Abstract
Genetic analysis of limiting quantities of genomic DNA play an important role in DNA forensics, paleoarcheology, genetic disease diagnosis, genetic linkage analysis, and genetic diversity studies. We have tested the ability of degenerate oligonucleotide primed polymerase chain reaction (DOP-PCR) to amplify picogram quantities of human genomic DNA for the purpose of increasing the amount of template for genotyping with microsatellite repeat markers. DNA was uniformly amplified at a large number of typable loci throughout the human genome with starting template DNAs from as little as 15 pg to as much as 400 ng. A much greater-fold enrichment was seen for the smaller genomic DOP-PCRs. All markers tested were amplified from starting genomic DNAs in the range of 0.6–40 ng with amplifications of 200- to 600-fold. The DOP-PCR-amplified genomic DNA was an excellent and reliable template for genotyping with microsatellites, which give distinct bands with no increase in stutter artifact on di-, tri-, and tetranucleotide repeats. There appears to be equal amplification of genomic DNA from 55 of 55 tested discrete microsatellites implying near complete coverage of the human genome. Thus, DOP-PCR appears to allow unbiased, hundreds-fold whole genome amplification of human genomic DNA for genotypic analysis.
Whole genome amplification is a valuable technique to allow an increase of limiting DNA in a sequence independent fashion. Various strategies for whole genome amplification have been developed. Primer extension preamplification, which uses a random 15-mer to prime Taq DNA synthesis frequently throughout the genome, has been used to amplify genomic DNA from as little as a single haploid cell and demonstrates good coverage (1–3). Alternate strategies of whole genome amplification, including linker adaptor-PCR, tagged-PCR, and degenerate oligonucleotide primed-PCR (DOP-PCR), while likely generating higher yields, have not been adequately evaluated for coverage (4–7). Linker adaptor-PCR requires digestion with a frequent cutter, ligation to an oligonucleotide, and amplification of the ligated product with primers specific to the oligonucleotides. While there should be very little sequence selection bias with linker adaptor-PCR, except on the basis of distance between restriction sites, the technique is more challenging than the other whole genome amplification methods. Tagged-PCR primers are designed with a random sequence at the 3′ end to allow binding to a wide variety of target sequences and a constant 5′ tail. The first rounds of amplification are performed similarly to primer extension preamplification to allow many priming sites. After several rounds of non-specific interactions, the unbound primers are removed, and PCR amplification is carried out with a primer specific to the 5′ constant region to allow exponential amplification.
We have investigated DOP-PCR amplification of total human genomic DNA at various starting amounts for yield and coverage using microsatellite repeats. DOP-PCR was developed to allow an unselected amplification of virtually any source DNA, has been specifically applied to cytogenetic techniques, and results in a more uniform signal than interspersed repeat-based methods of whole genome amplification (7). In a single PCR tube several low temperature annealings and extensions are performed to allow many binding sites in the human genome. After several low temperature annealings and extensions, the annealing temperature is increased to allow more specific priming only at the fragments now tagged with the above primer sequence. DOP-PCR amplification of discrete plasmids and cosmids results in discrete bands, implying that only a portion of any arbitrary stretch of DNA sequence is amplified. From these data, it has been estimated, based on the number and sizes of DOP-PCR products, that there should be about one million DOP-PCR fragments generated from the entire human genome (7). Since the average size product is 500 bp and the human genome is about 3 × 109 bp, we had anticipated that an arbitrary stretch of DNA had only a one in six chance of being included in the DOP-PCR product. However, in the course of slightly modifying DOP-PCR to amplify total genomic DNA for genotypic analysis, we observed that all PCR-based markers tested to date can be amplified by DOP-PCR and can be accurately genotyped from DOP-PCR-amplified human genomic DNA. This offers a general method of amplification of small genomic DNA sample amounts to allow hundreds-fold more DNA for genetic analyses.
MATERIALS AND METHODS
DOP Amplification of Genomic DNA.
Genomic DNA was prepared from whole blood with a commercially available kit (Gentra Systems). DOP-PCR amplification was performed with genomic template amounts ranging from 24 pg to 400 ng. Most experiments used 1–40 ng of template and 50 μl final volumes. The PCR reactions contained 2 μM DOP primer (5′-CCGACTCGAGNNNNNNATGTGG-3′), 200 μM dNTPs, 10 mM Tris·Cl (pH 9.0), 50 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100, and 0.01% gelatin in a final volume of 10–100 μl. After an initial denaturation of 8 min at 96°C, 2.5 units of Amplitaq DNA polymerase (Perkin–Elmer/Cetus) was added followed by 8 cycles of 93°C for 1 min, 30°C for 1 min, and 72°C for 3 min, and then 28 cycles of 93°C for 1 min, 60°C for 1 min, and 72°C for 3 min. This is a slight modification from the originally published use of this primer (7). Most amplifications were carried out in the PTC 100 Thermal Cycler (MJ Research, Cambridge, MA). Each amplified sample was quantitated by OD260.
