Abstract
Background
Despite intensive efforts devoted to collecting human polymorphism data, little is known about the role of gene flow in the ancestry of human populations. This is partly because most analyses have applied one of two simple models of population structure, the island model or the splitting model, which make unrealistic biological assumptions.
Results
Here, we analyze 98-kb of DNA sequence from 20 independently evolving intergenic regions on the X chromosome in a sample of 90 humans from six globally diverse populations. We employ an isolation-with-migration (IM) model, which assumes that populations split and subsequently exchange migrants, to independently estimate effective population sizes and migration rates. While the maximum effective size of modern humans is estimated at ~10,000, individual populations vary substantially in size, with African populations tending to be larger (2,300–9,000) than non-African populations (300–3,300). We estimate mean rates of bidirectional gene flow at 4.8 × 10-4/generation. Bidirectional migration rates are ~5-fold higher among non-African populations (1.5 × 10-3) than among African populations (2.7 × 10-4). Interestingly, because effective sizes and migration rates are inversely related in African and non-African populations, population migration rates are similar within Africa and Eurasia (e.g., global mean Nm = 2.4).
Conclusion
We conclude that gene flow has played an important role in structuring global human populations and that migration rates should be incorporated as critical parameters in models of human demography.
Background
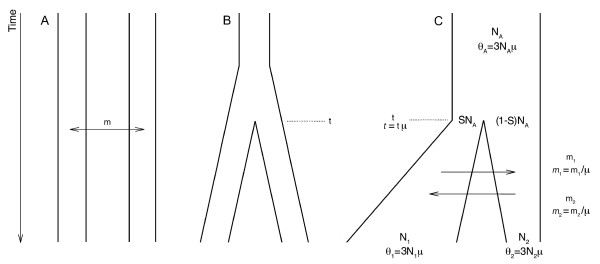
Reconstructing human history requires an accurate picture of global human population structure [1]. However, methods currently used to describe structure among human groups typically rely on very simple demographic models that make unrealistic biological assumptions. Two commonly used models include the island model, which assumes that populations have no shared ancestry and are related only through gene flow (Figure 1A), and the phylogenetic branching or splitting model, which assumes that populations diverged at some time in the past and have remained completely isolated ever since (i.e., no gene flow) (Figure 1B). Despite increasingly sophisticated genetic datasets, most contemporary studies still assume these unrealistic models to infer aspects of human demographic history [2-8].
Figure 1.
Models of population structure reflecting the (A) island, (B) splitting and (C) isolation-with-migration (IM) models. The island model assumes equilibrium gene flow (m) between subpopulations that have no shared ancestry. The divergence model describes an ancestral population, which splits at time t into two daughter populations that do not exchange genes in subsequent generations. The isolation-with-migration model describes a constant-sized ancestral population that splits into two daughter populations that can exchange genes and change in size. There are seven parameters in the isolation-with-migration model: effective population size of the ancestral deme (NA), effective population sizes of the two descendent demes (N1 and N2), unidirectional migration between the descendent populations (m1 and m2), proportion of the ancestral population founding deme 1 (S), and population divergence time (t).
Furthermore, human population structure is often considered only from the perspective of a single summary statistic, FST, which is a standardized measure of the genetic variation shared between populations. However, FST is dependent on the effective size (N), migration rate (m) and divergence time (t) of individual demes, and by itself has no straightforward demographic interpretation. A small FST between two populations may indicate large effective population sizes, high rates of gene flow since diverging from a common ancestral population, or recent population divergence. For instance, under Wright's [9] island model, N, m and FST are linked by a nonlinear relationship independent of t (equation 2) [10]. Assuming a global effective size for modern humans of ~104, an X chromosome FST of 0.2 [11,12] suggests that human populations have exchanged ~4 gene copies per generation. Conversely, under a simple divergence and isolation model, N, t and FST are related by a nonlinear relationship independent of m (equation 4) [13]. Assuming a mean generation time of 25 years, an X chromosome FST of 0.2 suggests that global human populations diverged on average ~84 Kya. However, despite being in common usage, gene flow-only and divergence-only models probably have little relevance to actual human demographic history.
Here, we examine the structure of human populations by means of the isolation-with-migration (IM) model [14], which incorporates both population splitting and gene flow (Figure 1C). The IM model describes two daughter populations, which can expand or contract in size, that diverge from a constant-sized ancestral population with continuing migration between the two demes. The IM model has seven demographic parameters: effective population size of the ancestral deme (NA), effective population sizes of the two descendent demes (N1 and N2), unidirectional migration rates between the descendent populations (m1 and m2), proportion of the ancestral population founding the first deme (S), and population divergence time (t). Unlike the island model, which assumes infinite divergence times, or the splitting model, which assumes zero migration, IM makes no a priori assumptions about these two demographic parameters. It is therefore a more flexible model system for inferring human history from genetic data.
Here, we apply a Bayesian inference framework together with a maximum likelihood algorithm [15] to determine demographic parameters for a range of human populations under the IM model. Marginal Bayesian posterior probabilities are calculated using Markov chain Monte Carlo, and best-fit parameterizations are inferred for each population pair (N = 15) for each of the seven demographic parameters. We analyze a multilocus resequencing dataset comprising 20 intergenic regions on the X chromosome, each of which represents ~5 kb of DNA [16]. To minimize any potential confounding effects of natural selection, the sequenced loci were chosen from single-copy non-coding (i.e., putatively non-functional) DNA in regions of medium or high recombination (r ≥ 0.9 cM/Mb). Such regions are recombinationally unlinked from genic regions. Because we use resequence data (i.e., all sites were sequenced in each individual) and not pre-ascertained single nucleotide polymorphisms (SNPs) [3,17,18], we avoid some biases in aspects of the data that rely on the site frequency spectrum (e.g., FST) [19,20]. These data are also well suited for testing the role of gene flow in the history of anatomically modern humans because multiple, diverse populations are represented in our survey (i.e., Biaka from Central African Republic, Mandenka from Senegal, San from Namibia, Basque from southern France, Han from northern China, and Melanesians from Papua New Guinea).
