Abstract
We have found that it is possible to use labeled peptide nucleic acid (PNA)-oligomers as probes in pre-gel hybridization experiments, as an alternative for Southern hybridization. In this technique, the PNA probe is hybridized to a denatured DNA sample at low ionic strength and the mixture is loaded directly on to an electrophoresis system for size separation. Ensuing gel electrophoresis separates the single-stranded DNA fragments by length. The neutral backbone of PNA allows for hybridization at low ionic strength and imparts very low mobility to excess PNA. Detection of the bound PNA is possible by direct fluorescence detection with capillary electrophoresis, or the DNA/PNA hybrids can be blotted onto a membrane and detected with standard chemiluminescent techniques. Efficient single bp discrimination was achieved routinely using both capillary and slab-gel electrophoresis.
Keywords: cystic fibrosis, capillary electrophoresis, fluorescence, chemiluminescence, single base-pair discrimination
The ability to rapidly identify specific sequences in DNA samples is becoming more important as links between phenotypes and genotypes are established. Two areas in which the correlation between phenotype and genotype is becoming increasingly well understood are oncology and genetic disorders (1–3). Rapid analysis methods for the presence or absence of a specific sequence will further help establish correlations between genotypes and phenotypes. Moreover, the rapidly expanding database of sequence information will lead to increased needs for genetic analysis methods in areas such as studies of gene expression, pathogen detection, and diagnostics.
Southern hybridization (4) is a technique often used to analyze genetic loci, using fragments of defined length derived from either PCR or restriction enzyme digestion. Southern hybridization derives its great utility from the ability to provide both size and sequence information. Sequence detection is supplemented with information regarding the genetic context. While Southern hybridization is a core technology in molecular biology, it has disadvantages that limit its application for screening large numbers of DNA samples. The Southern technique is a laborious multistep procedure, and sequence discrimination between closely related sequences is often difficult to obtain on a routine basis.
In our continued studies of the properties of peptide nucleic acid (PNA) (5, 6), a DNA mimic with a neutral peptide-like backbone, we have been working with labeled PNA oligomers in hybridization procedures. The hybridization of PNA oligomers to complementary DNA is essentially independent of the ionic strength due to the neutral backbone of PNA. Thus a PNA oligomer will hybridize to a complementary DNA oligomer under conditions where DNA/DNA hybridization is strongly disfavored, i.e., at low ionic strength (6). Also, since the backbone is neutral, the electrophoretic mobility of PNA oligomers will be determined by their size, and the charge (if any) contributed by the label. As such, they will generally migrate much more slowly than DNA in an electric field (6–8).
In an attempt to dramatically simplify the Southern procedure, we have studied PNA pre-gel hybridization—i.e., hybridization of a labeled PNA probe to a double-stranded DNA sample that has been denatured at low ionic strength prior to electrophoresis. Here the electrophoretic separation is performed on single-stranded DNA fragments with the target sequence carrying bound PNA. This procedure eliminates the postseparation, probing and washing steps that make the Southern procedure cumbersome. The affinity between long complementary DNA oligomers is very high even at low ionic strength (9). DNA reannealing will expel bound PNA probe that is much shorter. The success of these experiments is therefore dependent on a sufficiently slow reannealing rate for the DNA fragments.
In this paper we describe studies demonstrating the feasibility of the PNA pre-gel hybridization technique. We studied two different electrophoresis formats, slab-gel, and capillary electrophoresis (CE). DNA samples from pBR322, M13, and lambda were used as targets in our initial studies. The specificity and general utility of the PNA pre-gel hybridization technique was demonstrated by developing an analysis for the presence of the single bp mutation W1282X in cystic fibrosis carriers (10, 11).
MATERIALS AND METHODS
Reagents.
Reagents included casein (#C-5890, Sigma); pBR322 MspI digest (#303-2S, New England Biolabs) and BstNI digests (#301-4S, New England Biolabs); M13 DNA (#770704, United States Biochemical); control DNA (G304A 5491901, Promega); W1282X carrier DNA (NAO454OA, Coriell Cell Repository, Camden, NJ); and Immobilon-S (IMS02, Millipore).
