Abstract
In the luminal contents of metronidazole-treated rats, there was a dominant Bifidobacterium species. A strain has been isolated, its 16S rRNA gene has been sequenced, and the strain has been named Bifidobacterium pseudolongum strain Patronus. In this study, using an experimental model of healthy rats, the effects of metronidazole treatment and B. pseudolongum strain Patronus administration on the luminal and mucosa-associated microbiota and on gut oxidation processes were investigated. Metronidazole treatment and the daily gavage of rats with B. pseudolongum strain Patronus increased the numbers of bifidobacteria in cecal contents and in cecal mucosa-associated microbiota compared with those in control rats. Metronidazole reduced the colonic oxidative damage to proteins. This is the first evidence that B. pseudolongum strain Patronus exerts an effect on a biomarker of oxidative damage by reducing the susceptibility to oxidation of proteins in the colon and the small bowel. Antioxidant effects of metronidazole could be linked to the bifidobacterial increase but also to other bacterial modifications.
The colonic mucus barrier acts as the first defense against the wide range of potentially damaging agents occurring within the colon lumen (3). Among them, an excessive production of reactive oxygen species may be involved in the pathogenesis of inflammatory bowel diseases (IBD), leading to oxidative stress (12, 26, 41, 43) and altering antioxidant defense in the intestinal tissue of IBD patients (7, 21, 26, 49, 51).
Our gastrointestinal tracts are colonized by a vast community of symbiont and commensal bacteria that have important effects on immune function, nutrient processing, and a broad range of other host activities (16).
Results accumulated also suggest that the dynamic balance between commensal bacteria (or their products) and host defensive responses has a fundamental role in the initiation and pathogenesis of IBD (11). Furthermore, alteration in the mucosa-associated microbiota, e.g., a predominance of some potentially harmful bacterial groups or a decrease in potentially beneficial bacterial species, was observed in pediatric patients with IBD (2) and in adult patients with ulcerative colitis (35).
Metronidazole (MTZ), a nitroimidazole antibiotic effective against anaerobes, decreased the early recurrence of Crohn's disease lesions after ileal resection (44). MTZ was also effective in pouchitis (45) and provided immunomodulatory effects (8). For healthy rats, we have previously reported that this drug acts as an antioxidant in colonic tissue (42). This effect could be partly attributed to the modified composition of intestinal microbiota and particularly to the bifidobacterial population, which increased in the luminal and mucosa-associated microbiota of rats treated with MTZ compared to that of control rats. In the cecal luminal contents of MTZ-treated rats, there was a dominant Bifidobacterium strain. It was isolated and, based on the partial sequence of the 16S rRNA gene, corresponded to the species Bifidobacterium pseudolongum. This strain has been named B. pseudolongum strain Patronus.
Bifidobacteria are anaerobic, gram-positive, rod-like organisms inhabiting the intestines of humans and animals. The dominant species of bifidobacteria are generally different in humans and animals. For example, Bifidobacterium adolescentis and Bifidobacterium dentium have been isolated from humans and Bifidobacterium thermophilum, Bifidobacterium animalis subsp. animalis, and Bifidobacterium pseudolongum from animals (9). Bifidobacteria are generally considered to promote intestinal health by preventing the colonization of potential pathogens: they lower intestinal pH through increased fermentation products, e.g., acetate and lactate, produce inhibitory substances such as bacteriocins (10), and stimulate the immune system (53).
A few in vitro studies have reported that these bacteria or their extracts show a scavenging ability for lipid peroxidation products (malondialdehyde [MDA]) and also that they are able to protect plasma against lipid peroxidation (27-29).
The mechanisms of action of MTZ remain unclear. The relationship between the antioxidant effect of MTZ in rat intestines and bifidobacterial increases after MTZ treatment should receive special interest because these topics have not yet been investigated.
The aims of this study were, first, to investigate whether MTZ or bifidobacteria in healthy rats lead to similar modifications of luminal and mucosa-associated microbiota and, second, to evaluate, along the intestine, (i) the potential ability of bifidobacteria to exert antioxidant properties on the biomarkers of oxidative damage to proteins (protein carbonyls) and lipids (MDA) and (ii) the levels of antioxidant defense (glutathione [GSH]).
