Abstract
The dengue virus (DENV) has four distinct serotypes (DENV1, DENV2, DENV3, and DENV4) that require differentiation for effective prevention of morbid diseases. The recently developed DENV1-specific NS1 antigen capture enzyme-linked immunosorbent assay (ELISA) based on the monoclonal antibodies (MAbs) that recognize distinct epitopes on nonstructural protein 1 (NS1) of a specific DENV serotype is convenient and cost-effective, but assays have not yet been developed for DENV serotypes 2 to 4. This paper describes the development and validation of a DENV2-specific NS1 antigen capture ELISA by selection and optimization of the pair of well-characterized MAbs that recognized epitopes specific for DENV2 NS1 from a large panel of MAbs. The DENV2 NS1 ELISA displayed exclusive sensitivity with the DENV2 serotype and did not cross-react with the other three DENV serotypes. The sensitivity and specificity of the DENV2 NS1 ELISA were 83.3% (25/30) and 100% (504/504) when used to test 30 acute-phase serum samples from patients infected with DENV2 identified by virus isolation or reverse transcription-PCR serotyping and 504 serum samples from healthy individuals, respectively. The specificity of this assay was also evaluated using a panel of serum samples which were positive for DENV1, other flaviviruses, and nonflaviviruses; no cross-reactions were observed in these clinical samples. The DENV2 NS1 ELISA was eightfold more sensitive than a commercially available serotype-cross-reactive NS1 ELISA (Panbio Diagnostics, Brisbane, Australia) when the two assays were used to test the DENV2-infected cell culture supernatants in parallel. The Panbio NS1 ELISA displayed variation in sensitivity between DENV serotypes. The DENV2-specific NS1 antigen capture ELISA can be used as a tool for the rapid identification of DENV2 infections.
The mosquito-borne dengue disease is caused by infections with the dengue virus (DENV). It is a major public health problem that affects over 100 tropical and subtropical countries. Moreover, the geographical territory affected by this disease has increased markedly since the 1970s; contributing factors may include a lack of control over the mosquito vector, global warming, viral evolution, and increased ease of global travel (23). The DENV has four known serotypes (DENV1, DENV2, DENV3, and DENV4). An infection with any of these serotypes can cause dengue fever, dengue hemorrhagic fever (DHF), or dengue shock syndrome (DSS) (10). Subsequent heterologous infections may increase the risk of the developing DHF or DSS (19). Since protection against heterologous serotype infections is only partial and transient, people that live in epidemic areas are at risk for contracting an average of four DENV infections in their lives (14). Thus, the cocirculation of various DENV serotypes in a community is the most common risk factor associated with the emergence of the severe forms of disease (DHF or DSS) (13). Given the current absence of a vaccine that is effective against all four DENV serotypes (30), early diagnosis is the key to reducing the morbidity and mortality of DHF and DSS. Furthermore, symptoms of DENV infections are insufficiently specific for accurate clinical differentiation from other acute febrile illnesses, especially in areas where multiple tropical diseases such as malaria, yellow fever, West Nile disease, and St. Louis encephalitis are endemic. Case identification has become increasingly important in the differential diagnosis of the acute febrile illnesses and hemorrhagic fever of unknown flavivirus infections during the early symptomatic phase. Currently, molecular techniques (reverse transcription-PCR [RT-PCR]) have widely replaced traditional virus isolation methods in the early detection of DENV infections and differentiation of the four DENV serotypes and other related flavivirus infections (12, 21). Although molecular detection provides a promising sensitivity rate and rapid diagnosis, the molecular approach is costly, as it requires specialized laboratory equipment and experienced technicians; these are notable limitations in many developing countries where dengue disease is endemic (29).
An “ideal” simple, cost-effective, rapid, and accurate diagnostic tool is necessary to overcome the main disadvantages of current molecular techniques. Viral antigen detection methods that are convenient and cost-effective have been successfully used with various infectious diseases for the diagnosis and monitoring of disease activity (5, 28). DENV contains three structural (C, prM, and E) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (30). Among these proteins, NS1 is a relatively conserved 45- to 50-kDa glycoprotein that is highly expressed in infected cells in both membrane-associated and -secreted forms (8, 31). Although the precise function of the NS1 is unknown, it represents an interesting target antigen for diagnosis (1, 27, 35). Recently, two DENV NS1 antigen capture immunoassays have become commercially available for the early diagnosis of dengue virus (2, 3, 7, 18). The two NS1 antigen capture assays are suitable and sensitive and provide a rapid and reliable diagnosis of acute primary dengue disease and acute secondary dengue disease. However, one limitation of these assays is that they are unable to distinguish between the DENV serotypes, as the antibodies used in the assay are directed against cross-reactive antigenic determinants shared by all four DENV serotypes and other flaviviruses. NS1 has been identified as being a both group-specific and type-specific determinant (9, 15, 27); thus, the production of epitope-specific monoclonal antibodies (MAbs) holds potential for developing either group-specific or type-specific NS1 antigen assays. In a previous study, we prepared a panel of MAbs that recognized epitopes specific to DENV1 NS1 and successfully developed a DENV1 NS1 antigen capture ELISA (33). This DENV1-specific NS1 antigen assay has proved to be a valuable tool for the early detection and rapid identification of DENV1 infections. In the present study, we prepared a panel of MAbs that recognized epitopes specific to DENV2 NS1 and described the development of a DENV2-specific NS1 antigen capture ELISA.
MATERIALS AND METHODS
Serum specimens.
Clinical serum specimens used in this study were collected from patients with laboratory-confirmed DENV infections and other flavivirus or nonflavivirus infections. A total of 30 acute-phase (i.e., days 1 to 7 after onset) serum specimens from DENV2-infected patients were collected during an outbreak of DENV2 from August to November 2001 in Jiangmen, Guangdong Province, China (25). A total of 301 acute-phase serum specimens from DENV1-infected patients were collected during a DENV1 epidemic between 2002 and 2003 in Guangzhou, Guangdong Province, China (34). Laboratory diagnosis of DENV infections was performed at the Center for Disease Control and Prevention of Guangzhou, China, with DENV isolation in Aedes albopictus C6/36 cells (ATCC CRL-1660), followed by an immunofluorescence assay (IFA) with serotype-specific MAbs for DENV serotypes 1 to 4. Detection of viral RNA in serum specimens was carried out by a conventional RT-PCR with the Qiagen OneStep RT-PCR kit, using consensus primers targeting the C/pre-M genes of DENV, followed by a nested PCR with serotype-specific primers for DENV serotypes 1 to 4, as described previously (20). DENV-specific immunoglobulin M (IgM) and IgG antibodies were used with the commercial Panbio dengue IgM and IgG capture ELISA kits (Panbio, Brisbane, Australia) according to the manufacturer's instructions. In addition, clinical serum specimens from other flavivirus or nonflavivirus infections were used in this study, including 50 collected from Hantaan virus infections, 13 from Japanese encephalitis virus infections, 49 from measles virus infections, and 20 specimens from 18 patients with leptospirosis (33). Normal control serum specimens were obtained from 504 healthy humans. Aliquots of all serum specimens were stored at −80°C until they were used.
DENV propagation and plaque assay.