PNA Synthesis.
PNA oligomers were synthesized on an Expedite Nucleic Acid Synthesis system (PerSeptive Biosystems) according to a published solid-phase methodology (12). Biotin or fluorescein labeling of the resin-bound PNA was carried out using dimethoxy trityl-biotin-phenyl pyrazolanine ester or 5,6-carboxyfluorescein N-hydroxy-succinimide (Molecular Probes), respectively. The labeled PNAs were then cleaved from the resin, and protecting groups were removed using trifluoromethane sulfonic acid/trifluoracetic acid/m-cresol/thioanisole (2:6:1:1) for 2 hr at room temperature. The PNA was precipitated from the filtrate by addition of anhydrous ether. The crude PNA samples were purified by HPLC on a Deltapak C18 column (Waters) and their mass confirmed by matrix-assisted laser desorption ionization-time of flight (Voyager, PerSeptive Biosystems). The samples used for CE were further purified on Sephadex G-25 to remove fluorescent impurities.
The sequences synthesized were (N → C terminus); an O denotes the insertion of the spacer unit, 8-amino-3,6-dioxaoctanoic acid. pBR322: biotin-OO-ATG CAG GAG TCG CAT. Lambda: fluorescein-OO-GGT CAC TAT CAG TCA. M13: fluorescein-OO-TTT TCC CAG TCA CGA, fluorescein-OO-TTT TCC CAG GCA CGA, and fluorescein-OO-TTT TCA CAG GCA CGA. W1282 (wild type): fluorescein-OO-CTT TCC TCC ACT GTT. W1282X (mutant): fluorescein-OO-CTT TCC TTC ACT GTT.
DNA Oligomers.
All DNA oligomers (cartridge purified) were obtained from Genosys (The Woodlands, TX). Primer sequences for the cystic fibrosis assay: forward primer 5′-TCA CTT TTA CCT TAT AGG TGG GC and reverse primer 5′-TTC TGG CTA AGT CCT TTT GCT CA as suggested in Mashal et al. (13).
PCR.
A 30-cycle touchdown PCR (14) was used with a beginning annealing temperature of 70°C and an ending temperature of 61°C; with twenty additional cycles that anneal at 61°C. The PCR products were desalted using G50 TBE columns from Boehringer Mannheim (no. 1523 015) before hybridization for CE experiments.
PNA stock solutions were in 0.1% trifluoracetic acid. These stock solutions were then diluted with water to the appropriate concentration.
Slab Gel Analysis: Pre-Gel Hybridization.
In a typical experiment 0.5–1.0 μl of target DNA from a PCR (≈25 ng or 0.2 pmol of DNA) or plasmid (≈2–50 ng DNA) was transferred to an Eppendorf tube. Fluoresceinated PNA (0.5–1.0 pmol), and 1 μl of 250 mM Tris (pH 8.0) was added and the total volume was brought to either 10 or 20 μl with MilliQ water. The tube was placed in a heating block set at 95°C for 10 min. The tubes were left at ambient temperature, 10 min to 2 hr as specified in the text. Loading dye was added and samples were electrophoresed in a 1.5% TBE agarose gel for 25 min to 3 hr depending on the required resolution. The gel was immediately set up for a Southern transfer to a nylon membrane. The following day the membrane was dried and UV crosslinked at 254 nm.
Detection.
Chemiluminescence detection of biotinylated PNAs: Crosslinked membranes were placed in heat-sealable bags and were detected using the Photope kit (no. 7006, New England Biolabs) according to the manufacturers directions. Chemiluminescent detection of fluoresceinated PNAs: Crosslinked membranes were placed in heat-sealable bags and were detected using an antifluorescein antibody directly conjugated with alkaline phosphatase Dako (no. K046) according to the manufacturer’s directions. Finally, the membranes were rinsed in five volumes of wash II (10 mM Tris, pH 9.5/10 mM NaCl/1 mM MgCl2) for 2 min, and Lumigen solution (Phototope #7006, New England Biolabs) was made and used according to the manufacturers directions. The membranes were exposed to Fuji RX film.