MATERIALS AND METHODS
Bacterial strains and media.
Bifidobacterium pseudolongum strain Patronus was isolated from the feces of MTZ-treated rats. The 16S rRNA gene for this isolate was amplified by PCR, cloned, and sequenced (see “Amplification, cloning, and sequencing,” below). Stock cultures were stored at −80°C in tryptone-glucose-yeast extract-hemin (TGYH; tryptone peptone, 30 g/liter; glucose, 5 g/liter; yeast extract, 20 g/liter; hemin, 5 mg/liter; and cysteine HCl, 0.5 g/liter) broth containing glycerol.
The bifidobacteria were anaerobically (GenBox Anaer; Biomérieux) cultured in TGYH broth at 37°C.
B. pseudolongum strain Patronus inoculum.
Stock cultures were transferred to TGYH liquid medium at 37°C 48 h before the assay to prepare fresh prestart cultures.
For each daily gavage, the bifidobacteria were cultured anaerobically for 6 h (stopped during exponential growth) at 37°C from a prestart culture. Bacterial cells were harvested by centrifugation at 4,000 × g for 15 min at 4°C and were resuspended in sterile physiological saline in order to obtain an inoculum of 1 × 1010 cells/ml. Bacterial counts were determined, using culture on TGYH agar plates after anaerobic incubation at 37°C for 48 h.
Animals and treatment.
Thirty-six male Wistar rats (Charles River, France) weighing between 200 and 250 g were accommodated in a room with controlled temperature, humidity, and light (12 h light/12 h dark cycle) and maintained for 1 week on standard rodent chow (UAR; Villemoisson, France) ad libitum. They were cared for in compliance with French Ministry of Agriculture regulations.
The rats were assigned randomly to three groups of 12 rats named the control group, MTZ group (80 mg/kg body weight/day in drinking water for 1 week), and Bif group (fed with 1 × 1010 Bifidobacterium pseudolongum strain Patronus organisms every day for a week using an intubation feeding tube without anesthesia).
Samples.
At the end of treatment, rats were weighed and anesthetized by administration of 5% sodium pentobarbital. Laparotomy was carried out on the anesthetized animals, which were killed by aortic puncture. Then, the different samples were collected.
Cecal contents.
Cecal contents were collected in sterile tubes, weighed, and immediately frozen at −80°C.
Cecal mucosa samples.
The entire cecum was removed immediately, put in sterile physiological saline (0.9% NaCl), and chilled on ice. It was gently rinsed with physiological saline several times. Then, under sterile conditions, the cecum was opened and the cecal mucosa was gently rinsed another time. A segment (2 cm2) was obtained and immediately frozen at −80°C.
Colon and small intestine.
The colon and small intestine were removed immediately and chilled on ice.
PCR and TTGE. (i) Extraction and purification of total DNA from cecal contents.
Total DNA was extracted from 125 mg of frozen cecal contents using a bead beating method as previously described (31). Briefly, bacterial cells were lysed mechanically, enzymatically, and chemically, and DNAs were purified using phenol-chloroform treatment and isopropanol precipitation. Dry pellets were suspended in 50 to 100 μl of sterile water and stored at −20°C.
(ii) Extraction and purification of total DNA from cecal mucosa.
Before extraction, a mucosal segment was homogenized in guanidium isothiocyanate for 10 min with a stomacher in order to separate all mucosa-associated bacteria from the mucosa. Then, total DNA was extracted and purified using the protocol described above. Dried pellets were suspended in 50 μl of sterile water and stored at −20°C.
(iii) PCR amplification for bifidobacterial TTGE.
The forward primer Bif164f and reverse primer Bif662r without a GC clamp (46) were used to amplify 16S rRNA genes of bifidobacteria from the samples, as previously described (34). The PCR products were separated by temporal temperature gradient gel electrophoresis (TTGE), using a Dcode universal mutation detection system (Bio-Rad Laboratories, Hercules, CA) as previously described (34). Additionally, references representing known bifidobacterial strains were loaded to allow standardization of band migration and gel curvature among different gels. This ladder consisted of the following strains, frequently isolated from the human fecal microbiota: Bifidobacterium adolescentis CIP64.59T, Bifidobacterium bifidum CIP56.7T, Bifidobacterium longum (Bifidobacterium longum subsp. longum) CIP64.62T, Bifidobacterium dentium CIP104176T, Bifidobacterium pseudocatenulatum CIP104168T, and Bifidobacterium lactis (Bifidobacterium animalis subsp. lactis) CIP105265T. The DNA fragments were visualized using Sybr green I staining (Interchim, Montluçon, France), and the gel was scanned using Gel Doc 2000 (Bio-Rad). The gel patterns were analyzed using Diversity Database 2.1 software, which is part of the Discovery series (Bio-Rad).