Results
Population differentiation
Wright's FST for our global sample averages to 0.25. When we calculate FST between all pairs of populations, we find the greatest FST values between sub-Saharan African and non-African groups (i.e., FST ranges from 0.160–0.450) (Table 1). However, there is also a substantial amount of differentiation within sub-Saharan African (mean FST of 0.137) and within non-African groups (mean FST of 0.174), with FST values as high as 0.226 between Melanesians and Basque [16].
Table 1.
Mean effective population sizes, migration rates, divergence times and FST.
| Pop 1 | Pop 2 | NA | N1 | N2 | m12/gen ×10-4 | m21/gen ×10-4 | Nm | t (kya) | FST | |
| African | ||||||||||
| BIA | MAN | 9,980 | 3,980 | 6,600 | 2.8 | 1.9 | 4.9 | 48.7 | 0.117 | |
| BIA | SAN | 6,620 | 5,560 | 5,340 | 0.86 | 1.9 | 3.0 | 50.0 | 0.169 | |
| MAN | SAN | 9,530 | 6,930 | 3,790 | 0.72 | 0.020 | 0.8 | 46.8 | 0.126 | |
| African/Non-African | ||||||||||
| BIA | BAS | 11,200 | 4,650 | 3,250 | 1.1 | 0.43 | 1.2 | 61.4 | 0.311 | |
| BIA | HAN | 12,800 | 2,330 | 2,600 | 0.30 | 0.075 | 0.2 | 27.7 | 0.374 | |
| BIA | MEL | 11,600 | 6,900 | 1,570 | 0.48 | 0.71 | 1.0 | 88.5 | 0.331 | |
| MAN | BAS | 9,970 | 4,530 | 2,750 | 5.8 | 0.036 | 4.2 | 23.4 | 0.160 | |
| MAN | HAN | 11,000 | 2,750 | nd | 2.2 | 0.18 | nd | 15.5 | 0.236 | |
| MAN | MEL | 9,420 | 7,110 | 318 | 2.1 | 3.0 | 3.8 | 12.4 | 0.221 | |
| SAN | BAS | 10,400 | 5,820 | 2,490 | 0.42 | 0.0012 | 0.3 | 83.0 | 0.344 | |
| SAN | HAN | 10,200 | 7,270 | 1,880 | 0.24 | 0.46 | 0.6 | 151 | 0.450 | |
| SAN | MEL | 10,500 | 8,990 | 1,210 | <<0.001 | 1.6 | 1.6 | 68.5 | 0.390 | |
| Non-African/Non-African | ||||||||||
| BAS | HAN | 11,900 | 2,230 | 1,940 | 9.2 | 1.4 | 4.4 | 85.6 | 0.085 | |
| BAS | MEL | 11,600 | 2,120 | 283 | 0.26 | 12 | 3.0 | 62.5 | 0.226 | |
| HAN | MEL | 11,300 | 1,770 | 592 | 0.18 | 21 | 5.0 | 61.2 | 0.210 | |
| Mean | 10,500 | 3,700 | 2.4 | 2.4 | 59.1 | 0.25 | ||||
Abbreviations: BIA, Biaka; MAN, Mandenka; SAN, San; BAS, Basque; HAN, Han; MEL, Melanesians; nd: not determined.
Demographic inference under the isolation-with-migration model
Marginal Bayesian posterior probabilities were calculated using Markov chain Monte Carlo, and best-fit parameterizations were inferred for each unique population pair (e.g., see Figure 1 in Additional file 1). Results for the seven demographic parameters are discussed individually below (also see Table 1).
Effective population sizes
Modern effective population sizes (N) were inferred multiple times under the IM model (i.e., once for each paired population, Tables 1 and 2 in Additional file 1). Although individual estimates of N have some uncertainty, mean effective sizes are statistically larger for African populations (2,300–9,000) than non-African (300–3,300) populations (t20 = 6.9, P << 0.001). Some non-African groups have especially small effective sizes, such as Melanesians (N0 <1,500). Mean ancestral sizes (10,500, range 6,600–12,800) are generally larger than modern effective sizes, and are also often larger than the sum of their descendant populations (t19 = 3.42, P = 0.0029).
Population split proportions
Our dataset has little power to infer how ancestral effective sizes were apportioned among descendent demes. Most estimates of the split proportion, S, have large confidence intervals (Table 3 in Additional file 1). The few informative estimates indicate substantial retention of ancestral effective size in African populations; for instance, less than ~10% of the ancestral population size of Mandenka and Papua New Guineans is retained in the latter population today. Similarly, among non-Africans, the Han Chinese retain less than ~10% of the diversity carried when their ancestral deme still included the ancestors of modern Papua New Guineans (upper bounds of the 95% confidence interval for S).
Migration rates
Stationary estimates of long-term unidirectional migration rates average to 2.4 × 10-4/generation (range: 8.7 × 10-8 – 2.1 × 10-3/generation), thereby suggesting that gene flow between global populations is relatively frequent (Table 4 in Additional file 1). The highest bidirectional migration rate (2.1 × 10-3; lowest rate: 3.8 × 10-5) implies a movement of ~1 X chromosome every 2 years. Bidirectional migration rates within and between continents vary significantly (F2,12 = 21.5, P = 0.0001, η = 0.78), with continental assignment accounting for 61% of the variation in migration rates. Mean bidirectional migration rates within Africa (2.7 × 10-4), and between Africans and Eurasians (2.1 × 10-4), are relatively low compared to migration rates within Eurasia (1.5 × 10-3). Furthermore, migration patterns between populations are largely symmetric. Han Chinese and Melanesians provide a key exception; migration from China to the Pacific (2.1 × 10-3/generation) has significantly exceeded migration in the opposite direction (1.8 × 10-5/generation) (see 95% confidence intervals in Table 4 in Additional file 1).