CE.
CE–LIF experiments were performed on a P/ACE 2050 CE instrument with a Laser Module 488 argon ion laser (Beckman).
Free Solution CE.
A 50-μm i.d. polyacrylamide-coated capillary (27 cm in length and 20 cm to detector) was used with the cathode on the injection side and a run voltage of 10 kV. Laser-induced fluorescence detection was performed with excitation at 488 nm and emission at 520 nm. The buffer contained Tris·borate (pH 8.0) with 7 M urea buffer. Injection was by 10-sec gravity injection.
Restriction Fragment Analysis.
UV absorbance was measured at 260 nm. Separations were performed with the cathode on the injection side and a run voltage of 5 kV. The capillary was 75 μm i.d. uncoated (39 cm long, 32 cm to detector). The buffer contained Tris·borate with 7 M urea with 0.5% polyethylene oxide (PEO) (8 MDa). pBR322 experiments with LIF detection were performed with excitation at 488 nm and emission at 520 nm. The capillary was 75 μm i.d. uncoated capillary with 38 cm total length and 31 cm to detector. Buffer [1× TBE buffer/1% PEO (4 MDa)] was injected in a 20-sec electrokinetic injection; voltage was 5 kV.
W1282X Experiments.
Performed with LIF detection as in the pBR322 restriction fragment experiments. The capillary was 75 μm i.d. uncoated (27 cm total length, 20 cm to detector).
RESULTS
In earlier work we observed that PNA hybridizes very quickly to membrane bound DNA at low ionic strength (data not shown). Therefore, we expected that under conditions of low ionic strength, PNA would hybridize to a complementary sequence (if present) in heat denatured DNA, before the DNA could reanneal. Furthermore, we expected that these conditions would provide sufficient time for analysis before the DNA reannealed.
Another important observation made early in our study was that the high stability of the PNA/DNA duplex allows electrophoretic analysis of DNA with bound PNA under conditions that disrupt DNA/DNA binding. That is, dissociation of the PNA/DNA duplex occurs slowly under the conditions required for size separation of single-stranded DNA. This is illustrated in Fig. 1, showing the free solution CE separation of hybrid from excess fluorescently tagged PNA probe in a medium containing 7 M urea. Single-stranded M13 target was used in conjunction with a fluorescein-labeled PNA probe. Hybrid is specifically detected under these conditions and a large mobility difference is observed between the hybrid and the excess PNA.
Figure 1.
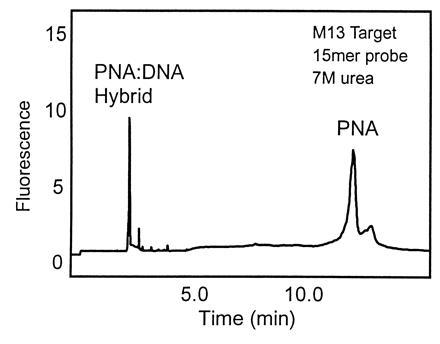
Free solution CE separation of PNA/DNA hybrid from excess PNA probe. M13 mp18 ssDNA 4.2 × 10−8 M and PNA probe 1.3 × 10−7 M. Detection: LIF 488/520 nm. Buffer: TBE, 7 M urea (pH 8.0).
Based on these results, we chose pBR322 as a model system to demonstrate the feasibility of the pre-gel hybridization technique (Fig. 2A). BstNI-digested pBR322 was heated for 10 min at 94°C with excess biotinylated PNA or DNA probes at low ionic strength. The solutions were moved to ambient temperature for 30 min, electrophoresed through a native agarose gel, and blotted to a nylon membrane. The presence of biotin was detected with standard chemiluminescent methods (Fig. 2B). Very little target sequence was detected with the DNA probe. In addition, because the DNA probe is charged, all of the excess probe is visualized in the assay. This is not the case with the PNA probe. Since neither the PNA nor the biotin carry any charge the excess probe does not migrate from the starting position.