(iv) Quantitative PCR.
The quantitative PCR method was used to quantify total bacteria as well as bifidobacteria in cecal contents and cecal mucosa.
The forward primer S-D-Bact-339-a-S-20 (5′-CTC CTA CGG GAG GCA GCA GT-3′) and the reverse primer S-D-Bact-788-a-A-19 (5′-GGA CTA CCA GGG TAT CTA A-3′) were used to amplify variable regions 3 and 4 of the bacterial 16S rRNA genes (32).
The forward primer Bif164f and reverse primer Bif662r without a GC clamp (46) were used to amplify 16S rRNA genes of bifidobacteria.
Quantitative PCR was performed using the Mastercycler ep realplex (Eppendorf SARL, Le Pecq, France). The amplification reactions were carried out in a total volume of 25 μl containing 11.25 μl Sybr green (20×)-RealMaster mix (2.5×) (0.05 U/μl HotMaster Taq DNA polymerase, 10 mM magnesium acetate, 1 mM deoxynucleoside triphosphates) (Eppendorf), 0.4 μl of each primer (0.3 μM), 10.95 μl of sterile water, and 2 μl of diluted DNA samples. The reaction conditions for amplification were 96°C for 2 min for enzyme activation and 40 cycles consisting of 96°C for 15 s, the specific hybridization temperature for 1 min (55°C for total bacteria and 62°C for bifidobacteria), and 68°C for 4 min. Each assay was performed in duplicate in the same run. The fluorescent product was detected at the last step of each cycle.
To determine the specificity of amplification, the melting curve (96°C for 30 s, slow heating from 60°C to 96°C for 20 min, and 96°C for 30 s) was analyzed after the last cycle of amplification. Analysis of the melting curve gave different melting temperatures between specific and nonspecific products based on GC content, amplicon length, and secondary structure. Negative and positive controls were included on each plate. For this positive control, the coefficient of variation was 2.1%.
Standard curves were constructed using the cloned 16S rRNA gene of Escherichia coli CIP 54127T and Bifidobacterium catenulatum strain H12 (GenBank accession no. AY856700) isolated from humans feces. Primers and PCR conditions have been described above. The PCR product was purified and concentrated using a QIAquick spin PCR purification kit (Qiagen S.A., Courtaboeuf, France). The purified product was cloned into pGEM-T (Promega Corp., Madison, WI) as recommended by the manufacturer. Plasmids of a single clone were extracted using a Qiagen plasmid maxi kit (Qiagen, S.A.) and eluted with 200 μl of pure water. The plasmid concentration was then determined spectrophotometrically using absorbances at 260 nm (A1) and 280 nm (A2) according to the formula absorbance rate = A1/A2, with a DNA concentration of 62.9 times the A1 minus 36 times the A2 (Shimadzu Corp.). The plasmid copy number was the DNA concentration over the plasmid mass, with the plasmid mass being equal to 3.82 × 10−18 g. Serial dilutions were performed, and 102 to 109 copies per reaction mixture were used for calibration. E. coli was used for quantification of the total number of bacteria and B. catenulatum for the quantification of bifidobacteria.
During the exponential phase of repeated PCR amplification cycles, the threshold cycle was calculated as the cycle number at which the fluorescence was higher than background. A standard curve was constructed by plotting the threshold cycles of samples containing known numbers of copies. The quantity of target copies in a sample was determined using the standard curve.
Amplification, cloning, and sequencing.