The four DENV serotype strains (DENV1, Hawaii; DENV2, New Guinea-C; DENV3, Guanxi-80-2; and DENV4, H241) used in this study were obtained from the Center for Disease Control and Prevention of Guangzhou, China. The viruses were propagated in C6/36 cells in Eagle's minimal essential medium at 33°C for 3 to 5 days. After cytopathic effects were observed, the cell culture supernatants were collected and clarified by centrifugation. The titers of each virus pool were determined by a plaque assay protocol in Vero-E6 cells (ATCC CRL-1586) as previously reported (26). Stocks of viruses were prepared at known plaque titers (PFU per milliliter) and kept frozen at −80°C until they were used.
Preparation of recombinant NS1 protein.
The gene encoding the NS1 protein was amplified from total viral RNA extracted from DENV2-infected C6/36 cells by RT-PCR with a forward primer (5′-CGGGATCCGATAGTGGTTGCGTTGTGAGCTGGAAAAAC-3′) and a reverse primer (5′-GGGGTACCTTAGGCTGTGACCAAGGAGTTGACCAAATTC-3′). The amplified NS1 gene was then ligated into the BamHI and KpnI sites of the prokaryotic expression vector pQE30 (Qiagen, Hilden, Germany) in frame and downstream of the six-His tag coding sequence. The six-His-tagged recombinant DENV2 NS1 protein (rDENV2-NS1) was expressed in Escherichia coli as previously described (33) with some modifications in the purification. In brief, the rDENV2-NS1 protein was denatured in 8 M urea and was further purified by Ni-nitrilotriacetic acid affinity chromatography (Qiagen) according to manufacturer's instructions. The solubilized rDENV2-NS1 protein was then refolded by slowly dialyzing with refolding buffers as previously described (16). The identification of rDENV2-NS1 was achieved by Western blot analysis with both DENV2-immunized rabbit serum (Abcam, Cambridge, United Kingdom) and the mouse anti-His MAb (Novagen, North Ryde, New South Wales, Australia). The concentration of protein was determined by the Coomassie Plus protein assay (Pierce Biotechnology, Rockford, IL) according to the manufacturer's instructions. The other three DENV serotype recombinant NS1 proteins were prepared with the same protocol in our laboratory for use in this study (data not shown). The four serotype NS1 proteins were used for characterizing MAb specificity and cross-reactivity determination.
Preparation and identification of MAbs.
The preparation and identification of MAbs against the NS1 protein were performed as previously described with some modifications (33). Briefly, 4- to 6-week-old female BALB/c mice were first immunized with subcutaneous injections of 1 dose of formalin-inactivated DENV2-infected culture supernatants administered at 10-day intervals. This was followed by 1 dose of refolded rDENV2-NS1 (10 μg). The mice were then given two boosters with inactivated DENV2 or rDENV2-NS1 at 10-day intervals. Ten days later, all mice were bled, and the serum samples were tested by ELISA with rDENV2-NS1 as the coating antigen to determine which animal had the greatest response to rDENV2-NS1. This mouse was given a further intravenous inoculation of 10 μg of rDENV2-NS1 in phosphate-buffered saline, and its splenocytes were fused with NS1 myeloma cells 3 days later. The hybridoma cell lines were screened primarily by an indirect ELISA with both rDENV2-NS1 and DENV2-infected cell lysates as coating antigens. The screening was further confirmed by an indirect IFA that detected MAb binding on C6/36 cells infected with each DENV serotype, according to our previously described protocol (4, 33). The specificities and cross-reactivities of the MAbs were evaluated by Western blot analysis with the four DENV serotype recombinant NS1 proteins as well as the infected cell lysates from each DENV serotype. Positive hybridoma cells were cloned by limiting dilution. The isotypes of each MAb were determined by use of a commercially available mouse MAb isotyping kit (Zymed Laboratories, Carlsbad, CA). Purified MAbs from ascitic fluids were conjugated by EZ-Link sulfo-N-hydroxysuccinimide-biotin reagents (Pierce) in accordance with the manufacturer's instructions.
Analysis of MAb-binding epitopes.
The binding epitopes of MAbs were analyzed by a competition ELISA with rDENV2-NS1 as the coating antigen, as described previously with modification (11, 33). Microwell plates (Costar Corning Inc., Corning, NY) were coated with 100 μl/well of the rDENV2-NS1 protein at a concentration of 1 μg/ml in coating buffer. After the blocking steps were performed, a constant concentration of one of the nonbiotinylated MAbs (50 μl) was incubated with an optimal concentration of different biotinylated MAbs (50 μl) for 1 h at 26°C. After the plates were washed, streptavidin conjugated with horseradish peroxidase (HRP; Zymed) was added, and plates were incubated for 30 min at 26°C. The plates were then washed, and binding of the biotinylated MAb was detected by the addition of tetramethylbenzidine (KPL, Gaithersburg, VA). The reaction was stopped after 10 min by the addition of 1 N sulfuric acid, and the plates were then examined in an ELISA plate reader (Bio-Tek, Winooski, VT). An irrelevant, unlabeled MAb was used as a control. The percentage of inhibition was calculated by the following formula: [1 − (OD450 of the test well/OD450 of the control well)] × 100 (where OD450 is the optical density at 450 nm). The results were described as follows: inhibition of >75% represented competitive binding, inhibition from 25 to 75% represented relatively competitive binding, and inhibition of <25% represented noncompetitive binding (33).
Development of MAb-based DENV2 NS1 antigen capture ELISA.
To select the best combination of capture and detector antibodies for an antigen capture ELISA, MAbs were paired according to their abilities to recognize different epitopes on NS1. The procedure was modified from that previously described (33). In brief, microwell plates (Costar) were coated with 100 μl/well of each capture MAb at a concentration of 10 μg/ml overnight at 4°C. After the blocking steps were performed, a series of known rDENV2-NS1 concentrations, virus-infected culture supernatants, and uninfected controls were added to duplicate wells (100 μl/well) and incubated for 1 h at 37°C. After the plates were washed, the biotinylated MAb was added (100 μl/well), and the plates were then incubated for 30 min at 26°C. Following incubation, streptavidin-HRP and tetramethylbenzidine were added, color development was terminated, and absorbance measurements were performed as described above.
Detection of NS1 antigen in viral culture supernatants and clinical samples by using the DENV2 NS1 antigen capture ELISA.
For testing viral culture supernatants, the inactivated viral culture supernatants that had been quantified by plaque assay from the four DENV serotypes were diluted in phosphate-buffered saline. For testing clinical serum samples, all serum samples were treated with a dissociation buffer (1.5 M glycine, pH 2.8) to dissociate the immune complexes, followed by neutralization with a neutralization buffer (1.5 M Tris-HCl, pH 9.7) as previously described (17). The process for assaying NS1 was performed by the DENV2 NS1 antigen capture ELISA as described above. The end point dilution of detectable NS1 in viral culture supernatants was defined as the highest culture dilution that resulted in an OD450 value twofold higher than that of uninfected culture supernatants. Normal serum specimens obtained from 504 healthy humans were included as controls to establish the normal range of the assay for clinical evaluation.
Detection of NS1 antigen by using a commercial DENV NS1 antigen capture ELISA.