Population divergence times
Marginal posterior distributions for t indicate that divergence times between African populations all occur ~50 kya, with the largest upper confidence interval for any two African populations at ~140 kya (Table 5 in Additional file 1. Estimates of non-African split times are made difficult by diffuse posterior distributions with very high variance, and thus caution is advised in interpreting the higher mean non-African divergence time (~67 kya). Estimates of the lower bounds for non-African divergence times, with a mean of ~30 kya, are more consistent with previous estimates. The mean divergence time between African and non-African populations is ~58 kya (or ~2,100 generations), which is very similar to previous estimates of the out-of-Africa expansion inferred from other genetic data [21-23].
Validation of inferred demographic parameters
The inference system employed here has been validated elsewhere [15], and our own tests indicate that known demographic parameters (including m) for data simulated under an IM model are recovered within 95% confidence intervals (unpublished data). However, to further assess the suitability of the IM model to explain observed patterns of variation, we used coalescent simulation to model each population pair (N = 15) using the parameter values of NA, N1, N2, m1, m2 and t inferred above. Due to poor estimates of the split proportion, we assumed S = 0.5. To check whether these coalescent simulations return data similar to the empirical loci, we compared observed summary statistics for all twenty X chromosome loci with summary distributions from these parameterized simulation models. We focused on four summaries of the data: i) FST, which describes the genetic distance between populations; ii and iii) θW and θπ, which are unbiased estimators of the population mutation rate θ (= 3Neμ); and iv) Tajima's D, which summarizes the site frequency spectrum.
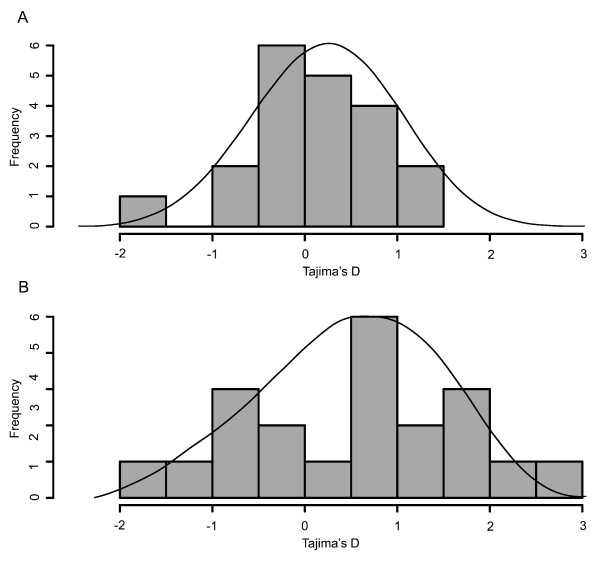
Observed FST values are correlated with FST values simulated under these 15 simulation models (Mantel test, r = 0.49, P = 0.039). Although this only explains 24% of the variance, we obtain mean FST values that are just slightly lower than those actually observed (i.e., mean FST of 0.21 versus 0.25, not significantly different). The simulation models also provide good fits to observed data for the remaining summaries, all of which reflect aspects of the population site frequency spectrum. A Bonferroni correction holding the experiment-wise type-I error rate constant at α = 0.05 was applied to accommodate multiple tests for each population and each summary. We observe 2% of loci as outliers under the corrected 95% confidence intervals for the 360 tests performed on our parameterized simulation models (i.e., 6 populations, 20 loci, and 3 summary statistics) (Table 6 in Additional file 1). This degree of consistency between observed and simulated data is similar for both African and non-African populations (e.g., Figure 2). This similarity suggests that our estimates of demographic parameters are not strongly biased by the inference method, and that the IM model is capable of portraying the evolutionary processes underlying human population structure, at least at the geographic scale examined here [24].
Figure 2.
Observed values (bars) of Tajima's D for 20 X chromosome loci in (A) Biaka pygmies and (B) Melanesians compared to simulated distributions for each population (curves). Tajima's D values for the majority of empirical loci are consistent with simulated distributions that are obtained from an isolation-with-migration model parameterized with the inferred demography of these two populations.
Discussion
It is generally appreciated that migration affects many important ecological and evolutionary properties of populations [25]. Yet there is little consensus in the literature on the role of gene flow in shaping patterns of human population structure. Equilibrium models, like the island model [9], were favored historically because analytical expectations are relatively simple to obtain. On the other hand, while it is accepted that migration occurs frequently among human populations [26-31], gene flow is often ignored in models of human demography. Recent examples ignoring gene flow include studies of African replacement models [32] and serial-founder models for the settlement of Eurasia [5]. Other studies of human demographic history analyze data separately for each population sample and simply neglect any effects of shared common ancestry or gene flow [7].
As pointed out by Whitlock and McCauley [25], both direct and indirect measures of migration and gene flow are fraught with difficulty. The use of summary statistics such as FST is limiting because it provides no insight into which historical processes are responsible for the observed genetic differences between populations. It is therefore left up to individual investigators to choose which model of population structure is used to interpret data, and inferences depend heavily upon the assumptions inherent in each model. We set out to directly estimate rates of gene flow between human populations through the use of the isolation-with-migration (IM) model, which incorporates both population splitting and gene flow. For this purpose, we have analyzed a large DNA sequence database collected with the expressed purpose of constructing models of human demographic history, and hence, is focused on intergenic (i.e., putatively neutral) regions on the X chromosome [16]. Values of FST for this dataset were found to be slightly higher than those estimated from other large X chromosome resequence datasets [11,12]. This difference is probably driven, at least in part, by the inclusion of more isolated groups than were included in other DNA sequence surveys of X-linked loci (i.e., the San from Namibia, Biaka from Central African Republic, and Melanesians from Papua New Guinea) [16].