Specific Microsatellite Analysis.
DOP-PCR-amplified genomic DNA was tested for the presence of specific microsatellite sequences. Microsatellite primers were selected at random to analyze a variety of sizes of di-, tri-, and tetranucleotide repeats from the Weber version 6.0 screening set (Research Genetics, Huntsville, AL). Genotyping was performed with 40 ng DOP-amplified DNA, fluorescent primers (0.8 μM each), 200 μM dNTP, 2.5 units Amplitaq DNA polymerase (Perkin–Elmer), 10 mM Tris·Cl (pH 9.0), 50 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100, and 0.01% gelatin in a final volume of 10 μl. Amplification was carried out by an initial denaturation for 3 min at 94°C followed by 30 cycles of 93°C for 45 sec, 56°C for 45 sec, and 72°C for 45 sec and a final extension at 72°C for 3 min. Each reaction was diluted 50- to 100-fold. One microliter was mixed with 12 μl formamide and 1 μl GeneScan 500 size standards (Perkin–Elmer), denatured, and loaded onto an Applied Biosystems 310 Genotyper. Multiplexing with up to four primers was typically performed in the microsatellite PCR. The samples were capillary electrophoresed on an Applied Biosystems 310 Genotyper with 3% polymer/6.6 M urea solution on the short denatured C protocol. Fluorescent quantitation was performed by the Genotyper software (Applied Biosystems). Quantitation of specific alleles in the DOP-amplified product was attained by mixing twice as much of another individual’s genomic DNA with the DOP sample. PCR amplification of the mixed sample with a specific microsatellite repeat allows direct comparison of the fluorescent peaks, which accurately reflects the relative amounts of template DNA in a manner analogous to competitive PCR (10).
RESULTS
Our first experiment was designed to test how often a given microsatellite sequence is present within the DOP-PCR-amplified portion of human genomic DNA to measure the coverage of DOP-PCR. Initially, we expected only a minority of microsatellite sequences to be amplified by DOP-PCR. However, the first 20 primer sets yielded accurate genotyping data with amplifications in the range of 200- to 400-fold in terms of amplifiability of the microsatellites (the total DNA yield was generally 2- to 3-fold higher). If 40 ng of total genomic DNA typically yielded a fluorescent peak height of 1 arbitrary unit, a one:four hundred dilution of the DOP-amplification product from 40 ng of genomic DNA would yield a 1 unit peak as well. All of the initial experiments amplified 1–40 ng of genomic DNA in 50–100-μl reactions. The resulting DOP product typically yielded 5–15 μg of product that ranged in size from 200-1000 bp on ethidium bromide-stained agarose gels, and 40 ng of this DNA was used as a template for genotyping.
After this initial observation, we wished to see if the accuracy of genotyping was affected by DOP-PCR. Microsatellite repeats (D1S2130, D2S1360, D3S2418, D4S2366, D5S816, D6S1006, D7S1802, D8S1132, D12S372, and D17S1290) were randomly selected from the Weber screening set for analysis to give a variety of sizes. Forty nanograms of genomic DNA was DOP-PCR amplified to yield on average 8 μg by OD260. The fluorescent microsatellite repeat primers were used for amplification of 40 ng of the starting genomic DNA and 40 ng of the DOP-amplified DNA from a single individual. Accurate allele scoring was attained at all loci. Furthermore, the electropherograms indicated that there was no increase in stutter bands with tri- and tetranucleotide repeats, and the peak heights from the genomic DNA and the DOP-PCR DNA were roughly equivalent (Fig. 1). As is apparent from Fig. 1, the electropherograms at both heterozygous and homozygous loci on the DOP-amplified DNA had no higher background noise than genomic DNA, and the determination of genotypes was clear. Dinucleotide repeats tend to give more problematic shadow banding, which can interfere with accurate allele scoring (9). Nine of nine dinucleotide repeat markers tested were accurately scored without an increase in the number of shadow bands (not shown). To date, 55 of 55 microsatellites that have been tested on the DOP-PCR products were effectively amplified, which indicates that a large proportion of the genome is amplified at a starting template range of 1–40 ng of genomic DNA.