The NS1 antigens in viral culture supernatants and clinical serum samples were measured using a commercially available Pan-E dengue early ELISA test kit (Panbio Diagnostics, Brisbane, Australia) according to the manufacturer's instructions. Briefly, the diluted positive and negative controls, calibrator, and serum samples of dengue patients were added to microwell plates coated with anti-NS1 antibody, and the plates were incubated for 1 h at 37°C. This was followed by the addition of HRP-conjugated anti-NS1 MAb and a second incubation for 1 h at 37°C; finally, the substrate reaction was performed. An index value was calculated, according to the manufacturer's instructions, by the following formula (absorbance values supplied with the kit): [sample absorbance (OD450)/mean calibrator absorbance (OD450)] × the calibration factor. The results were calculated by the following formula: Panbio units = index value × 10. Results of <9, 9 to 11, and >11 Panbio units were considered negative, equivocal, and positive, respectively.
Data analysis.
For evaluation of clinical serum samples, the sensitivity of the assay was calculated as the ratio of the number of clinical samples that tested positive to the total number of samples that were positively identified by virus isolation or RT-PCR; the specificity of the assay was calculated as the ratio of the number of samples that tested negative to the total number of negative samples. The derived data in the figures were tabulated in Microsoft Office Excel 2003 worksheets. Statistically significant differences (P < 0.05) in NS1 detection rates between acute primary infections and acute secondary infections were calculated using McNemar's chi-square test.
RESULTS
Characterization of MAb specific reactivity with DENV2 NS1 and development of a DENV2 NS1 antigen capture ELISA.
A total of 37 hybridoma cell lines that stably produced MAbs were initially established on the basis of their strong positive reactivity with both the rDENV2-NS1 protein and DENV2-infected cell lysates in ELISA (Table 1). The immunoglobulin isotype determinations revealed that the 37 MAbs comprised IgG2a (n = 1), IgG2b (n = 3), and IgG1 (n = 33). Western blot analysis with the rDENV2-NS1 protein and DENV2-infected cell culture supernatants revealed that all MAbs reacted to the same immunoreactive protein band with an approximate molecular mass of 45 kDa (data not shown). The serotype specificity of the MAbs was further identified by testing their reactivity to the four DENV serotypes in the IFA and the rDENV2-NS1-based ELISA. Of the 37 MAbs, 20 MAbs specifically reacted to DENV2 with no cross-reactivity to the other three DENV serotypes in either the ELISA or the IFA. The remaining 17 MAbs had cross-reactivity to the four DENV serotypes in different cross-reactivity patterns. The results demonstrated that 20 MAbs specific for DENV2 could be useful for developing a serotype-specific DENV2 NS1 antigen capture ELISA according to our previously described protocol (5, 33). Next, the distinct binding epitopes of the 20 DENV2 NS1-specific MAbs were determined by competition ELISA using rDENV2-NS1 as the immobilized antigen. The results showed that the 20 MAbs bound to at least four different epitopes on the DENV2 NS1 protein. Among the 20 MAbs, each was paired according to their abilities to recognize distinct epitopes on NS1 (Table 2). Then, a sandwich ELISA was performed to select the optimal capture-detector MAb pair. Pairing between eight MAbs in epitope group III and nine MAbs in epitope group IV displayed high sensitivity in DENV2 NS1 protein detection. Ultimately, the pairing of MAb M6 as a solid-phase immobilized capture antibody and MAb M14 as a labeled detecting antibody gave the highest combination of sensitivity and specificity in the detection of DENV2-specific NS1 in serial dilutions of DENV2-infected cell culture supernatants (based on infectious titer) and a panel of serum specimens from healthy humans.
TABLE 1.
Characteristics of MAbs raised against the NS1 proteina
| Hybridoma (original clone no.) | Isotype | Reactivity with rDENV2-NS1
|
IFA result from cells infected by:
|
||||
|---|---|---|---|---|---|---|---|
| ELISA | WB | DENV1 | DENV2 | DENV3 | DENV4 | ||
| M1 (1E22A3) | IgG2b | + | ++++ | − | +++ | − | − |
| M2 (1F7A5) | IgG1 | ++++ | ++++ | − | ++ | − | − |
| M3 (3D34A16) | IgG1 | ++++ | ++++ | − | ++ | − | − |
| M4 (4B41A1) | IgG1 | +++ | ++ | − | ++++ | − | − |
| M5 (4D26A2) | IgG1 | ++++ | ++ | − | ++++ | − | − |
| M6 (4D41A6) | IgG1 | +++ | ++ | − | ++++ | − | − |
| M7 (5A62A12) | IgG1 | ++++ | +++ | − | + | − | − |
| M8 (5C15A13) | IgG1 | ++++ | + | − | ++++ | − | − |
| M9 (5E22A7) | IgG1 | ++++ | ++ | − | ++++ | − | − |
| M10 (5E27A12) | IgG1 | ++++ | ++ | − | ++++ | − | − |
| M11 (5E48A15) | IgG1 | +++ | + | − | ++ | − | − |
| M12 (4D35A16) | IgG1 | ++++ | + | − | ++ | − | − |
| M13 (4E93A3) | IgG1 | ++++ | + | − | ++ | − | − |
| M14 (5A47A8) | IgG2b | ++++ | +++ | − | +++ | − | − |
| M15 (5A70A4) | IgG1 | ++++ | ++ | − | +++ | − | − |
| M16 (5C29A9) | IgG1 | ++++ | + | − | ++++ | − | − |
| M17 (5D5A2) | IgG1 | ++++ | +++ | − | ++ | − | − |
| M18 (5D21A1) | IgG1 | ++++ | + | − | +++ | − | − |
| M19 (5E19A5) | IgG1 | ++++ | + | − | ++ | − | − |
| M20 (5E30A5) | IgG1 | ++++ | ++ | − | ++++ | − | − |
| M21 (1D36A3) | IgG1 | ++++ | ++ | ++ | + | ++ | ++ |
| M22 (1F2A27) | IgG1 | ++++ | ++ | + | + | + | + |
| M23 (1F75A4) | IgG1 | ++++ | + | + | + | + | + |
| M24 (4C69A19) | IgG1 | ++++ | +++ | ++ | ++++ | ++++ | ++++ |
| M25 (4D42A1) | IgG1 | ++++ | ++ | ++ | +++ | ++++ | ++++ |
| M26 (5A7A5) | IgG1 | ++++ | ++ | +++ | +++ | ++++ | ++++ |
| M27 (5A11A5) | IgG1 | ++++ | ++ | +++ | +++ | ++++ | ++++ |
| M28 (5A27A3) | IgG1 | ++++ | ++ | +++ | +++ | ++++ | ++++ |
| M29 (3D5A1) | IgG1 | ++++ | + | +++ | ++ | +++ | ++ |
| M30 (5C22A5) | IgG2b | ++++ | + | + | + | + | + |
| M31 (1A63A20) | IgG1 | ++++ | ++ | ++ | + | − | + |
| M32 (1D1A1) | IgG1 | ++++ | + | + | + | + | − |
| M33 (3D9A8) | IgG1 | ++++ | ++ | + | + | + | + |
| M34 (4A61A8) | IgG1 | ++++ | ++ | ++ | +++ | ++ | + |
| M35 (4B17A12) | IgG1 | ++++ | ++ | ++ | +++ | ++ | + |
| M36 (4D67A6) | IgG1 | ++++ | +++ | +++ | ++++ | +++ | +++ |
| M37 (4E58A12) | IgG2a | ++++ | +++ | + | ++++ | + | + |
WB, Western blotting; −, none; +, weak; ++, intermediate; +++, strong; ++++, very strong.