To disentangle the evolutionary processes underlying FST in real human populations, we inferred N, m and t separately for the six populations in our survey using the IM model. While the mean global value for ancestral population size, ~104, is consistent with previous estimates of the global population size of modern humans [33-37], no individual population approaches the effective size of modern humans as a whole. Although sizes of individual populations estimated from nuclear loci remain sparse, other studies have produced similar results [2,38-40], including previous applications of the IM model to human genomic sequences [41]. Non-African population sizes (N0 ≈ 3000) estimated from linkage disequilibrium (LD) [42] are particularly noteworthy because recombination information is independent of the site frequency spectrum, which we used to infer effective sizes here. That similar results are obtained using unrelated subsets of the genetic information, and different populations, reinforces our confidence in these low estimates of effective population size.
This finding is important because many studies make the simplifying assumption that individual human populations have an effective population size of 104 [e.g., [43]]. We note that estimates of N based on the standard neutral model (i.e., θ = 3Nμ) are 2–4 times higher than those returned by IM for individual African and Eurasian samples, and are even higher for our Melanesian sample (see Table 1 in Wall et al. [16]). We suspect that this difference is largely due to violations of assumptions of the Wright-Fisher model rather than a systematic bias in parameters returned by IM (see Results). For example, population substructure and gene flow are expected to upwardly bias estimates of within population diversity (e.g., θ) when applying models that do not incorporate these variables.
Indeed, we demonstrate here that rates of gene flow between subdivided human populations are non-zero. For unidirectional migration rates (m1, m2), ~87% of pairwise comparisons (i.e., 42 of 48) showed gene flow in at least one direction (i.e., their 95% confidence intervals exclude zero migration; Table 4 in Additional file 1). When bidirectional migration rates (m = m1 + m2) are considered, lower bounds of 95% confidence intervals are greater than zero for all 15 pairwise comparisons. Furthermore, mean bidirectional migration rates within Africa (2.7 × 10-4/generation), and between Africans and Eurasians (2.1 × 10-4), are significantly lower than migration rates within Eurasia (1.5 × 10-3).
Correspondingly, estimates of the population migration rate range from 0.2–4.9 (mean Nm = 2.4). This implies that 2–3 X chromosome copies, on average, move between human populations every generation, although this stationary estimate does not explain how migration events are distributed through time. We infer slightly higher population migration rates within Africa and within Eurasia than between continents (Figure 3). Interestingly, because effective sizes and migration rates are inversely related in African and non-African populations, values of Nm are similar within Africa and Eurasia (Figure 4). Furthermore, migration rates are not associated with geographic proximity in any simple fashion. For instance, Basque and Han are located far apart geographically, but exhibit one of the highest rates of gene flow. This presumably reflects factors affecting historical mobility that are more complex than simple isolation-by-distance.
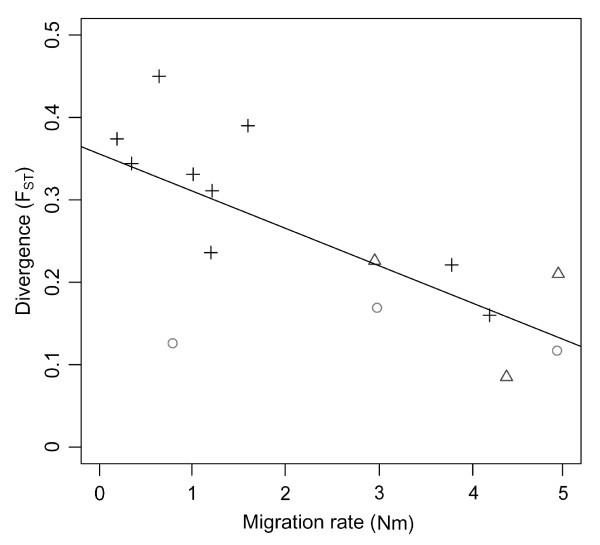
Figure 3.
Correlation between population divergence (FST) and inter-deme migration (Nm). African population pairs are indicated by circles, non-African population pairs by triangles, and African/non-African population pairs by crosses.
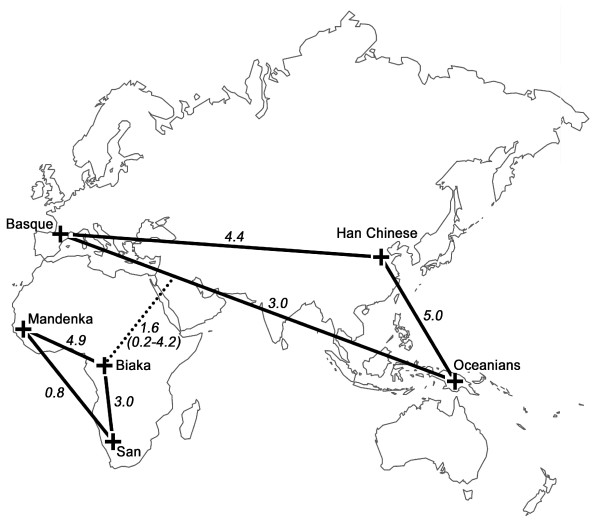
Figure 4.
Geographic representation of population migration rates Nm. Mean and range of Nm are provided for African/non-African population pairs.
Clearly, our results cannot be interpreted under either the pure splitting or island models, which assume no gene flow and no shared ancestry, respectively. In any case, under an island model, a population migration rate Nm greater than ~0.25 would be too high to explain the value of FST observed here (cf. Figure 2A in Additional file 1). Indeed, no further differentiation of local island populations is expected when Nm > 1 [44,45]. Note, however, that the axiom whereby exchanging one gene copy per generation inhibits population divergence does not hold for the IM model; populations can continue to diverge, albeit slowly, even when Nm = 3.3 (Figure 2C in Additional file 1). Importantly too, FST depends on interactions between a suite of demographic parameters, including N, m and t (Figure 3 in Additional file 1). These results highlight the limited power of FST to yield insights into human demographic processes without further knowledge of population divergence times, effective sizes and migration rates (i.e., the very parameters that we often attempt to infer from FST). Although FST is often considered directly proportional to divergence time, where migration is assumed to be absent and all population sizes identical [46:29–30], these assumptions do not hold for the human populations examined here. Thus, caution is warranted when interpreting FST as a simple proxy for population history [e.g., [5,46]].