Figure 1.
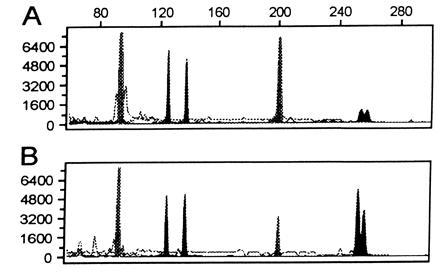
Genotypic analysis of genomic DNA as compared with DOP-PCR-amplified genomic DNA. The individual is homozygous at two loci, D3S2418 (97 bp, 97 bp) and D6S1006 (200 bp, 200 bp) and heterozygous at two loci, D4S2366 (127 bp, 136 bp) and D1S2130 (252 bp, 256 bp). (A) An electropherogram of a multiplex PCR at these four microsatellite loci using 40 ng of genomic DNA as template. (B) An electropherogram of a multiplex PCR at the same four loci using 40 ng of DOP-PCR-amplified genomic DNA as template. Comparison of A and B shows matching genotypes and corresponding peak areas from the two templates. The y-axis represents arbitrary fluorescent units, and the x-axis represents the size of fragments in base pairs.
To quantify the effective yield for genotyping from DOP-PCR at a series of starting genomic DNA amounts, we performed DOP-PCR on serial dilutions of purified human genomic DNA. A serial 5-fold dilution generated genomic DNA of 400 ng, 80 ng, 16 ng, 3.2 ng, 640 pg, 128 pg, and 26 pg, which were amplified in 50-μl final volumes. A second serial 4-fold dilution generated genomic template amounts of 60 ng, 15 ng, 3.8 ng, 0.94 ng, 230 pg, 59 pg, and 15 pg, which were DOP-PCR amplified in 10 μl. The samples were analyzed at locus D8S1132. Each sample was quantitated by OD260 and, based on this measurement, 40 ng was mixed with 80 ng of another individual’s DNA, who shares neither allele in common with the DNA, which was DOP-PCR amplified. Thus, by comparing the relative fluorescent peak heights, we could determine the amount of amplification of each individual’s DNA at this locus in a manner analogous to competitive PCR (8). Fig. 2 shows the electropherograms of the mixed samples from the first dilution series at which the two individuals are both heterozygotes. The shorter peaks at 158 and 168 bp are from the DOP-PCR-amplified genomic DNA, and the 162- and 172-bp taller peaks are from total genomic DNA. The DOP-PCR product is generally one-fourth to one-half as effectively amplified as genomic DNA, and there is a trend toward lower amplifiability of the DOP-PCR product with lower starting genomic DNAs. Based on the amount of microsatellite product generated, we corrected the yield of DNA from the DOP-PCR amplification (Table 1). As much as a 23,000-fold effective amplification is seen from as little as 15–26 pg of genomic DNA. However, the reliability of the microsatellite scoring of the DOP-PCR product was lower from the smaller genomic amounts. We were able to amplify 6 of 12 and 9 of 12 microsatellites in the lowest two dilutions. Nonetheless, all markers tested on 40 ng of DOP-PCR-amplified DNA from greater than 128 pg of genomic DNA have allowed accurate genotyping at all markers tested.
Figure 2.