TABLE 2.
Analysis of distinct epitope binding of MAbs by competition ELISAa
| MAb | Epitope group | % Inhibition of the biotin-labeled MAbb:
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M1 | M2 | M3 | M4 | M5 | M6 | M7 | M8 | M9 | M10 | M11 | M12 | M13 | M14 | M15 | M16 | M17 | M18 | M19 | M20 | ||
| M1 | I | 97 | 98 | 13 | 19 | 5 | 0 | 0 | 0 | 11 | 5 | 11 | 0 | 0 | 0 | 0 | 21 | 0 | 22 | 18 | 15 |
| M2 | I | 97 | 97 | 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 3 | 0 |
| M3 | II | 39 | 0 | 94 | 35 | 38 | 30 | 38 | 18 | 23 | 14 | 35 | 20 | 9 | 7 | 22 | 27 | 0 | 22 | 29 | 23 |
| M4 | III | 31 | 0 | 10 | 96 | 94 | 84 | 67 | 89 | 94 | 91 | 84 | 0 | 0 | 0 | 0 | 29 | 2 | 18 | 26 | 6 |
| M5 | III | 8 | 0 | 4 | 92 | 93 | 79 | 67 | 82 | 92 | 87 | 78 | 4 | 0 | 0 | 0 | 19 | 0 | 20 | 16 | 13 |
| M6 | III | 36 | 0 | 12 | 93 | 90 | 90 | 78 | 86 | 89 | 81 | 84 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| M7 | III | 3 | 0 | 29 | 96 | 96 | 94 | 97 | 94 | 96 | 96 | 93 | 0 | 0 | 0 | 5 | 32 | 3 | 20 | 25 | 19 |
| M8 | III | 17 | 0 | 13 | 96 | 95 | 90 | 74 | 92 | 96 | 95 | 90 | 5 | 5 | 0 | 0 | 29 | 3 | 26 | 30 | 23 |
| M9 | III | 20 | 7 | 23 | 95 | 95 | 81 | 76 | 92 | 95 | 94 | 84 | 25 | 32 | 15 | 36 | 10 | 0 | 23 | 24 | 14 |
| M10 | III | 0 | 1 | 6 | 95 | 95 | 82 | 71 | 92 | 96 | 95 | 87 | 21 | 17 | 12 | 13 | 20 | 3 | 22 | 29 | 13 |
| M11 | III | 9 | 0 | 31 | 83 | 83 | 73 | 75 | 78 | 81 | 71 | 89 | 24 | 0 | 0 | 8 | 10 | 0 | 12 | 18 | 4 |
| M12 | IV | 28 | 0 | 17 | 7 | 18 | 0 | 0 | 0 | 28 | 36 | 34 | 92 | 93 | 94 | 91 | 93 | 94 | 90 | 92 | 84 |
| M13 | IV | 41 | 2 | 0 | 23 | 24 | 4 | 0 | 12 | 23 | 28 | 0 | 63 | 85 | 81 | 74 | 80 | 60 | 69 | 73 | 63 |
| M14 | IV | 33 | 0 | 17 | 16 | 21 | 0 | 0 | 0 | 21 | 13 | 0 | 95 | 90 | 96 | 95 | 96 | 87 | 96 | 96 | 94 |
| M15 | IV | 0 | 0 | 17 | 14 | 15 | 5 | 42 | 0 | 24 | 15 | 0 | 89 | 90 | 91 | 95 | 95 | 88 | 82 | 86 | 89 |
| M16 | IV | 17 | 0 | 22 | 26 | 5 | 2 | 0 | 0 | 14 | 14 | 11 | 90 | 90 | 94 | 94 | 96 | 85 | 87 | 89 | 93 |
| M17 | IV | 0 | 5 | 34 | 37 | 42 | 8 | 0 | 3 | 29 | 40 | 0 | 94 | 90 | 94 | 96 | 97 | 97 | 89 | 92 | 96 |
| M18 | IV | 0 | 0 | 19 | 14 | 4 | 0 | 36 | 0 | 22 | 39 | 0 | 92 | 92 | 95 | 92 | 95 | 81 | 96 | 96 | 91 |
| M19 | IV | 27 | 0 | 19 | 27 | 27 | 0 | 0 | 0 | 20 | 35 | 0 | 90 | 92 | 94 | 90 | 92 | 69 | 92 | 92 | 83 |
| M20 | IV | 6 | 5 | 38 | 5 | 0 | 3 | 0 | 8 | 28 | 21 | 0 | 93 | 91 | 95 | 96 | 97 | 91 | 90 | 92 | 95 |
The competition ELISA was performed to analyze the binding epitopes of the different MAbs. By use of the criteria described in Materials and Methods, MAbs M1 and M2 effectively competed (≥97% inhibition) with each other, which suggested that these two MAbs recognized the same epitope. Similarly, the 60 to 97% inhibition of each other's binding of MAbs M4, M5, M6, M7, M8, M9, M10, and M11 and MAbs M12, M13, M14, M15, M16, M17, M18, M19, and M20 suggested that they form two other distinct groups. MAb M3 did not interfere with heterologous MAb binding (≤39% inhibition), suggesting that this MAb may recognize a fourth discrete epitope. Therefore, these 20 MAbs recognized four distinct epitopes, and MAbs in each group reacted with the same epitope or with sterically overlapping epitopes on the DENV2 NS1 protein. The experiment was repeated with similar results.
Competitive inhibition values of ≥75% are in boldface type.
Specificity, sensitivity, and reproducibility of the DENV2 NS1 antigen capture ELISA.
To evaluate the sensitivity of the antigen capture assay, a series of known rDENV2-NS1 concentrations were analyzed. A standard curve for the NS1 protein was constructed, as shown in Fig. 1. Bovine serum albumin (BSA) was used to establish the baseline for the assay, and a sample was considered positive if the OD450 value was twice that of BSA. With these criteria, the minimal amount of rDENV2-NS1 detection with this assay was approximately 0.3 to 0.5 ng/well. The linear portion of the standard curve ranged from 10 to 100 ng/ml and could be used to estimate the NS1 levels in clinical samples (Fig. 1).
FIG. 1.
Standard curve for NS1 binding in the DENV2 NS1 antigen capture ELISA determined with purified rDENV2-NS1. Various concentrations of rDENV2-NS1 were analyzed. BSA was used to establish the baseline. Data points represent the mean ± the standard deviation of the results for four replicates.