The forward primer S-D-Bact-0008-a-S-20 (5′ AGA GTT TGA TCC TGG CTC AG 3′ [14]), which targets the domain Bacteria, and the reverse primer S-*-Univ-1492-b-A-21 (5′ ACG GCT ACC TTG TTA CGA CTT 3′ [20]), which targets all living organisms, were used to amplify 16S rRNA genes by PCR. Reaction tubes contained 1 μl of bacterial suspension, 0.5 U of Taq DNA polymerase (AmpliTaq Gold; Perkin-Elmer Corporation, Foster City, CA), 1× AmpliTaq Gold reaction buffer, 2.5 mM MgCl2, 200 μM each deoxyribonucleoside triphosphate, and 0.44 μM each primer in a final volume of 20 μl. Initial DNA denaturation and enzyme activation steps were performed at 94°C for 10 min, followed by 35 cycles of denaturation at 97°C for 1 min, annealing at 48°C for 1 min, and elongation at 72°C for 1.5 min, which was followed by a final elongation at 72°C for 15 min, in a personal Mastercycler (Eppendorf).
Three primers were used for complete sequence determination: S-D-Bact-0008-a-S-20, S-D-Bact-339-a-S-20 (5′ CTC CTA CGG GAG GCA GCA GT 3′), and S-*-Univ-1492-b-A-21. The sequencing reaction was run by Cogenics Genome Express SA, Meylan, France.
The sequence was edited to exclude the PCR primer binding sites. It was compared to sequences available in public databases (Ribosomal Database Project [RDP] and GenBank) in order to ascertain their closest relatives. ClustalX version 1.81 (54) was used for sequence analysis.
Alignments were visualized using Bioedit (13). The SSU_Prok alignment of 16S rRNA gene sequences from the RDP (7.1 release) was used as a baseline (33). The phylogenetic inferences were based on a distance method with the neighbor-joining algorithm of Saitou and Nei (44a) using unambiguously aligned nucleotides.
The stability of branches was assessed by the bootstrap method with clustalX (54) for 1,000 replications.
Oxidative-stress markers. (i) Preparation of tissue homogenates.
The colons and the small intestines of the rats were gently rinsed with 0.9% NaCl and longitudinally cut. The mucosa was scraped away with the blunt edge of a glass slide washed in 10 mmol/liter Tris-HCl, 150 mmol/liter KCl, 1 mmol/liter EDTA buffer (pH 7.4) and centrifuged for 5 min at 700 × g. The pellets were homogenized using a Teflon pestle/glass homogenizer in the same buffer containing 0.25 mmol/liter phenylmethylsulfonyl fluoride (1:9, wt/vol, for the colon and 1:3, wt/vol, for the small intestine). All homogenates were aliquoted and frozen at −80°C.
(ii) Measurement of oxidative-stress markers and antioxidant defense.
Lipid peroxidation is assessed by the measure of the thiobarbituric acid-reactive substances, mainly, MDA. MDA was formed after the decomposition of polyunsaturated fatty acids, which are quantified by the 2-thiobarbituric acid test, generating a 1:2 MDA-thiobarbituric acid adduct at low pH and elevated temperature.
For this, we used a modified version of the method initially described by Ohkawa et al. (39). A 3.14-ml mixture containing homogenate of small intestine or colon in 0.45-mmol/liter (final concentrations) butylated hydroxytoluene in glacial acetic acid (to prevent ex vivo lipid peroxidation), 0.25 mmol/liter EDTA, 7.6 mmol/liter sodium dodecyl sulfate, 1.8 mmol/liter phosphotungstic acid, and 6.8 mol/liter HCl was maintained at 4°C for 30 min. In control blanks, no homogenate was added. Centrifugation (5,000 × g for 15 min) yielded a supernatant (2 ml) to which 1 ml of 2-thiobarbituric acid (35 mmol/liter thiobarbituric acid in 20 mmol/liter Tris-HCl buffer, pH 7.0) was added (final pH, 3.5). The mixture was heated at 100°C for 45 min. After cooling, the mixture was extracted with 2 ml of n-butanol. The amount of red pigment produced was determined by absorbance at 532 nm using the extinction coefficient of 1.56 × 105 mol−1 cm−1 for the 1:2 MDA-thiobarbituric acid adduct.