Figure 2.
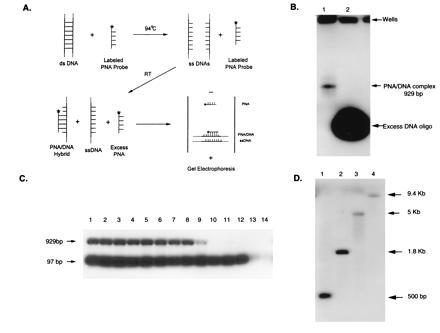
Pre-gel hybridization parameters. (A) Schematic diagram of pre-gel hybridization technique. (B) Comparison of 1 pmol each of biotinylated PNA and DNA pBR322 15-mer probes hybridized to 2 ng of BstNI-digested pBR322. Lanes: 1, PNA probe; 2, DNA probe. (C) The effect of different sodium ion concentrations on a mixture of MspI (50 ng) and BstNI (15 ng) digests of pBR322. One picomole of fluorescently labeled PNA was used. Lanes: 1, no salt; 2, 0.1 mM; 3, 0.5 mM; 4, 1 mM; 5, 5 mM; 6, 10 mM; 7, 20 mM; 8, 30 mM; 9, 40 mM; 10, 50 mM; 11, 75 mM; 12, 100 mM; 13, 250 mM; and 14, 500 mM. (D) Size separation hybridizing 1 pmol lambda fluorescein-tagged PNA probe and 50 ng of each digest. Lanes: 1, HindIII/BamHI; 2, EcoRI/BamHI; 3, BstEII; 4, HindIII.
We investigated three of the parameters critical to DNA reannealing: temperature, sodium ion concentration, and fragment length. It is critical that the DNA reannealing rate be sufficiently slow for pre-gel hybridization to work. The effect of the sodium ion concentration was investigated using a mixture of MspI and BstNI digests with two target fragments, 97 and 929 bp, respectively (Fig. 2C). Increasing the sodium ion concentration to 50 mM extinguished the signal from the 929-bp fragment whereas the signal from the 97-bp fragment was extinguished at 500 mM NaCl. The reannealing rate increases with increasing sodium ion concentration for both fragments and varies with fragment length.
We demonstrated that size information can be obtained, for single-strand DNA fragments, by using a lambda-specific PNA probe, to hybridize to different lambda DNA digests. Many different fragment lengths were generated for each digest, however only one specific band was recognized in each case. These include 500-bp 1.8-, 5-, and 9.4-kb bands (Fig. 2D).
The pBR322 pre-gel hybridization experiments were also performed with CE (15). For size separation, a sieving medium of 1% polyethylene oxide was used. To demonstrate the size separation capability of the sieving system, a mixture of pBR322 MspI and BstNI digests were analyzed with UV detection (Fig. 3A). The pre-gel hybridization method was used in conjunction with fluorescence detection to analyze these restriction digests individually and as a mixture. With fluorescence detection, only the specific fragments hybridizing with the PNA probe are detected (Fig. 3B). Both size and specific sequence information are obtained.
Figure 3.