In sum, we have independently inferred effective population sizes, times of divergence and rates of migration under an IM model of population structure based on the analysis of a large X chromosome DNA sequence database. The parameters that we have estimated for six globally distributed populations indicate relatively high levels of migration (e.g., mean Nm = 2.4) (Figure 4). A simple interpretation of our results is that human populations have exchanged migrants at a constant high rate during the entire history since their divergence from a common ancestral population. A more realistic interpretation is that they have experienced changing migration rates through time. Unfortunately, we cannot currently discriminate between these two scenarios; IM can only infer a stationary rate of gene flow (i.e., a time averaged rate). Our initial explorations permitting migration rates to change through time suggest that this process can have significant effects on the value of FST (e.g., see Figure 4 in Additional file 1). There is also independent evidence that rates of gene flow may be increasing towards the present. For example, the very large and asymmetric gene flow that we observe between Chinese Han and Melanesians may well result from the late Holocene (3–4 kya) expansion of agricultural populations recorded not only by haploid genetic loci [47,48], but also archaeologically and linguistically [49]. This is similar to Wakeley's [50] inference based on a maximum likelihood analysis of restriction fragment length polymorphisms (RFLPs) from across the genome that human populations have transitioned from a period of higher to lower population structure through an increased rate of migration towards the present. We also acknowledge that long-term mean rates of migration may be affected by both recurrent gene flow restricted by geographical distance [51] and long range dispersals by groups of migrants (i.e., range expansions), which can occur sporadically and still have large effects on global values of FST [29,52,53]. Interestingly, population genetic models with spatial and temporal fluctuation in the population migration rate (averaging around Nm ~1 in an island model) have been shown to be conducive both to the rapid spread of beneficial mutations throughout the species and local population differentiation [54]. We note that theoretical predictions based upon other models of subdivision (such as metapopulation models) [8,52] would be worth developing to further explore the nature and extent of human population structure.
Finally, the finding of high rates of gene flow among human populations has important implications for how we interpret the distribution of SNPs associated with disease [55] and the role of natural selection in shaping patterns of diversity [31,52,53]. For instance, recent migration produces haplotypes with extended linkage disequilibrium that could be misinterpreted as recent selection [56]. Gene flow can also significantly skew the site frequency spectrum within populations [8], and may therefore lead to erroneous inferences regarding demography.
Conclusion
While the maximum effective size of modern humans is estimated at ~10,000, individual populations vary substantially in size. African populations tend to be larger (2,300–9,000) than non-African populations (300–3,300). We independently estimate mean rates of bidirectional gene flow at 4.8 × 10-4/generation, and these rates are higher among non-African populations (1.5 × 10-3) than among African populations (2.7 × 10-4). Interestingly, because effective sizes and migration rates are inversely related in African and non-African populations, effective migration rates are similar globally (e.g., mean Nm = 2.4). While significant theoretical challenges remain in disentangling the evolutionary factors that structure human populations, it is clear that migration can no longer be treated as a simple, equilibrium parameter – or ignored – as it often is in reconstructions of human history.
Methods
Genomic data
Our database comprises 20 loci from intergenic regions on the X chromosome. Each region chosen for sequencing spans ~20 kb of primarily single-copy non-coding (i.e., putatively non-functional) DNA in regions of medium or high recombination, which are at least 50 kb away and recombinationally unlinked from the nearest gene [16]. No two regions are within one megabase of each other. Within each region, there are ~4–6 kb of sequence data from 3 discrete subsections that span most of the distance of each region. 10–24 X chromosomes are sampled from each population, which include 3 from Africa (10 San from Namibia, 16 Biaka pygmies from Central African Republic, 18 Mandenka from Senegal) and 3 from Eurasia/Oceania (16 French Basque, 16 Han Chinese, 24 Melanesians) (see Wall et al. [16] for a complete description of the sequencing strategy).
FST estimates
FST can be calculated using several different algorithms. Here, we adopt the approach of Hudson et al. [57], defined in terms of polymorphic site heterozygosity, which we have amended to accommodate unequal sample sizes [58] and missing data.
| (1) |
where Hw (≡ πw) is the mean distance per polymorphic site sampled from the same population, and Hb (≡ πb) is the mean distance per polymorphic site sampled from both populations. Reported values represent the mean FST at all segregating sites across all 20 X chromosome loci.
The expected value of FST for X chromosome loci under the island model with an infinite number of demes depends only on the product of the effective population size N, and the migration rate per generation m [10: 294-5]
| ⟨FST⟩ ≈ (1 + 3 Nm)-1 | (2) |
FST estimates must be corrected if a finite number of demes d are intended instead [59]
| (3) |
The population-scaled rate of gene flow Nm can be derived by simple rearrangement of equation 3.