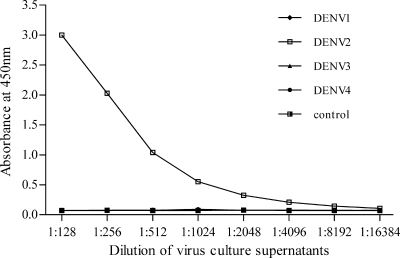
The specificity and sensitivity of the DENV2 NS1 ELISA were further analyzed by testing the presence of NS1 in the culture supernatants from the four DENV serotypes and comparing them with those of a commercially available dengue NS1 antigen capture ELISA (Panbio NS1 ELISA). Twofold serial dilutions of each DENV serotype that had been quantified by plaque assay were subjected to the analysis. The DENV2 NS1 ELISA demonstrated exceptional sensitivity toward the DENV2-infected culture supernatants, and no positive results were observed for the other three DENV serotypes (Fig. 2). In contrast, the Panbio NS1 ELISA revealed marked variations in cross-reactivity with the four DENV serotypes; the highest dilutions for detecting NS1 were 1:2,048 (13.2 PFU/0.1 ml) for DENV1, 1:512 (46.8 PFU/0.1 ml) for DENV2, 1:128 (367.2 PFU/0.1 ml) for DENV3, and 1:16 (10,000 PFU/0.1 ml) for DENV4 (Table 3). We also compared the sensitivities of the DENV2 NS1 ELISA with those of the Panbio NS1 ELISA in the detection of NS1 in the serial dilutions of DENV2-infected culture supernatants. Based on the cutoff values of the two assays, the highest dilution for detection of NS1 was 1:4,096 (5.8 PFU/0.1 ml) in the DENV2 NS1 ELISA, while the highest dilution for detection of NS1 was 1:512 (46.8 PFU/0.1 ml) in the Panbio NS1 ELISA (Table 4). Thus, the DENV2 NS1 ELISA demonstrated a sensitivity approximately eightfold higher than that of the Panbio NS1 ELISA.
FIG. 2.
Evaluation of the sensitivity and specificity of the DENV2 NS1 antigen capture ELISA for the detection of NS1 in cell cultures. Serial dilutions of supernatants obtained from virus cultures infected with the four DENV serotypes were subjected to the DENV2 NS1 antigen capture ELISA. Positive OD450 values were only observed for the DENV2 serotype (the data points for the other serotypes overlap those of the control).
TABLE 3.
Detection of the presence of NS1 in infected cell culture supernatants from the four DENV serotypes with the Panbio NS1 ELISA
| DENV (original concn) | NS1 detected at indicated serial twofold dilutiona
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1:2 | 1:4 | 1:8 | 1:16 | 1:32 | 1:64 | 1:128 | 1:256 | 1:512 | 1:1,024 | 1:2,048 | 1:4,096 | 1:8,192 | 1:16,384 | |
| DENV1 (2.7 × 105 PFU/ml) | 133/+ | 133/+ | 133/+ | 133/+ | 133/+ | 122/+ | 89.6/+ | 56.3/+ | 37.6/+ | 20.6/+ | 13.2/+ | 8.9/− | 7.5/− | 6.4/− |
| DENV2 (2.4 × 105 PFU/ml) | 133/+ | 133/+ | 133/+ | 104/+ | 71.5/+ | 48.3/+ | 29.2/+ | 19.1/+ | 12.4/+ | 8.6/− | 7.2/− | 6.4/− | 5.8/− | 5.6/− |
| DENV3 (4.7 × 105 PFU/ml) | 133/+ | 118/+ | 96.7/+ | 67.5/+ | 43.5/+ | 25.2/+ | 16.0/+ | 8.9/− | 7.4/− | 6.4/− | 6.2/− | 5.7/− | 5.6/− | 5.4/− |
| DENV4 (1.6 × 106 PFU/ml) | 40.8/+ | 25.2/+ | 17.8/+ | 11.2/+ | 8.9/− | 7.5/− | 6.7/− | 5.6/− | 5.8/− | 5.5/− | 5.5/− | 5.2/− | 5.2/− | 5.2/− |
| Control (uninfected culture) | 5.8/− | 5.8/− | 5.6/− | 5.6/− | 5.4/− | 5.6/− | 5.6/− | 5.6/− | 5.5/− | 5.4/− | 5.2/− | 5.4/− | 5.2/− | 5.2/− |
Results are given as Panbio units/results, with results of <9, 9 to 11, and >11 defined as negative (−), equivocal (±), and positive (+), respectively, according to the manufacturer's instructions. The positive results are in boldface type. The experiment was repeated twice with similar results.
TABLE 4.
Comparison of sensitivities of the DENV2 NS1 ELISA with the Panbio NS1 ELISA in detection of the presence of NS1 in DENV2-infected cell culture supernatants
| NS1 ELISA | ELISA results at indicated serial twofold dilution of DENV2-infected cell culture supernatantsa
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1:2 | 1:4 | 1:8 | 1:16 | 1:32 | 1:64 | 1:128 | 1:256 | 1:512 | 1:1,024 | 1:2,048 | 1:4,096 | 1:8,192 | 1:16,384 | |
| DENV2 | 3.00/+ | 3.00/+ | 3.00/+ | 3.00/+ | 3.00/+ | 3.00/+ | 3.00/+ | 2.03/+ | 1.04/+ | 0.55/+ | 0.32/+ | 0.21/+ | 0.14/− | 0.11/− |
| Panbio | 133/+ | 133/+ | 133/+ | 104/+ | 71.5/+ | 48.3/+ | 29.2/+ | 19.1/+ | 12.4/+ | 8.6/− | 7.2/− | 6.4/− | 5.8/− | 5.6/− |
Supernatants were infected with DENV2 at 2.4 × 105 PFU/ml prior to dilution. For the DENV 2 NS1 ELISA, the results are given as OD450 value/results; positive detection of NS1 in viral culture supernatants was defined as an OD450 value twofold higher than that of uninfected culture supernatants. For the Panbio NS1 ELISA, the results are given as Panbio units/results; results of <9, 9 to 11, and >11 were defined as negative (−), equivocal (±), and positive (+) detection of NSI, respectively, according to the manufacturer's instructions. Positive results are in boldface. The experiment was repeated twice with similar results.
Furthermore, the intra- and interassay variation of the DENV2 NS1 ELISA was assessed with mock clinical samples. Four normal human serum samples were spiked with DENV2 of a known plaque titer at different dilutions of 1:100, 1:200, 1:400, and 1:800 to mock different infection levels. The intra-assay coefficients of variation were determined by measuring 10 replicates of the four mock clinical samples in the same assay run; they were 3% (1:100), 2.8% (1:200), 2.7% (1:400), and 2.9% (1:800). The interassay coefficients of variation were determined by measuring the same samples daily over a 10-day period; they were 4.9% (1:100), 5.6% (1:200), 5.1% (1:400), and 4.3% (1:800).
Examination of clinical serum samples by the DENV2 NS1 antigen capture ELISA.