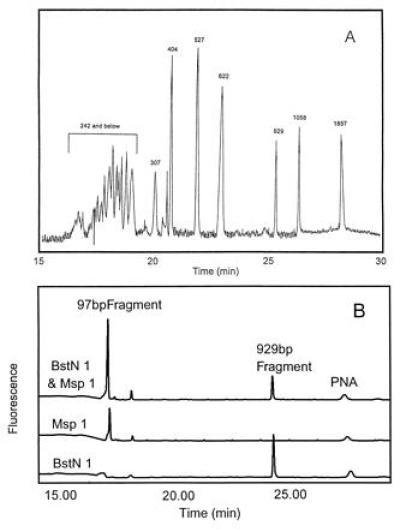
CE separation of a mixed MspI- and Bst NI-digested pBR322. (A) UV detection. Voltage: 5 kV. Detection: 260 nm. Buffer: TBE/7 M urea/0.5% PEO (8 MDa). (B) LIF detection. Voltage: 5 kV. Detection: LIF 488 nm/520 nm. Buffer: 1× TBE/1% PEO (4 MDa). Temperature 50°C.
The CE system permits the stringency of the analysis to be controlled by adjusting the temperature of the capillary and by selecting the concentration of the denaturants such as urea and formamide. To determine the discriminatory power of the probe assay in the CE-based system a medium containing 30% formamide was used. The fluorescence intensity of the hybrid signal was determined as a function of the capillary temperature when the above M13 probe was incubated with single-stranded M13. The same experiment was conducted with two other PNA sequences, containing one and two mismatches, respectively. The plots are overlaid in Fig. 4 and demonstrate very clear discrimination. Most significantly there is a 13°C interval where there is 100% signal from the perfectly matched probe with only baseline signal from the target containing a single mismatch.
Figure 4.
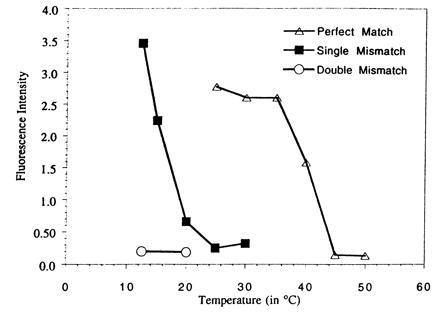
Fluorescence intensity of the PNA/DNA hybrid vs. separation temperature. fluorescein-labeled PNA probes with complementary (3′) Voltage: 20 kV. Detection: LIF 488/520 nm. Buffer: 1× TBE/30% formamide (pH 8.3). The following M13 probes were used: 5′ fluorescein-OO-TTT TCC CAG TCA CGA (perfect match), 5′-fluorescein-OO-TTT TCC CAG GCA CGA (single mismatch), 5′-fluorescein-OO-TTT TCA CAG GCA CGA (double mismatch).
The W1282X mutation in cystic fibrosis carriers was chosen to rigorously test the specificity of the PNA pre-gel hybridization technique. The W1282X is a single bp mutation where a C·G pair in position 4041 has been replaced with an T·A bp. Wild-type and mutant PNA probes labeled with fluorescein were compared in their ability to bind either DNA from an unaffected individual, or DNA from a heterozygous carrier of the W1282X mutation. Following PCR, the PNA wild-type probe and the mutant probe were incubated separately with the desalted PCR product. The analysis (Fig. 5 A and B) shows the DNA from an unaffected individual and a carrier of the W1282X mutation (one affected allele). The mutant PNA probe gives a positive signal only with the DNA sample from the individual known to be heterozygous for the mutation. When DNA from an unaffected individual is used, only the fluorescent impurities present in the PNA were detected with the mutant probe. The wild-type probe gave positive signals with both samples, with the signal from the heterozygote being approximately half that from the unaffected individual.
Figure 5.
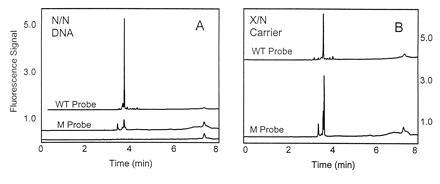
CE analysis with fluorescence detection with a wild-type (WT) W1282 and mutant (M) W1282X PNA probes of DNA from an unaffected individual (A) and DNA from a heterozygous individual (B). The lower trace in A is the probe background (no DNA). Voltage: 5 kV. Detection: LIF 488/520 nm. Buffer: 1× TBE/1% PEO (4 MDa).