Correspondingly, the expected value of FST for X chromosome loci under a divergence model depends only on the divergence time t, in generations, scaled by the effective population size N [13]
| (4) |
Demographic inference
Genetic diversity at twenty X chromosome loci is applied to determine the most likely parameterization for a series of paired population isolation-with-migration models. Seven demographic parameters are inferred from the genomic data under each two-deme IM model: effective population size of the ancestral deme (NA), effective population sizes of the two descendent demes (N1 and N2), unidirectional migration rates between descendent populations (m1 and m2), proportion of the ancestral population founding the first deme (S), and population divergence time (t). Populations are analyzed in all pairwise combinations using the Markov chain Monte Carlo Bayesian/maximum likelihood framework implemented in the 31 July 2006 version of IM http://lifesci.rutgers.edu/~heylab/heylabsoftware.htm#IM. More complete descriptions of this method are available elsewhere [14,15,41,60,61]. The IM software infers all coalescent parameters, except S, as mutation-scaled rates – θ1 = 3N1μ, θ2 = 3N2μ, θA = 3NAμ, m1 = m1/μ, m2 = m2/μ, and t = tμ – which, if mutation rates are known, can subsequently be transformed to real world values (i.e., actual population sizes, migration rates and chronological dates). Mutation rates per year are estimated for each locus from mean Homo/Pan sequence divergence assuming a split time of 6 × 106 years. Per generation rates assume a mean generation interval of 28 years, as estimated from cross-cultural ethnographic data [62]. A 1:1 mating ratio is also assumed throughout. Migration rates m1 and m2 are inferred in the coalescent (i.e., backward in time), and bidirectional migration rates m are simply the summation of m1 and m2.
Because the IM algorithm has most power with perfectly treelike data (an infinite sites implementation), datasets with no evidence of recombination were extracted from each locus using the four-gamete approach of Hudson and Kaplan [63]. Individuals and segregating sites are given equal weighting, and the largest non-recombining block maximizing overall information content is selected for each locus, a practice that minimizes any bias in the resulting dataset [64]. Analyses are run at a mean CPU speed of 2.5 GHz on an ~100 core Condor grid at the University of Arizona Computer Science Department. All datasets are initially parameterized from a single run with bounded uniform priors: θ1, θ2, θA ∈ U(0, 40), m1, m2 ∈ U(0, 20), S ∈ U(0, 1) and t ∈ U(0, 3). Ranges are raised or lowered in subsequent runs to incorporate complete marginal posterior probability densities. Once these bounds are established, a minimum of four replicate jobs each of >5 chains are run for a minimum of 107 steps. Chain mixing by Metropolis-Hasting coupling, long run times and multiple independent runs allow us to identify convergence on each parameter's underlying stationary distribution.
Because we observe little variation among multiple independent runs (e.g., Figure 1 in Additional file 1), marginal posterior probability densities for a given parameter are combined and analyzed as a single probability distribution. Maximum probability estimates and 95% confidence intervals are reported as modes and {0.025, 0.975} quantiles of posterior probability densities, respectively. We estimate all seven parameters of the model (Tables 1–5 in Additional file 1) for all population pairs. Uninformative posterior distributions are sometimes encountered for some population pairs; in particular, the ancestral population split time parameter t often proves difficult to infer (Table 5 in Additional file 1). However, the seven demographic parameters are inferred as marginal densities, and therefore uncertainty in one parameter has no impact on the remaining parameters. Parameter values are treated as unknown unless we observe clear maximum likelihood peaks that are replicated across multiple independent IM jobs.
Coalescent simulations for demographic parameter validation
The nonlinear relationship between gene flow, divergence time and FST under the isolation-with-migration model is explored using coalescent simulation with the software ms [65]. An IM model conditioned on mean values for the seven demographic parameters, as inferred from the empirical dataset (Tables 1–5 in Additional file 1), is implemented for each population pair. A suite of summary statistics is explored under these models: θW and θπ, which are unbiased estimators of the population mutation rate; Tajima's D, which summarizes the population site frequency spectrum; and FST, which summarizes the joint site frequency spectrum. Observed values of these four summary statistics, calculated from the empirical dataset, are compared to the summary statistic distributions returned by coalescent simulation. A Bonferroni correction holding the experiment-wise type-I error rate constant at α = 0.05 is applied to account for the use of multiple per-population tests. A Mantel test is used to determine the correlation for FST, a between-population (i.e., matrix) test.
Authors' contributions
MPC participated in the design of the study, contributed to data collection, ran analyses, and wrote the manuscript. AEW contributed to data collection. JDW advised on data analysis, and provided comments on the manuscript. MFH participated in the design of the study, advised on data analysis, and helped revise the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Supplemental Materials for "Intergenic DNA sequences from the human X chromosome reveal high rates of global gene flow". This document contains tables and figures showing additional results referenced in the main text.
Acknowledgments
Acknowledgements
We are grateful to J Hey (Rutgers University) and F Mendez (University of Arizona) for helpful discussion; and S Kobourov for access to the dispersed-computing cluster in the Department of Computer Science (University of Arizona). The National Science Foundation helped fund genetic data collection and analysis via the grant BCS-0423670 to M.F.H. and J.D.W, as well as providing computational support via the San Diego Supercomputing Center under TeraGrid grant DBS060002T to M.P.C.
Contributor Information
Murray P Cox, Email: mpcox@u.arizona.edu.
August E Woerner, Email: augustw@u.arizona.edu.
Jeffrey D Wall, Email: wallj@humgen.ucsf.edu.
Michael F Hammer, Email: mfh@u.arizona.edu.