Protein carbonyl content is currently the best overall marker of oxidative-stress-mediated protein oxidation. The carbonyl contents in oxidized proteins was assessed by the method of Levine et al. (25). Aliquots of homogenates were incubated for 1 h at room temperature with 10 mmol/liter diphenylhydrazine (DPNH) in 2 mol/liter HCl. DPNH was omitted in the control blanks. Proteins were precipitated with 500 μl of 500-g/liter trichloroacetic acid and washed three times with 1 ml of 1:1 (vol/vol) ethanol-ethyl acetate. The final precipitate was solved in 6 mol/liter guanidine. The spectrum of the DPNH versus HCl controls was monitored at 350 to 375 nm. The concentration of carbonyl groups was calculated using 21.5 nmol−1 cm−1 as the extinction coefficient for aliphatic hydrazones.
The levels of nonprotein sulfhydryl groups, mostly GSH, were determined as previously described (48). The results were expressed in relation to protein concentration by using the method of Lowry et al. (30).
Statistical analysis.
Data were analyzed using the Statgraphics Plus software package version 5.1 (Rockville, MD). Analysis of variance was performed using the general linear model procedure or the Kruskal-Wallis test. Comparisons of group means were made using Duncan's multiple-range test. Results were expressed as mean values ± standard errors of the means (SEM), with the number of animals indicated.
For the microbial analysis, the bacterial quantification of the cecal contents was expressed as the mean log number of 16S rRNA gene copies/g (wet weight) ± the SEM, and the quantification of mucosa-associated bacteria was expressed as the mean log number of 16S rRNA gene copies/mm2 ± the SEM.
Nucleotide sequence accession number.
The 16S rRNA gene of strain Patronus was deposited in GenBank under accession number FJ356014.
RESULTS
Rats.
Rat weights were similar in the control and Bif groups (281 ± 5 g and 276 ± 4 g, respectively) but decreased with MTZ treatment (252 ± 6 g; P < 0.05).
Detection of bifidobacteria in cecal contents using PCR-TTGE.
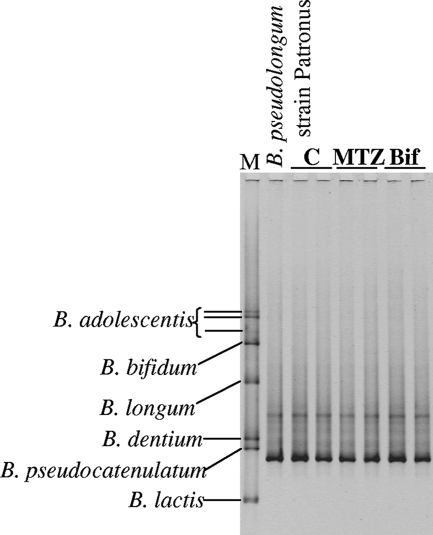
Bifidobacterial PCR-TTGE profiles from the cecal contents of six rats are shown in Fig. 1. These profiles were identical for the other 30 rats. Each profile was composed of a unique dominant band, which comigrated with the B. pseudolongum strain Patronus band; the cecal contents of all rats seem to harbor in dominance a single bifidobacterial species.
FIG. 1.
PCR-TTGE patterns of bifidobacteria from the cecal contents of six rats in the control (lanes C), MTZ, and Bif groups. Lane M, molecular markers.
Quantification of total bacteria and bifidobacteria in cecal contents.
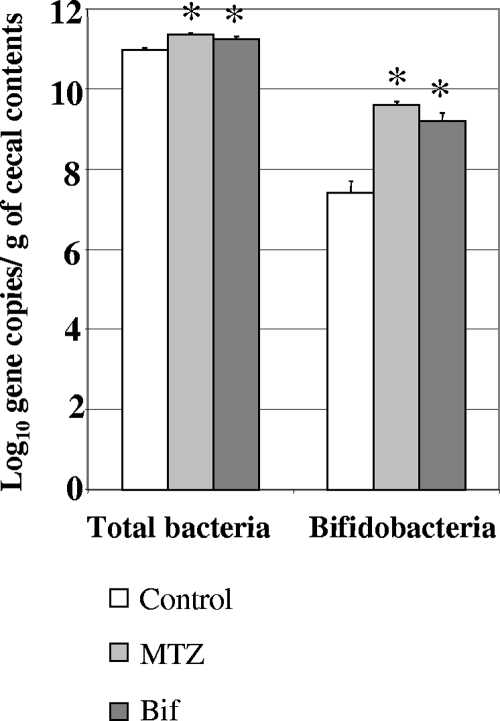
Numbers of total bacteria and of bifidobacteria in cecal contents are presented in Fig. 2. The number of total bacteria in the cecal contents of the MTZ and Bif groups (11.37 ± 0.03 and 11.25 ± 0.06 log10 gene copies/g, respectively) were significantly higher than the values for the control group (10.97 ± 0.06 log10 gene copies/g) (P < 0.05).