We performed the same experiment in the slab-gel format. Fig. 6 demonstrates the power of discrimination even in this simple format. An increased PNA concentration led to an increase in the signal from the unaffected individual with the mutant probe (Fig. 6B). We also found there was no need to desalt the PCR product prior to analysis; a 20-fold dilution with distilled water was sufficient to bring the salt concentration into a workable range.
Figure 6.
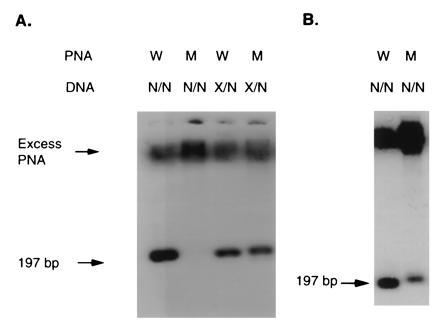
Analysis of W1282X mutation using slab gel format. (A) PCR DNA samples (25 ng) of either wild-type homozygous (N/N) or carrier heterozygous (X/N) were denatured and hybridized with 0.5 pmol of fluorescently labeled wild-type PNA, or mutant PNA. (B) The same DNA samples were hybridized with 10 pmol of either PNA probe (wild type, W) or (mutant, M) as indicated.
DISCUSSION
The initial experiments with BstNI-digested pBR322 suggested that pre-gel hybridization with PNA was feasible—i.e., that the reannealing rate of the DNA fragments was sufficiently slow for the PNA/DNA duplex to remain intact for the duration of the assay (Fig. 2B).
The rate of hybridization of PNA to DNA is high (unpublished data) and like the melting temperature of the PNA/DNA duplex can be presumed to be relatively independent of ionic strength (3). It is therefore reasonable to assume that an excess of PNA probe binds to its target sequence before the DNA can reanneal under the conditions employed here. Even at low ionic strength, however, the affinity between long complementary DNA oligonucleotides is very high and reannealing of the single-stranded target DNA will expel the bound PNA probe (7, §), Thus, the success of pre-gel hybridization is dependent on a sufficiently slow reannealing rate for the DNA fragments. The effect of increasing the sodium ion concentration provides us with insight on the reannealing rate of the DNA fragments (Fig. 2C). For example, the signal from a longer fragment is eliminated at lower salt concentration than for a shorter fragment. These results are in accordance with the general observation that longer DNA fragments reanneal faster than shorter fragments (9). In other words this is a nonequilibrium system, where a slow reannealing process competes with a fast binding reaction between the PNA probe and the target DNA.
In Southern-type analyses, size and sequence information are simultaneously obtained. To illustrate that the same information can be obtained by our method we analyzed various lambda DNA digests with target fragment sizes ranging from 500 bp to 9.4 kb (Fig. 2D). It is important to note that the separation is performed on single-strand DNA fragments so that single-stranded standards should be used to obtain accurate fragment length determination. Note that the PNA probe will not significantly alter the mobility of the fragments it binds to, and for example the binding of a 15-mer PNA to a 197-bp fragment only changes the mass/charge ratio by ≈6% and that of a 1-kb fragment by ≈1%. This is less than the resolving power of the agarose gels employed here. Standards that take this into account could be developed if accurate size determination is desired.
CE offers real-time detection of the separated species. We performed the pBR322 experiments with fluorescently tagged PNA probes using CE. A mixture of MspI and BstNI digests of pBR322 were resolved on a gel-filled capillary. Fig. 3A shows the separation of the fragments with UV detection. The equivalent of a Southern hybridization is obtained when pre-gel hybridization and fluorescence detection are employed. Here the two target fragments are detected individually and in combination (Fig. 3B). Under these conditions the slowly migrating PNA probes eluted much later. The analysis can be terminated when the DNA has eluted and the excess probe purged from the capillary when the sieving medium is replaced.