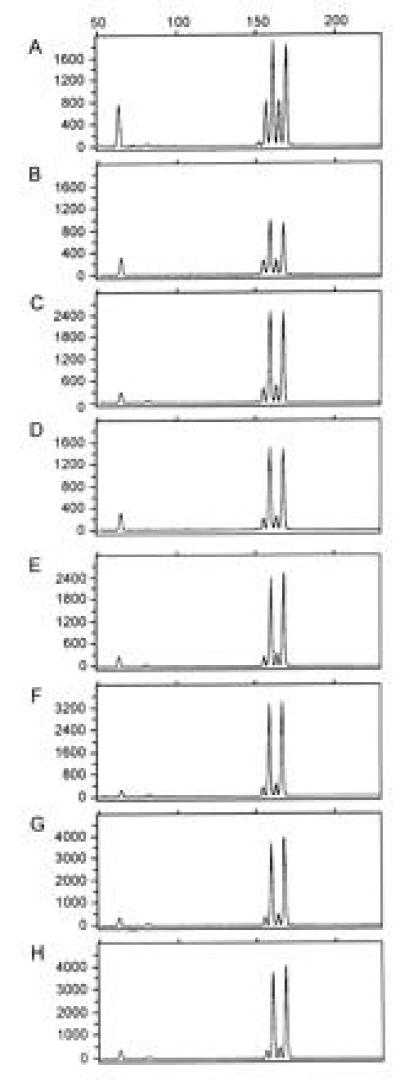
Comparison of yield from the DOP-PCR product generated from serially diluted genomic DNAs. All panels show electropherograms from genotyping of mixtures of DNA with fluorescently labeled D8S1132 primers as discussed. (A) An electropherogram from a mixture of total genomic DNA from two heterozygous individuals: 40 nanograms of genomic DNA from individual 1 (allele sizes 152 bp and 162 bp) were mixed with 80 ng of genomic DNA from individual 2 (allele sizes 158 bp and 168 bp). (B–H) Electropherograms at D8S1132 on 40 ng of DOP-PCR product derived from a DOP-PCR of a serial dilution of starting genomic DNA of individual 1 mixed with 80 ng of genomic DNA from individual 2, which allow measurement of relative yield. DOP-PCR product used in B–H are derived from starting genomic DNA amounts of 400 ng, 80 ng, 16 ng, 3.2 ng, 640 pg, 128 pg, and 26 pg, respectively. Comparison of the peak areas on the electropherograms reflects how the DOP-PCR products amplify relative to genomic DNA. These data were used to determine the corrected yield in Table 1. The y-axis represents arbitrary fluorescent units, and the x-axis represents fragment size in base pairs.
Table 1.
Yield of DOP-PCR from varying amounts of starting genomic DNA
| Set | Amount of starting DNA | Corrected yield from DOP-PCR, μg | Fold amplification |
|---|---|---|---|
| A | 400 ng | 5 | 13 |
| 80 ng | 2.3 | 29 | |
| 16 ng | 2.0 | 125 | |
| 3.2 ng | 1.5 | 469 | |
| 640 pg | 0.8 | 1,250 | |
| 128 pg | 0.8 | 6,250 | |
| 26 pg | 0.6 | 23,077 | |
| B | 60 ng | 0.7 | 12 |
| 15 ng | 0.6 | 40 | |
| 3.8 ng | 0.4 | 105 | |
| 900 pg | 0.2 | 222 | |
| 200 pg | 0.2 | 1,000 | |
| 60 pg | 0.4 | 6,667 | |
| 15 pg | 0.3 | 20,000 |
DISCUSSION
In this paper we demonstrate that DOP-PCR can be used for whole genome amplification to give high yields and excellent coverage on the basis of microsatellite repeat amplification of heterozygous and homozygous loci. Because there is no reason to assume that microsatellites should be any more likely to be amplified than any other sequence in the human genome, our data suggest that DOP-PCR can be used to achieve several hundred-fold amplification of virtually all sequences in the human genome with sufficient starting template genomic DNA. We have not, however, tested the DOP-PCR-amplified samples for their use in other PCR-based genetic analyses such as sequencing, single-stranded conformation polymorphism, and denaturing gradient gel electrophoresis. If for some unclear and unlikely reason microsatellites are more frequently represented in the DOP-PCR-amplified portion of the DNA, then some of these other fragments may not be reliably amplified.
The DOP-PCR product that we generate never amplifies better than the starting genomic DNA and usually amplifies one-third as well. This may simply indicate that nonspecific priming is occurring and in effect creating non-specific DNA, which has no relation to the human genome. Alternatively highly repetitive DNAs could be over-represented by the PCR process, but not tested by the genotyping. A third explanation may be that overlapping sets of DNA fragments are formed such that only one in three contain both primer sites for each microsatellite. It is likely that all three explanations are occurring to varying degrees in DOP-PCR, although we have little direct evidence. There should be some more-abundant priming sites in the human genome, and we would expect an overabundance of these fragments in the DOP-PCR amplified pool. If priming at the initial low temperature annealings is occurring because of all six specific base pairs on the 3′ end, we would expect to have amplified only about 1 of every 10 200-1000-bp stretches of the human genome. The ATGTGG site would be expected to occur on average every 4630 bp [(0.2)3(0.3)3], or occur in a head-to-head arrangement in the genome separated by 200-1000 bp to allow amplification in our product size range about every 10–20 kb. We thus assume that priming is occurring from a variety of partially hybridized primers at the specific 3′ end to generate sufficient tagged-genomic DNA. The subsequent PCR amplification at higher annealing temperatures makes use of the specific 6-bp 3′ end and specific 10-bp 5′ end to allow specific hybridization to only the tagged-genomic sequences and exponential increase in DNA until Taq DNA polymerase is limiting. DOP-PCR on simple sequences like plasmids yield a reproducible banding pattern of dominant products and a smear of DNA fragments indicating that both reactions are occurring (7).