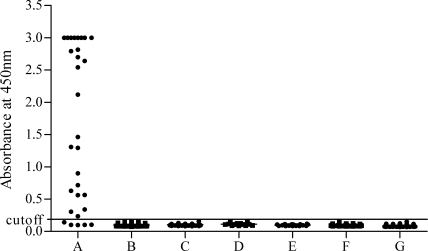
Serum specimens from 504 healthy humans were analyzed to establish the background signals of the DENV2 NS1 ELISA before further clinical evaluation. The mean OD450 value for these 504 serum samples with the DENV2 NS1 ELISA was 0.087, with a standard deviation of 0.02. The cutoff detection limit for the clinical samples was set as the average value of the control serum samples, plus 3 standard deviations; this produced a cutoff OD450 value of 0.147. With these criteria, we found that no false-positive results were obtained with any of the 504 samples (Fig. 3), indicating that the specificity of the DENV2 NS1 ELISA was 100% (0 of 504). In order to eliminate any potential false-positive results, we selected a higher cutoff value of 0.187 based on the average value of the control serum samples, plus 5 standard deviations. The result was considered positive if a sample yielded an OD450 value above the cutoff value.
FIG. 3.
Evaluation of the sensitivity and specificity of the DENV2 NS1 antigen capture ELISA for detection of the NS1 antigen in serum samples. Data represent the OD450 values of serum samples tested at a dilution of 1:10. Serum samples were taken from the following sources: patients infected with DENV2 identified by virus isolation or RT-PCR serotyping (n = 30) (A); patients with documented DENV1 infections (n = 301) (B); patients infected with Hantan virus (n = 50) (C); patients infected with Japanese encephalitis virus (n = 13) (D); patients infected with leptospirosis (n = 20) (E); patients infected with measles virus (n = 49) (F); and healthy humans (n = 504) (G).
To evaluate the accuracy of the DENV2 NS1 ELISA in detecting NS1 in clinical serum samples, we assessed a total of 30 acute-phase serum specimens from patients with laboratory-confirmed DENV2 infection. Among these specimens, 10 were confirmed by both DENV2 isolation and RT-PCR, and 20 were confirmed by both RT-PCR and Panbio dengue IgM or IgG capture ELISAs. According to World Health Organization criteria (32), 17 serum samples were serologically classified as having acute primary infections, and 13 were classified as having secondary infections. Based on a sample dilution of 1:10, the DENV2 NS1 ELISA detected NS1 in 25 of the 30 acute-phase serum specimens. The overall sensitivity of DENV2 NS1 antigen detection with respect to virus isolation and/or RT-PCR was 83.3% (25 of 30). These results were consistent with the results from the Panbio NS1 ELISA when the assays were performed in parallel at a 1:10 dilution; the sensitivity for both assays was 83.3% (25 of 30), and five samples did not have detectable NS1 with either assay. Both NS1 ELISAs detected 88.2% (15/17) of the acute primary infections and 76.9% (10/13) of the acute secondary infections. There was no statistically significant difference in NS1 detection rate between samples with acute primary infections and those with acute secondary infections (P = 0.410).
To further evaluate the sensitivities of both NS1 detection assays, serial dilutions of the NS1-positive serum samples were analyzed. From a total of 25 serum samples in which the NS1 antigen was positively detected by both assays, only 19 samples had sufficient residual volume to allow us to further compare the sensitivities of both NS1 detection assays. Table 5 shows a summary comparing the sensitivities of the two NS1 detection assays. Since the DENV2 NS1 assay displayed a markedly higher sensitivity in the DENV2-infected culture supernatants, as described above, it was not unexpected that DENV2 NS1 remained detectable even when a patient's serum was at a 1:10,000 dilution.
TABLE 5.
Comparison of sensitivities of the DENV2 NS1 ELISA and the Panbio NS1 ELISA for the detection of serial dilutions of serum samples from DENV2-infected patients
| Serum dilution | No. of samples positive by indicated ELISA/total no. of samples tested (%)
|
|
|---|---|---|
| DENV2 | Panbio | |
| 1:10 | 25/30 (83.3) | 25/30 (83.3) |
| 1:40 | 19/19 (100) | 15/19 (78.9) |
| 1:80 | 19/19 (100) | 13/19 (68.4) |
| 1:160 | 17/19 (89.5) | 10/19 (52.6) |
| 1:320 | 15/19 (78.9) | 5/19 (26.3) |
| 1:640 | 12/19 (63.2) | 4/19 (21.1) |
| 1:1,280 | 6/19 (31.6) | 2/19 (10.5) |
| 1:2,560 | 4/19 (21.1) | 1/19 (5.3) |
| 1:5,120 | 2/19 (10.5) | 0/19 (0.0) |
| 1:10,240 | 1/19 (5.3) | 0/19 (0.0) |
The DENV2 NS1 ELISA was also evaluated on a panel of clinical serum samples from patients infected with DENV1 or other flaviviruses and nonflaviviruses. We analyzed a total of 301 acute-phase serum samples that had previously tested positive for NS1 by the DENV1 NS1 antigen capture ELISA (33). Additionally, we tested a total of 132 acute-phase serum samples, comprising 50 samples from 50 cases of hemorrhagic fever with Hantaan virus infections, 13 samples from 13 cases of Japanese encephalitis, 49 samples from 49 cases of measles, and 20 samples from 18 cases of leptospirosis. We found that none of these samples were positive for NS1 by the DENV2 NS1 ELISA (Fig. 3).
DISCUSSION
Rapid laboratory diagnosis of DENV infections and the differentiation of multiple DENV infections in the acute phase of illness are important for timely clinical management and epidemiological control in areas where multiple flaviviruses are endemic. However, currently, speed and accuracy of diagnosis must be balanced against the cost of testing, especially in developing countries that have limited laboratory facilities. An appropriate test that offers convenience and cost-effectiveness is needed for the rapid diagnosis of virus infections (6, 22, 28).
We previously developed a DENV1 NS1 antigen capture ELISA with MAbs specific for the DENV1 NS1 protein (33). To continue this line of development, in the current study, we raised hybridomas against the combination of rDENV2-NS1 protein and native NS1 from the DENV2-infected culture. We screened the MAbs to identify those that recognized specific epitopes on DENV2 NS1, and we simultaneously eliminated clones that cross-reacted to the other three DENV serotypes. Because selecting well-characterized MAbs is a crucial element in the development of antigen detection assays, we evaluated pairs of MAbs that recognized distinct epitopes of NS1, and we used the optimal pair to design the NS1 antigen capture ELISA. The optimal pair was MAbs M6 and M14, and we found that the sensitivity of the resulting DENV2 NS1 ELISA was eightfold greater than that of a commercial Panbio NS1 ELISA tested in parallel on DENV2-infected cell culture supernatants. This reason for the dramatic increase in sensitivity was that the DENV2 NS1 ELISA used MAbs that recognized predominant epitopes on NS1 that were distinct for the DENV2 serotype, while the Panbio NS1 ELISA used MAbs that recognized epitopes on NS1 that cross-reacted among different serotypes. The latter had difficulty finding MAbs with equally high affinity targeting the predominant epitopes on NS1 in the presence of all four DENV serotypes; thus, its sensitivity appeared to vary between the different serotypes, which may reduce detection. Our observation is in agreement with another published evaluation of the Panbio NS1 ELISA, which found sensitivities with the highest rates in DENV1-positive sera and the lowest rates in DENV4-positive sera (2).