Using free-solution CE, PNA/DNA, and PNAs are readily resolved as illustrated with single-stranded M13 (Fig. 1). This mode has the advantage of very rapid analysis—i.e., analysis in minutes, but only limited information is obtained about length of the DNA fragment analyzed. We optimized the system for single bp mutation analysis. Using a second PNA probe with a single mismatch, and a third probe with two noncontiguous mismatches we studied the specificity as a function of separation temperature with 30% formamide in the medium (Fig. 4). We observed a difference of 25°C between the perfectly matched and single mismatched PNA probe. More significantly there was a 13°C interval where there was optimal signal from the perfectly matched PNA probe with only baseline signal from the PNA probe containing a single mismatch. This feature will allow for the construction of very robust assays for clinical research and diagnostics. In addition to offering very good control of the hybridization parameters, this format offers unique opportunities for automation.
Discrimination between alleles differing in only one or a few base pairs is inherently difficult and assays that are single bp specific operate with extremely narrow ranges of hybridization and washing temperatures (16). Often clinical researchers have to rely on the presence of an overlapping restriction site to consistently determine single bp mutations (2, 3). The W1282X mutation in cystic fibrosis carriers was used to test the utility of the method for a single base mutation in a clinically relevant assay.
The carrier in this example is heterozygous—i.e., there is one allele with a single base mutation and one unaffected allele. The fluorescein-labeled PNA probe for the unaffected allele (wild type) will hybridize with the target in both samples. The mutant probe, on the other hand, only hybridizes to the mutation containing allele of the carrier but not to either of the alleles from the unaffected individual (Fig. 5 A and B). This experiment was done under nondenaturing conditions at 50°C. For the carrier sample, both wild-type and mutant probes give signals with roughly half the magnitude of the signal from the homozygous normal with the wild type probe—i.e., the assay is quantitative.
While the stringency on the CE platform can be carefully controlled with the column temperature, this is not possible in an agarose gel. Even so, employing the standard conditions established for the analysis of pBR322, very efficient discrimination was observed as seen in Fig. 6. Increasing PNA probe concentration led to a decline in the specificity as expected, although a significant difference in the binding to unaffected and carrier DNA for the mutant probe was maintained. The intensities of the bands were also as predicted—i.e., the heterozygous samples gave about half the signal of the homozygous sample.
We believe that the efficient single bp discrimination observed here is due to two factors. First, PNAs are intrinsically more specific than the corresponding DNA probes (6, 17). Of equal importance is the short probe length. This highlights the significance of the ability to use shorter PNA probes than the 25–40 nucleotide units typically used for DNA probes. As a general rule, shorter probes are less unique but are inherently more specific, both in the case of PNA and DNA probes. Combined, these two effects provide the remarkable power of discrimination observed in this case.
Finally, as demonstrated in Fig. 6A, our method enables the use of multiple probes in a single blot since the PNA probe is hybridized prior to loading on the gel. In principle this can be combined with multiplex PCR, so that, with for example, a 60-lane gel with 10 PCR reactions in each, 600 different mutations could be screened in a single gel run. The number of mutations analyzed could further be enhanced by the use of multiple fluorophores and a gel scanner.
CONCLUSION
In summary, we have demonstrated the PNA pre-gel hybridization technique with seven different PNA probes in several systems using two different electrophoresis formats. The strong binding of PNA to DNA allows PNA/DNA duplexes to remain intact under nonequilibrium conditions in media selected to disrupt DNA/DNA binding. This permits specific sequence detection with simultaneous size separation of the target DNA. High specificity was obtained with single bp discrimination being demonstrated for several systems. The key features of this method are rapid analysis and a straightforward protocol. The technique eliminates the tedious and time-consuming hybridization and washing steps of Southern hybridization with a significant increase in specificity. We currently are working toward extending these findings to the analysis of single copy genes in genomic samples by optimizing the sample preparation and detection techniques.