FIG. 2.
Quantification of total bacteria and bifidobacteria in the cecal contents of rats in the control, MTZ, and Bif groups. Results are expressed as mean values ± SEM. *, P was <0.05 versus the values of the control group.
A significant increase in bifidobacterial populations was observed either in the MTZ or in the Bif group (9.6 ± 0.1 and 9.2 ± 0.2 log10 gene copies/g, respectively) compared with the level in the control group (7.4 ± 0.3 log10 gene copies/g) (P < 0.05).
Quantification of mucosa-associated microbiota in the cecum.
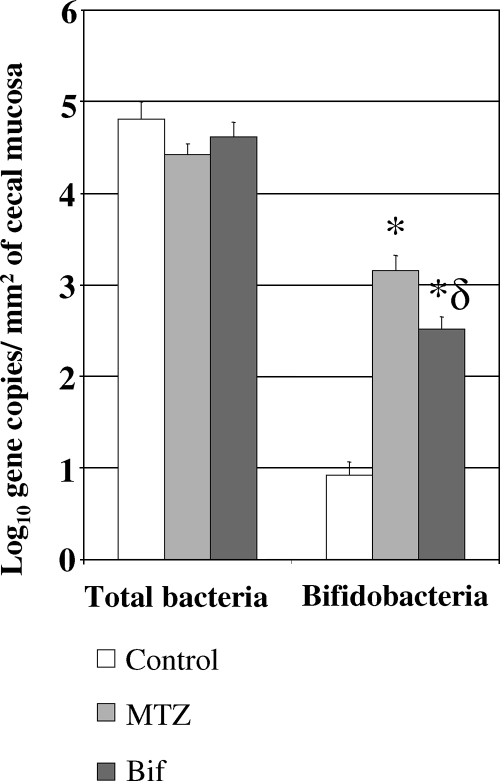
The results were expressed in log10 numbers of gene copies/mm2 of cecal mucosa and are presented in Fig. 3. The numbers of total bacteria were similar in the three groups: 4.42 ± 0.12 (MTZ group), 4.61 ± 0.16 (Bif group), and 4.81 ± 0.19 (control group) log10 gene copies/mm2.
FIG. 3.
Quantification of total bacteria and bifidobacteria in the cecal mucosa of the control, MTZ, and Bif groups. Results are expressed as mean values ± SEM. *, P was <0.05 versus the values of the control group; δ, P was <0.05 versus the values of the MTZ group.
Treatments significantly increased bifidobacterial populations in the mucosal bacteria of the cecum compared with control values (3.2 ± 0.2 and 2.5 ± 0.1 log10 gene copies/mm2 for the MTZ and Bif groups, respectively, versus 0.9 ± 0.1 log10 gene copies/mm2 for the control group; P < 0.05). Moreover, in cecal mucosa, the bifidobacterial populations were significantly (P < 0.05) more increased by MTZ treatment than by bifidobacteria.
Effect of treatments on oxidative stress and antioxidant defense.
As shown in Table 1, in the colonic tissue, MTZ treatment increased the thiobarbituric acid-reactive substance contents of the colon (by 19%; P < 0.05). Nevertheless, GSH levels were not altered by MTZ.
TABLE 1.