We determined that the sensitivity of our NS1 antigen capture ELISA was as low as 3 ng/ml (0.3 ng/well) when purified rDENV2-NS1 was used. These results indicated that the DENV2 NS1 ELISA is well suited for serum NS1 measurement, as the level of circulating NS1 in acute-phase serum samples is within the range of 10 ng/ml to 50 μg/ml (1). A further clinical assessment of the sensitivity and specificity of the DENV2 NS1 ELISA was performed with well-characterized clinical serum specimens. Our DENV2 NS1 ELISA detected NS1 antigen in 25 of 30 (83.3%) virus culture and/or viral RNA-positive acute-phase serum samples from laboratory-confirmed DENV2-infected patients. The positive results were consistent with those of the Panbio NS1 ELISA performed in parallel. However, our DENV2 NS1 assay displayed higher sensitivity than did the Panbio NS1 ELISA when tested with serial dilutions of patients’ sera with the detection of a very low antigen titer (1:10,000 dilution) in the specimen, suggesting that our DENV2 NS1 assay can ensure that clinical samples with a low antigen titer would be recognized as being DENV positive. Although the NS1 antigen detection assay was less sensitive than molecular detection of DENV genomic RNA by RT-PCR, there are some advantages of the NS1 antigen assay over molecular detection of viral RNA. First, the antigen test can be performed with simple procedures that do not require expensive equipment, and it also allows simultaneous processing of multiple samples. Second, the viral antigen is considerably more stable than viral RNA in clinical specimens. On the other hand, a diagnostic strategy is a combination of NS1 antigen and IgM antibody tests, which could increase the overall diagnostic sensitivity (7). This strategy could particularly allow a medical center lacking sophisticated equipment to identify DENV serotype infections early enough to adjust patient management and enhancement of epidemiological control measures.
The development of the NS1 antigen capture ELISAs has been attributed to several groups. Alcon et al. and Young et al. used rabbit and mouse anti-NS1 polyclonal antibodies, respectively, raised against immunoaffinity-purified NS1 to construct an NS1 antigen capture ELISA (1, 35). However, both these antigen assays were based on polyclonal antibodies and, thus, were subject to intra- and interlaboratory variability due to batch-to-batch variations in antisera. The MAb-based assay has several advantages over the polyclonal antibody-based assay; primarily, MAbs from stable hybridoma clones ensure a continuous supply of large quantities of well-characterized antibodies that can be easily standardized among different laboratories. Recently, two commercialized MAb-based NS1 antigen capture ELISAs have been developed (3, 7, 18). Both assays demonstrated that the NS1 antigen capture ELISA might be a promising tool for the diagnosis of acute primary dengue and acute secondary dengue; but both assays had the limitation that they were unable to distinguish among the four DENV serotypes. In a previously published study, we reported a DENV1 NS1 antigen capture ELISA (33). We demonstrated that this DENV1 NS1 assay can specifically identify DENV1 infections. In the present study, we described the development and validation of a DENV2 NS1 antigen capture ELISA that uses well-characterized MAbs specific for DENV2 NS1. The present results confirm that NS1 is an effective marker for facilitating early detection and serotyping of DENV infections. One limitation of the present study is the relatively low number of clinical serum specimens. Further work is needed to determine the sensitivity of the assay with a larger number of patients with acute primary and secondary DENV infections at different times in the course of DENV infection. In the future, the work will assess the clinical value of the assay, with the aim of developing a full test combining the four individual serotype assays, perhaps based on an immunochip, which can be performed at the bedside in daily routine screening of suspect patients. It may also serve as a surveillance tool for monitoring natural DENV infections in mosquitoes in the field (24).
Acknowledgments
This work was supported by grant 2006AA02Z446 from the National High Technology Research and Development Program of China, grant 30671874 of the Research Program of the National Natural Science Foundation of China, and grant 30725031 of the National Outstanding Young Scientist Foundation of China.
Footnotes
Published ahead of print on 19 November 2008.
REFERENCES
- 1.Alcon, S., A. Talarmin, M. Debruyne, A. Falconar, V. Deubel, and M. Flamand. 2002. Enzyme-linked immunosorbent assay specific to dengue virus type 1 nonstructural protein NS1 reveals circulation of the antigen in the blood during the acute phase of disease in patients experiencing primary or secondary infections. J. Clin. Microbiol. 40376-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bessoff, K., M. Delorey, W. Sun, and E. Hunsperger. 2008. Comparison of two commercially available dengue NS1 capture enzyme-linked immunosorbent assays using a single clinical sample for diagnosis of acute DENV infection. Clin. Vaccine Immunol. 151513-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blacksell, S. D., M. P. Mammen, Jr., S. Thongpaseuth, R. V. Gibbons, R. G. Jarman, K. Jenjaroen, A. Nisalak, R. Phetsouvanh, P. N. Newton, and N. P. Day. 2008. Evaluation of the Panbio dengue virus nonstructural 1 antigen detection and immunoglobulin M antibody enzyme-linked immunosorbent assays for the diagnosis of acute dengue infections in Laos. Diagn. Microbiol. Infect. Dis. 6043-49. [DOI] [PubMed] [Google Scholar]
- 4.Che, X. Y., L. W. Qiu, Z. Y. Liao, Y. D. Wang, K. Wen, Y. X. Pan, W. Hao, Y. B. Mei, V. C. Cheng, and K. Y. Yuen. 2005. Antigenic cross-reactivity between severe acute respiratory syndrome-associated coronavirus and human coronaviruses 229E and OC43. J. Infect. Dis. 1912033-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Che, X. Y., L. W. Qiu, Y. X. Pan, K. Wen, W. Hao, L. Y. Zhang, Y. D. Wang, Z. Y. Liao, X. Hua, V. C. Cheng, and K. Y. Yuen. 2004. Sensitive and specific monoclonal antibody-based capture enzyme immunoassay for detection of nucleocapsid antigen in sera from patients with severe acute respiratory syndrome. J. Clin. Microbiol. 422629-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crafford, J. E., A. J. Guthrie, M. van Vuuren, P. P. Mertens, J. N. Burroughs, P. G. Howell, and C. Hamblin. 2003. A group-specific, indirect sandwich ELISA for the detection of equine encephalosis virus antigen. J. Virol. Methods 112129-135. [DOI] [PubMed] [Google Scholar]
- 7.Dussart, P., B. Labeau, G. Lagathu, P. Louis, M. R. Nunes, S. G. Rodrigues, C. Storck-Herrmann, R. Cesaire, J. Morvan, M. Flamand, and L. Baril. 2006. Evaluation of an enzyme immunoassay for detection of dengue virus NS1 antigen in human serum. Clin.Vaccine Immunol. 131185-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falconar, A. K., and P. R. Young. 1990. Immunoaffinity purification of native dimer forms of the flavivirus non-structural glycoprotein, NS1. J. Virol. Methods 30323-332. [DOI] [PubMed] [Google Scholar]
- 9.Falconar, A. K., and P. R. Young. 1991. Production of dimer-specific and dengue virus group cross-reactive mouse monoclonal antibodies to the dengue 2 virus non-structural glycoprotein NS1. J. Gen. Virol. 72961-965. [DOI] [PubMed] [Google Scholar]
- 10.George, R., and L. C. S. Lum. 1997. Clinical spectrum of dengue infection. p. 89-114. In D. J. Gubler and G. Kuno (ed.), Dengue and dengue hemorrhagic fever. CAB International, London, United Kingdom.
