 1991;5:1935–1945. doi: 10.1101/gad.5.11.1935. [DOI] [PubMed] [Google Scholar]
- 15.Hahn S, Buratowski S, Sharp P A, Guarente L. Cell. 1989;58:1173–1181. doi: 10.1016/0092-8674(89)90515-1. [DOI] [PubMed] [Google Scholar]
- 16.Horikoshi M, Wang C K, Fujii H, Cromlish J A, Weil P A, Roeder R G. Nature (London) 1989;341:299–303. doi: 10.1038/341299a0. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt M C, Kao C C, Pei R, Berk A J. Proc Natl Acad Sci USA. 1989;86:7785–7789. doi: 10.1073/pnas.86.20.7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cavallini B, Faus I, Matthes H, Chipoulet J M, Winsor B, Egly J M, Chambon P. Proc Natl Acad Sci USA. 1989;86:9803–9807. doi: 10.1073/pnas.86.24.9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dubois E, Bercy J, Descamps F, Messenguy F. Gene. 1987;55:265–275. doi: 10.1016/0378-1119(87)90286-1. [DOI] [PubMed] [Google Scholar]
- 20.Mengus G, May M, Jacq X, Staub A, Tora L, Chambon P, Davidson I. EMBO J. 1995;14:1520–1531. doi: 10.1002/j.1460-2075.1995.tb07138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams N. Science. 1996;272:481. doi: 10.1126/science.272.5261.481. [DOI] [PubMed] [Google Scholar]
- 22.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 23.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1991. [Google Scholar]
- 24.Guthrie C, Fink G R. Methods Enzymol. 1991;194:1–933. [Google Scholar]
- 25.Wilson I, Niman H, Houghten R, Cherenson A, Connolly M, Lerner R. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- 26.Boeke J, Truehart J, Natsoulis B, Fink G R. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 27.Sayre M H, Tschochner H, Kornberg R D. J Biol Chem. 1992;267:23376–23382. [PubMed] [Google Scholar]
- 28.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 29.Hoey T, Weinzerl R O J, Gill G, Chen J L, Dynlacht B D, Tjian R. Cell. 1993;72:247–260. doi: 10.1016/0092-8674(93)90664-c. [DOI] [PubMed] [Google Scholar]
- 30.Kokubo T, Gong D W, Roeder R G, Horikoshi M, Nakatani Y. Proc Natl Acad Sci USA. 1993;90:5896–5900. doi: 10.1073/pnas.90.13.5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiang C M, Roeder R G. Science. 1995;267:531–536. doi: 10.1126/science.7824954. [DOI] [PubMed] [Google Scholar]
- 32.Lavigne A C, Mengus G, May M, Dubrovskaya V, Tora L, Chambon P, Davidson I. J Biol Chem. 1996;271:19774–19780. doi: 10.1074/jbc.271.33.19774. [DOI] [PubMed] [Google Scholar]
- 33.May M, Mengus G, Lavigne A C, Chambon P, Davidson I. EMBO J. 1996;15:3093–3104. [PMC free article] [PubMed] [Google Scholar]
- 34.Yokomori K, Chen J L, Admon A, Zhou S, Tjian R. Genes Dev. 1993;7:2587–2597. doi: 10.1101/gad.7.12b.2587. [DOI] [PubMed] [Google Scholar]
- 35.Jacq X, Brou C, Lutz Y, Davidson I, Chambon P, Tora L. Cell. 1994;79:107–117. doi: 10.1016/0092-8674(94)90404-9. [DOI] [PubMed] [Google Scholar]
- 36.Hoffmann A, Roeder R G. J Biol Chem. 1996;271:18194–18202. doi: 10.1074/jbc.271.30.18194. [DOI] [PubMed] [Google Scholar]
- 37.Elion E A, Grisafi P L, Fink G R. Cell. 1990;60:649–664. doi: 10.1016/0092-8674(90)90668-5. [DOI] [PubMed] [Google Scholar]
- 38.Dubrovskaya V, Lavigne A C, Davidson I, Acker J, Staub A, Tora L. EMBO J. 1996;15:3702–3712. [PMC free article] [PubMed] [Google Scholar]
- 39.Eisenmann D M, Arndt K M, Ricupero S L, Rooney J W, Winston F. Genes Dev. 1992;6:1319–1331. doi: 10.1101/gad.6.7.1319. [DOI] [PubMed] [Google Scholar]
- 40.Ponticelli A S, Pardee T S, Struhl K. Mol Cell Biol. 1995;15:983–988. doi: 10.1128/mcb.15.2.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffmann A, Chiang C M, Oelgeschlager T, Xie X, Burley S K, Nakatani Y, Roeder R G. Nature (London) 1996;380:356–359. doi: 10.1038/380356a0. [DOI] [PubMed] [Google Scholar]