References
- Garrigan D, Hammer MF. Reconstructing human origins in the genomic era. Nat Rev Genet. 2006;7:669–680. doi: 10.1038/nrg1941. [DOI] [PubMed] [Google Scholar]
- Frisse L, Hudson RR, Bartoszewicz A, Wall JD, Donfack J, Di Rienzo A. Gene conversion and different population histories may explain the contrast between polymorphism and linkage disequilibrium levels. Am J Hum Genet. 2001;69:831–843. doi: 10.1086/323612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinan A, Mullikin JC, Patterson N, Reich D. Measurement of the human allele frequency spectrum demonstrates greater genetic drift in East Asians than in Europeans. Nat Genet. 2007;39:1251–1255. doi: 10.1038/ng2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prugnolle F, Manica A, Balloux F. Geography predicts neutral genetic diversity of human populations. Curr Biol. 2005;15:R159–160. doi: 10.1016/j.cub.2005.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S, Deshpande O, Roseman CC, Rosenberg NA, Feldman MW, Cavalli-Sforza LL. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc Natl Acad Sci USA. 2005;102:15942–15947. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray N, Currat M, Excoffier L. Intra-deme molecular diversity in spatially expanding populations. Mol Biol Evol. 2003;20:76–86. doi: 10.1093/molbev/msg009. [DOI] [PubMed] [Google Scholar]
- Voight BF, Adams AM, Frisse LA, Qian Y, Hudson RR, Di Rienzo A. Interrogating multiple aspects of variation in a full resequencing data set to infer human population size changes. Proc Natl Acad Sci USA. 2005;102:18508–18513. doi: 10.1073/pnas.0507325102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley J, Aliacar N. Gene genealogies in a metapopulation. Genetics. 2001;159:893–905. doi: 10.1093/genetics/159.2.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. Breeding structure of populations in relation to speciation. Amer Nat. 1940;74:232–248. doi: 10.1086/280891. [DOI] [Google Scholar]
- Wright S. The Theory of Gene Frequencies. Vol. 2. Chicago: University of Chicago Press; 1969. Evolution and the Genetics of Populations. [Google Scholar]
- Akey JM, Zhang G, Zhang K, Jin L, Shriver MD. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 2002;12:1805–1814. doi: 10.1101/gr.631202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Nielsen R, Wakeley J. Distinguishing migration from isolation: A Markov chain Monte Carlo approach. Genetics. 2001;158:885–896. doi: 10.1093/genetics/158.2.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J, Nielsen R. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics. 2004;167:747–760. doi: 10.1534/genetics.103.024182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD, Cox MP, Mendez FL, Woerner A, Severson T, Hammer MF. A novel DNA sequence database for analyzing human demographic history. Genome Res. 2008;18:1354–1361. doi: 10.1101/gr.075630.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M, Scholz SW, Scheet P, Gibbs JR, VanLiere JM, Fung H-C, Szpiech ZA, Degnan JH, Wang K, Guerreiro R, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman MW, Cavalli-Sforza LL, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- Clark AG, Hubisz MJ, Bustamante CD, Williamson SH, Nielsen R. Ascertainment bias in studies of human genome-wide polymorphism. Genome Res. 2005;15:1496–1502. doi: 10.1101/gr.4107905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Cardon LR, Anderson AD, Nielsen R, Hill WG. Measures of human population structure show heterogeneity among genomic regions. Genome Res. 2005;15:1468–1476. doi: 10.1101/gr.4398405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Prugnolle F, Manica A, Balloux F. A geographically explicit genetic model of worldwide human-settlement history. Am J Hum Genet. 2006;79:230–237. doi: 10.1086/505436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingman M, Kaessmann H, Pääbo S, Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature. 2000;408:708–713. doi: 10.1038/35047064. [DOI] [PubMed] [Google Scholar]
- Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 2008;18:830–838. doi: 10.1101/gr.7172008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins JF, Marlowe FW. Sex-biased migration in humans: what should we expect from genetic data? Bioessays. 2006;28:290–300. doi: 10.1002/bies.20378. [DOI] [PubMed] [Google Scholar]
- Whitlock MC, McCauley DE. Indirect measures of gene flow and migration: FST ≠ 1/(4Nm + 1) Heredity. 1999;82:117–125. doi: 10.1038/sj.hdy.6884960. [DOI] [PubMed] [Google Scholar]
- Eller E. Population substructure and isolation by distance in three continental regions. Am J Phys Anthropol. 1999;108:147–159. doi: 10.1002/(SICI)1096-8644(199902)108:2<147::AID-AJPA2>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Fix AG. Migration and Colonization in Human Microevolution. Cambridge: Cambridge University Press; 1999. [Google Scholar]
- Jorde LB. Human genetic distance studies: Present status and future prospects. Ann Rev Anthropol. 1985;14:343–373. doi: 10.1146/annurev.an.14.100185.002015. [DOI] [Google Scholar]
- Relethford JH. Genetics and modern human origins. Evol Anthropol. 1995;4:53–63. doi: 10.1002/evan.1360040207. [DOI] [Google Scholar]
- Santos EJM, Epplen JT, Epplen C. Extensive gene flow in human populations as revealed by protein and microsatellite DNA markers. Hum Hered. 1997;47:165–172. doi: 10.1159/000154405. [DOI] [PubMed] [Google Scholar]
- Wijsman EM, Cavalli-Sforza LL. Migration and genetic population structure with special reference to humans. Ann Rev Ecol Syst. 1984;15:279–301. doi: 10.1146/annurev.es.15.110184.001431. [DOI] [Google Scholar]
- Fagundes NJR, Ray N, Beaumont M, Neuenschwander S, Salzano FM, Bonatto SL, Excoffier L. Statistical evaluation of alternative models of human evolution. Proc Natl Acad Sci USA. 2007;104:17614–17619. doi: 10.1073/pnas.0708280104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlich HA, Bergstrom TF, Stoneking M, Gyllensten U. HLA sequence polymorphism and the origin of humans. Science. 1996;274:1552–1554. doi: 10.1126/science.274.5292.1552. [DOI] [PubMed] [Google Scholar]
- Haigh J, Maynard Smith J. Population size and protein variation in man. Genet Res. 1972;19:73–89. doi: 10.1017/s0016672300014282. [DOI] [PubMed] [Google Scholar]