DOP-PCR yields 30-to 40-fold amplification of specific product from 100 ng of genomic DNA with 1.25 units Amplitaq DNA polymerase (7). The authors proposed that the limiting reagent in the DOP-PCR is Taq DNA polymerase. This may account for our lower effective amplification at higher genomic DNA amounts (80–400 ng). If by using a different buffer and eight instead of five primer extension reactions there is an increase in the number of priming sites in the genome, the polymerase may be exhausted with less total starting genomic DNA in our experiments. Thus, less than linear amplification (12-fold) is seen at 400 ng of starting DNA, whereas linear amplification is seen at 80 ng genomic DNA, and increasing exponential yield is observed with lower starting DNAs. Our experiments all use 2.5 units of Amplitaq DNA polymerase. The striking feature is that as DNA becomes more limiting there is only a minor effect on the total DOP-PCR product. Thus, DOP-PCR is robust in regards to the yield of starting genomic source DNA. However, as genomic DNA is reduced below 640 pg, there is an increased likelihood that a given locus will not be represented in the DOP-PCR product. This is consistent with theoretical consideration of the tagged-PCR as compared with primer extension preamplification, which indicates that primer extension preamplification should offer more complete coverage at lower genomic DNA copy numbers (10). While it is possible that higher amounts of Taq DNA polymerase may improve the overall yield, higher Taq DNA polymerase can increase the amount of primer-related products (7). We have not attempted to use other thermophilic polymerases or other preparations of Taq DNA polymerase to see if similar yields are attained as Amplitaq DNA polymerase was sufficient for our purposes.
The observation that DOP-PCR can be used to amplify all the sequences of human genomic DNA for genotypic analyses has several implications. Principally, a potentially enormous benefit of DOP-PCR is that very small samples of precious material can be expanded to allow increased numbers of analyses. For instance, the need to immortalize cell lines from subjects participating in genomic mapping projects would be lessened. Typically, 300 μg genomic DNA can be routinely extracted from 10 ml of whole blood. Thus, even for individuals critical to the study a vast number of genotypic analyses can be performed after DOP-PCR of a small fraction of this genomic DNA. Because it is not known at the initiation of the analysis whose DNA will be limiting, frequently a cell line is created on all individuals, which is unnecessary. In addition, it may prove possible to use limiting quantities of genomic DNA for complete genome screens. For instance, in pedigrees where individual members are deceased (i.e., in lethal genetic diseases), but a biopsy sample remains, it may be possible to generate sufficient material for many hundreds of genotypings. We generally multiplex four primer sets in 10 μl with 40 ng of DOP-PCR template. Thus, 10 nanograms of total genomic DNA can be amplified by DOP-PCR to 4 μg, which creates sufficient template for 400 genotypic analyses. In forensic analysis of genomic DNA, limited and possibly partially degraded samples can be amplified to more reliably determine identity. In paleoarcheology, where precious little long DNA sequences may remain, the few longer strands may be able to be amplified significantly prior to analysis. For instance, we have DOP-PCR amplified a brain biopsy-derived DNA sample, which was degraded to a 100-bp average size and untypable with any PCR-based markers. The post-DOP-PCR amplification sample was sufficient for genotyping at many loci. The few longer strands of genomic DNA after purification were relatively enriched because they contained two primer-binding sites.
In instances where DNA sample collection is difficult, samples could be preserved as dried blood spots on Guthrie cards. Several studies indicate that purification of 1.5 μg is achievable from a 25-μl blood spot (about 10 mm) (11, 12). Less than one-hundredth of this amount can be amplified by DOP-PCR to yield sufficient template for hundreds of analyses. For instance, the human genome diversity project will need to travel to remote locations, collect cellular samples, and preserve that material until returning to genetic laboratories (13). Combining DOP-PCR on dried blood spots may create a convenient cost-effective means for genomic DNA collection and analysis. DOP-PCR of genomic DNA prepared from other easily collected cellular material like buccal smears, mouthwash, and hair follicles would allow many PCR-based analyses (14, 15).