Acknowledgments
This paper is dedicated to the memory of Professor Ole Buchardt, University of Copenhagen. Ms. Shelagh Casey is thanked for skilled technical assistance in generating the PNAs; Dr. Wassim Nashabeh is thanked for developing the PNA CE probe purification method. Drs. Mark Roskey and Larry Haff are thanked for comments on the manuscript.
Footnotes
Abbreviations: PNA, peptide nucleic acid; CE, capillary electrophoresis; PEO, polyethelen oxide.
Certain PNA constructs have been shown to be very efficient at binding to intact double-stranded DNA by strand invasion. This binding mode is, however, limited to homopyrimidine PNA oligomers, and it has been shown that mixed sequence PNA oligomers like the probes used in this paper cannot be used for this binding mode for persistent binding to target sequences (P. E. Nielsen, personal communication).
References
- 1.Karp J E, Broder S. Nat Med. 1996;1:309–320. doi: 10.1038/nm0495-309. [DOI] [PubMed] [Google Scholar]
- 2.Dean M. Nat Genet. 1995;9:103–104. doi: 10.1038/ng0295-103. [DOI] [PubMed] [Google Scholar]
- 3.Forrest S, Cotton R, Landegren U, Southern E. Nat Genet. 1995;10:375–376. doi: 10.1038/ng0895-375. [DOI] [PubMed] [Google Scholar]
- 4.Southern E M. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 5.Egholm M, Buchardt O, Nielsen P E, Berg R H. J Am Chem Soc. 1992;114:1895–1897. [Google Scholar]
- 6.Egholm M, Buchardt O, Christensen L, Behrens C, Freier S, Driver D A, Berg R H, Kim S K, Norden B, Nielsen P E. Nature (London) 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 7.Rose D J. Anal Chem. 1993;65:3545–3549. doi: 10.1021/ac00072a003. [DOI] [PubMed] [Google Scholar]
- 8.Carlsson C, Jonsson M, Nordén B, Dulay M T, Zare R N, Noolandi J, Nielsen P E, Tsui L-C, Zielenski J. Nature (London) 1996;380:207. doi: 10.1038/380207a0. [DOI] [PubMed] [Google Scholar]
- 9.Britten R J, Graham D E, Neufeld B R. Methods Enzymol. 1974;29E:363–405. doi: 10.1016/0076-6879(74)29033-5. [DOI] [PubMed] [Google Scholar]
- 10.Riordan J R, Rommens J M, Kerem B, Alon N, Rozmahel R, Zbyszko G, Zielenski J, Lok S, Plavsic N, Chou J, Drumm M L, Ianuzzi M C, Collins F M, Tsui L. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 11.Kerem B, Rommens J M, Buchanan J A, Markiewicz D, Cox T, Chakravarti A, Buchwald M, Tsui L. Science. 1989;245:1073–1080. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- 12.Christensen, L. Fitzpatrick, R., Gildea, B., Warren, B. & Coull, J. (1994) Innovation and Perspectives in Solid Phase Synthesis and Complementary Technologies-Biological and Biomedical Applications, SPS Oxford Symposia Book (Mayflower Worldwide, Birmingham, U.K.), p. 149.
- 13.Mashal R D, Koontz J, Sklar J. Nat Genet. 1995;9:177–183. doi: 10.1038/ng0295-177. [DOI] [PubMed] [Google Scholar]
- 14.Don R H, Cox P T, Wainwright B J, Baker K, Mattick J S. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fung E N, Yeung E S. Anal Chem. 1995;67:1913–1919. [Google Scholar]
- 16.Handelin B, Schuber A P. In: Current Protocols in Human Molecular Genetics. Dracopoli N C, Haines J L, Korf B R, Moir D T, Morton C C, Seidman C E, Seidman J G, Smith D R, editors. New York: Wiley; 1994. pp. 9.4.1–9.4.8. [Google Scholar]
- 17.Ørum H, Nielsen P E, Jørgensen M, Larsson C, Stanley C, Koch T. BioTechniques. 1995;19:472–480. [PubMed] [Google Scholar]