- Lahr MM, Foley RA. Towards a theory of modern human origins: Geography, demography and diversity in recent human evolution. Am J Phys Anthropol. 1998:137–176. doi: 10.1002/(sici)1096-8644(1998)107:27+<137::aid-ajpa6>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Sherry ST, Harpending HC, Batzer MA, Stoneking M. Alu evolution in human populations: Using the coalescent to estimate effective population size. Genetics. 1997;147:1977–1982. doi: 10.1093/genetics/147.4.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N. Allelic genealogy and human evolution. Mol Biol Evol. 1993;10:2–22. doi: 10.1093/oxfordjournals.molbev.a039995. [DOI] [PubMed] [Google Scholar]
- Garrigan D, Kingan SB, Metni Pilkington M, Wilder JA, Cox MP, Soodyall H, Strassmann B, Destro-Bisol G, de Knijff P, Novelletto A, et al. Inferring human population sizes, divergence times and rates of gene flow from mitochondrial, X and Y chromosome resequencing data. Genetics. 2007;177:2195–2207. doi: 10.1534/genetics.107.077495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataillon T, Mailund T, Thorlacius S, Steingrimsson E, Rafnar T, Halldorsson MM, Calian V, Schierup MH. The effective size of the Icelandic population and the prospects for LD mapping: Inference from unphased microsatellite markers. Eur J Hum Genet. 2006;14:1044–1053. doi: 10.1038/sj.ejhg.5201669. [DOI] [PubMed] [Google Scholar]
- Harris EE, Hey J. X chromosome evidence for ancient human histories. Proc Natl Acad Sci USA. 1999;96:3320–3324. doi: 10.1073/pnas.96.6.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J. On the number of New World founders: A population genetic portrait of the peopling of the Americas. PLoS Biol. 2005;3:e193. doi: 10.1371/journal.pbio.0030193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenesa A, Navarro P, Hayes BJ, Duffy DL, Clarke GM, Goddard ME, Visscher PM. Recent human effective population size estimated from linkage disequilibrium. Genome Res. 2007;17:520–526. doi: 10.1101/gr.6023607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpending H, Rogers A. Genetic perspectives on human origins and differentiation. Annu Rev Genomics Hum Genet. 2000;1:361–385. doi: 10.1146/annurev.genom.1.1.361. [DOI] [PubMed] [Google Scholar]
- Crow J, Kimura M. An Introduction to Population Genetics Theory. Minneapolis, MN: Burgess Publishing Company; 1970. [Google Scholar]
- Spieth PT. Gene flow and genetic differentiation. Genetics. 1974;78:961–965. doi: 10.1093/genetics/78.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A. The History and Geography of Human Genes. Princeton: Princeton University Press; 1994. [Google Scholar]
- Cox MP. Indonesian mitochondrial DNA and its opposition to a Pleistocene era origin of proto-Polynesians in Island Southeast Asia. Hum Biol. 2005;77:179–188. doi: 10.1353/hub.2005.0037. [DOI] [PubMed] [Google Scholar]
- Cox MP, Lahr MM. Y-Chromosome diversity is inversely associated with language affiliation in paired Austronesian- and Papuan-speaking communities from Solomon Islands. Am J Hum Biol. 2006;18:35–50. doi: 10.1002/ajhb.20459. [DOI] [PubMed] [Google Scholar]
- Bellwood P. The First Farmers: The Origins of Agricultural Societies. Oxford: Blackwell Publishing; 2005. [Google Scholar]
- Wakeley J. Nonequilibrium migration in human history. Genetics. 1999;153:1863–1871. doi: 10.1093/genetics/153.4.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. Isolation by distance. Genetics. 1943;28:114–138. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M. Gene flow in natural populations. Ann Rev Ecol Syst. 1985;16:393–430. doi: 10.1146/annurev.ecolsys.16.1.393. [DOI] [Google Scholar]
- Templeton AR. Human races: A genetic and evolutionary perspective. Am Anthropologist. 1999;100:632–650. doi: 10.1525/aa.1998.100.3.632. [DOI] [Google Scholar]
- Barton NH, Rouhani S. Adaptation and the 'shifting balance'. Genet Res Camb. 1993;61:57–74. [Google Scholar]
- Reich DE, Cargill M, Bolk S, Ireland J, Sabeti PC, Richter DJ, Lavery T, Kouyoumjian R, Farhadian SF, Ward R, et al. Linkage disequilibrium in the human genome. Nature. 2001;411:199–204. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]
- Hawks J, Wang ET, Cochran GM, Harpending HC, Moyzis RK. Recent acceleration of human adaptive evolution. Proc Natl Acad Sci USA. 2008;104:20753–20758. doi: 10.1073/pnas.0707650104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Slatkin M, Maddison WP. Estimation of levels of gene flow from DNA sequence data. Genetics. 1992;132:583–589. doi: 10.1093/genetics/132.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagnol V, Wall JD. Possible ancestral structure in human populations. PLoS Genet. 2006;2:e105. doi: 10.1371/journal.pgen.0020105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M, Voelm L. FST in a hierarchical island model. Genetics. 1991;127:627–629. doi: 10.1093/genetics/127.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J, Won YJ, Sivasundar A, Nielsen R, Markert JA. Using nuclear haplotypes with microsatellites to study gene flow between recently separated Cichlid species. Mol Ecol. 2004;13:909–919. doi: 10.1046/j.1365-294X.2003.02031.x. [DOI] [PubMed] [Google Scholar]
- Won YJ, Hey J. Divergence population genetics of chimpanzees. Mol Biol Evol. 2005;22:297–307. doi: 10.1093/molbev/msi017. [DOI] [PubMed] [Google Scholar]
- Fenner JN. Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies. Am J Phys Anthropol. 2005;128:415–423. doi: 10.1002/ajpa.20188. [DOI] [PubMed] [Google Scholar]
- Hudson RR, Kaplan NL. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics. 1985;111:147–164. doi: 10.1093/genetics/111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woerner AE, Cox MP, Hammer MF. Recombination-filtered genomic datasets by information maximization. Bioinformatics. 2007;23:1851–1853. doi: 10.1093/bioinformatics/btm253. [DOI] [PubMed] [Google Scholar]
- Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Materials for "Intergenic DNA sequences from the human X chromosome reveal high rates of global gene flow". This document contains tables and figures showing additional results referenced in the main text.