Because this technique allows virtually any small quantity of DNA (i.e., from biopsies) to be identifiable and useful for genetic mapping strategies or specific gene detection, it further highlights the ongoing discussion regarding the ethical issues surrounding the uses of banked-tissue samples (16, 17). Stored Guthrie cards, from neonatal screening programs, have been proposed as DNA “banks” and used to a certain degree for genetic analyses. With the capability to expand the number of discrete tests that can be performed on any one sample, the interest in these resources can be expected to increase. A recent survey of laboratories with stored Guthrie cards indicates variability in practices regarding the release of samples for research (18). As molecular diagnostic techniques improve, it becomes more critical to address the scope of permissible uses with stored DNA from biopsies and neonatal screens.
Acknowledgments
We thank Norman Arnheim, Edward McCabe, and Elizabeth Neufeld for comments on the manuscript. The work was supported by National Institutes of Health Grant R29 HG01141 and the Gwynne Hazen Cherry Memorial Laboratory.
Footnotes
Abbreviation: DOP-PCR, degenerate oligonucleotide primed PCR.
References
- 1.Zhang L, Cui X, Schmitt K, Hubert R, Navidi W, Arnheim N. Proc Natl Acad Sci USA. 1992;89:5847–5851. doi: 10.1073/pnas.89.13.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Snabes M C, Chong S S, Subramanian S B, Kristjansson K, DiSepio D, Hughes M R. Proc Natl Acad Sci USA. 1994;91:6181–6185. doi: 10.1073/pnas.91.13.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrett M T, Reid B J, Joslyn G. Nucleic Acids Res. 1995;23:3488–3492. doi: 10.1093/nar/23.17.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grothues D, Cantor C R, Smith C L. Nucleic Acids Res. 1993;21:1321–1322. doi: 10.1093/nar/21.5.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ludecke H J, Senger G, Claussen U, Horsthemke B. Nature (London) 1989;338:348–350. doi: 10.1038/338348a0. [DOI] [PubMed] [Google Scholar]
- 6.Saunders R D, Glover D M, Ashburner M, Siden-Kiamos I, Louis C, Monastirioti M, Savakis C, Kafatos F. Nucleic Acids Res. 1989;17:9027–9037. doi: 10.1093/nar/17.22.9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Telenius H, Carter N P, Bebb C E, Nordenskjold M, Ponder B A, Tunnacliffe A. Genomics. 1992;13:718–725. doi: 10.1016/0888-7543(92)90147-k. [DOI] [PubMed] [Google Scholar]
- 8.Siebert P D, Larrick J W. Nature (London) 1992;359:557–558. doi: 10.1038/359557a0. [DOI] [PubMed] [Google Scholar]
- 9.Ginot F, Bordelais I, Nguyen S, Gyapay G. Nucleic Acids Res. 1996;24:540–541. doi: 10.1093/nar/24.3.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun F, Arnheim N, Waterman M S. Nucleic Acids Res. 1995;23:3034–3040. doi: 10.1093/nar/23.15.3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Descartes M, Huang Y, Zhang Y H, McCabe L L, Gibbs R, Therrell B L, Jr, McCabe E R. Pediatr Res. 1992;31:217–221. doi: 10.1203/00006450-199203000-00005. [DOI] [PubMed] [Google Scholar]
- 12.Carducci C, Ellul L, Antonozzi I, Pontecorvi A. BioTechniques. 1992;13:735–737. [PubMed] [Google Scholar]
- 13.Kidd J R, Kidd K K, Weiss K M. Hum Biol. 1993;65:1–6. [PubMed] [Google Scholar]
- 14.Hayney M S, Dimanlig P, Lipsky J J, Poland G A. Mayo Clin Proc. 1995;70:951–954. doi: 10.4065/70.10.951. [DOI] [PubMed] [Google Scholar]
- 15.Lench N, Stanier P, Williamson R. Lancet. 1988;i:1356–1358. doi: 10.1016/s0140-6736(88)92178-2. [DOI] [PubMed] [Google Scholar]
- 16.Grody W W. Diagn Mol Pathol. 1995;4:155–157. doi: 10.1097/00019606-199509000-00001. [DOI] [PubMed] [Google Scholar]
- 17.Clayton E W, Steinberg K K, Khoury M J, Thomson E, Andrews L, Kahn M J, Kopelman L M, Weiss J O. J Am Med Assoc. 1995;274:1786–1792. [PubMed] [Google Scholar]
- 18.McEwen J E, Reilly P R. Am J Hum Genet. 1994;55:196–200. [PMC free article] [PubMed] [Google Scholar]