- 11.Graves, D. C., S. J. McNabb, M. H. Ivey, and M. A. Worley. 1986. Development and characterization of monoclonal antibodies to Pneumocystis carinii. Infect. Immun. 51125-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grobusch, M. P., M. Niedrig, K. Gobels, K. Klipstein-Grobusch, and D. Teichmann. 2006. Evaluation of the use of RT-PCR for the early diagnosis of dengue fever. Clin. Microbiol. Infect. 12395-397. [DOI] [PubMed] [Google Scholar]
- 13.Gubler, D. J. 1998. Dengue and dengue hemorrhagic fever. Clin. Microbiol. Rev. 11480-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guzmán, M. G., and G. Kouri. 2002. Dengue: an update. Lancet Infect. Dis. 233-42. [DOI] [PubMed] [Google Scholar]
- 15.Henchal, E. A., L. S. Henchal, and B. K. Thaisomboonsuk. 1987. Topological mapping of unique epitopes on the dengue-2 virus NS1 protein using monoclonal antibodies. J. Gen. Virol. 68845-851. [DOI] [PubMed] [Google Scholar]
- 16.Huang, J. L., J. H. Huang, R. H. Shyu, C. W. Teng, Y. L. Lin, M. D. Kuo, C. W. Yao, and M. F. Shaio. 2001. High-level expression of recombinant dengue viral NS-1 protein and its potential use as a diagnostic antigen. J. Med. Virol. 65553-560. [PubMed] [Google Scholar]
- 17.Koraka, P., C. P. Burghoorn-Maas, A. Falconar, T. E. Setiati, K. Djamiatun, J. Groen, and A. D. Osterhaus. 2003. Detection of immune-complex-dissociated nonstructural-1 antigen in patients with acute dengue virus infections. J. Clin. Microbiol. 414154-4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumarasamy, V., A. H. Wahab, S. K. Chua, Z. Hassan, Y. K. Chem., M. Mohamad, and K. B. Chua. 2007. Evaluation of a commercial dengue NS1 antigen-capture ELISA for laboratory diagnosis of acute dengue virus infection. J. Virol. Methods 14075-79. [DOI] [PubMed] [Google Scholar]
- 19.Kurane, I., and F. E. Ennis. 1992. Immunity and immunopathology in dengue virus infections. Semin. Immunol. 4121-127. [PubMed] [Google Scholar]
- 20.Lanciotti, R. S., C. H. Calisher, D. J. Gubler, G. J. Chang, and A. V. Vorndam. 1992. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J. Clin. Microbiol. 30545-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levi, J. E., A. F. Tateno, A. F. Machado, D. C. Ramalho, V. A. de Souza, A. O. Guilarde, V. C. de Rezende Feres, C. M. Martelli, M. D. Turchi, J. B. Siqueira, Jr., and C. S. Pannuti. 2007. Evaluation of a commercial real-time PCR kit for detection of dengue virus in samples collected during an outbreak in Goiania, Central Brazil, in 2005. J. Clin. Microbiol. 451893-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindsay, R., I. Barker, G. Nayar, M. Drebot, S. Calvin, C. Scammell, C. Sachvie, T. S. Fleur, A. Dibernardo, M. Andonova, and H. Artsob. 2003. Rapid antigen-capture assay to detect West Nile virus in dead corvids. Emerg. Infect. Dis. 91406-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mackenzie, J. S., D. J. Gubler, and L. R. Petersen. 2004. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat. Med. 10S98-S109. [DOI] [PubMed] [Google Scholar]
- 24.Philip Samuel, P., and B. K. Tyagi. 2006. Diagnostic methods for detection & isolation of dengue viruses from vector mosquitoes. Indian J. Med. Res. 123615-628. [PubMed] [Google Scholar]
- 25.Ren, R. W., M. Y. Fang, W. Y. Hong, B. M. Huang, L. H. Jiang, J. W. Liu, X. D. Tian, and G. F. Cheng. 2003. Isolation, identification and sequence analyses of dengue virus type 2 strain GD19/2001. Zhonghua Liu Xing Bing Xue Za Zhi 24288-290. [PubMed] [Google Scholar]
- 26.Roehrig, J. T., J. Hombach, and A. D. Barrett. 2008. Guidelines for plaque-reduction neutralization testing of human antibodies to dengue viruses. Viral Immunol. 21123-132. [DOI] [PubMed] [Google Scholar]
- 27.Shu, P. Y., L. K. Chen, S. F. Chang, Y. Y. Yueh, L. Chow, L. J. Chien, C. Chin, H. H. Yang, T. H. Lin, and J. H. Huang. 2002. Potential application of nonstructural protein NS1 serotype-specific immunoglobulin G enzyme-linked immunosorbent assay in the seroepidemiologic study of dengue virus infection: correlation of results with those of the plaque reduction neutralization test. J. Clin. Microbiol. 401840-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sutthent, R., N. Gaudart, K. Chokpaibulkit, N. Tanliang, C. Kanoksinsombath, and P. Chaisilwatana. 2003. p24 antigen detection assay modified with a booster step for diagnosis and monitoring of human immunodeficiency virus type 1 infection. J. Clin. Microbiol. 411016-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teles, F. R., D. M. Prazeres, and J. L. Lima-Filho. 2005. Trends in dengue diagnosis. Rev. Med. Virol. 15287-302. [DOI] [PubMed] [Google Scholar]
- 30.Whitehead, S. S., J. E. Blaney, A. P. Durbin, and B. R. Murphy. 2007. Prospects for a dengue virus vaccine. Nat. Rev. Microbiol. 5518-528. [DOI] [PubMed] [Google Scholar]
- 31.Winkler, G., S. E. Maxwell, C. Ruemmler, and V. Stollar. 1989. Newly synthesized dengue-2 virus nonstructural protein NS1 is a soluble protein but becomes partially hydrophobic and membrane-associated after dimerization. Virology 171302-305. [DOI] [PubMed] [Google Scholar]
- 32.World Health Organization. 1997. Dengue haemorrhagic fever: diagnosis, treatment, prevention and control. World Health Organization, Geneva, Switzerland.
- 33.Xu, H., B. Di, Y. X. Pan, L. W. Qiu, Y. D. Wang, W. Hao, L. J. He, K. Y. Yuen, and X. Y. Che. 2006. Serotype 1-specific monoclonal antibody-based antigen capture immunoassay for detection of circulating nonstructural protein NS1: implications for early diagnosis and serotyping of dengue virus infections. J. Clin. Microbiol. 442872-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ying, R. S., X. P. Tang, F. C. Zhang, W. P. Cai, Y. Q. Chen, J. Wang, W. X. Hong, and Y. Z. Long. 2007. Clinical characteristics of the patients with dengue fever seen from 2002 to 2006 in Guangzhou. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi 21123-125. [PubMed] [Google Scholar]
- 35.Young, P. R., P. A. Hilditch, C. Bletchly, and W. Halloran. 2000. An antigen capture enzyme-linked immunosorbent assay reveals high levels of the dengue virus protein NS1 in the sera of infected patients. J. Clin. Microbiol. 381053-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]