Oxidative stress and antioxidant defense in the colons and small intestines of rats in the control, MTZ, and Bif groups
| Oxidative stress parameter and localization | Amt of substance (nmol/mg protein)a in:
|
||
|---|---|---|---|
| Control group | MTZ group | Bif group | |
| Protein carbonyls | |||
| Small intestine | 9.03 ± 0.66 | 8.38 ± 0.9 | 6.21 ± 0.62*ϕ |
| Colon | 8.33 ± 0.69 | 6.67 ± 0.45 | 5.45 ± 0.74*ϕ |
| GSH | |||
| Small intestine | 13.32 ± 1.24 | 14.95 ± 1.31 | 10.2 ± 0.75ϕ |
| Colon | 24.78 ± 1.29 | 26.99 ± 1.64 | 17.5 ± 1.37*ϕ |
| Thiobarbituric acid-reactive substances | |||
| Small intestine | 0.84 ± 0.04 | 0.79 ± 0.03 | 0.75 ± 0.05 |
| Colon | 0.93 ± 0.02 | 1.11 ± 0.05* | 0.8 ± 0.04ϕ |
Values are presented as means ± SEM (n = 12). *, P was <0.05 versus the values for the control group; ϕ, P was <0.05 versus the values for the MTZ group.
In the small intestine, for the MTZ group, the levels of the two markers of oxidative stress and GSH levels were not different from those of the control group.
Bifidobacterial treatment significantly decreased protein carbonyl levels (by the same extent in the colon [−35%; P < 0.05] and in the small intestine [−31%; P < 0.05]) compared with levels in control rats. Lipid peroxidation in the Bif group was not different from that in the control group all along the intestine. Nevertheless, GSH levels along the gut in this group were lower than those in controls (−29% and −23%, respectively; P < 0.05).
DISCUSSION
In the present study, using an experimental model of healthy rats, we investigated the effects of MTZ treatment and bifidobacterial administration on, first, the luminal and mucosa-associated microbiota and, second, on the gut oxidation processes (assessed by protein carbonylation and lipid peroxidation) and the antioxidant defense.
We have confirmed that MTZ reduces the colonic level of protein oxidation to a lesser extent than in a previous study (42). Although colonic lipid peroxidation was enhanced by MTZ, the levels of GSH remained similar to control levels, suggesting that no adduct to GSH was formed. Consequently, the quick degradation of several aldehydes possibly plays an important role in this antioxidative defense system in order to protect proteins from modification by the lipid peroxidation products (50).
Moreover, this is the first evidence that Bifidobacterium pseudolongum strain Patronus exerts an effect on a biomarker of oxidative damage by reducing the susceptibility of proteins to oxidation either in the colon or in the small bowel. Bifidobacteria are dominant in the intestine and constitute part of the barrier effect that prevents intestine colonization by pathogenic bacteria. This commensal strain decreased cellular GSH levels, although oxidative modification of proteins was below the control level along the intestine. This result could be partly explained by previous data (15): GSH concentration in vivo is sufficient for various GSH-dependent enzymes and is well maintained by rapid resynthesis in patients undergoing endoscopy.
In previous studies, oral administration of Bifidobacterium bifidum strain Yakult effectively decreased colonic oxidative stress induced by an iron overload diet (17, 18). It has also been reported that Streptococcus thermophilus YIT 2001 protected the colonic mucosa against oxidative stress (17).
In the control, MTZ, and Bif groups, the cecal microbiota exhibited one dominant bifidobacterial species, corresponding to Bifidobacterium pseudolongum strain Patronus, as shown by the PCR-TTGE profiles of the cecal microbiota.
Therefore, we investigated the quantification of total bacteria and bifidobacteria in the cecal contents and mucosa-associated microbiota of the cecum using real-time quantitative PCR. The recent development of quantitative-PCR methods should allow a better knowledge of the composition of gastrointestinal communities, how these bacteria are related to diet, and their roles in health and disease (6, 19, 36).
Our present results for total bacteria in luminal contents were in accordance with previous results obtained using real-time quantitative PCR: 11.1 ± 0.9 log gene copies of total bacteria/g fresh matter in intestinal samples of pigs (1). Our quantifications of bifidobacteria in cecal contents were in agreement with results of other studies using real-time PCR (TaqMan probes) with control rat samples (average, 7.6 log of cell/g of feces) (5) and using a microbiological culture approach to determine levels of Bifidobacterium spp. in rat feces: 6.5 log CFU/g of raw material (4).
Moreover, mucosa-associated bacteria may be more reflective of a long-term stay in the colonic wall. As these microorganisms may strongly interact with host cells, they may be a better indicator of disease-related microbiotal modifications. In our laboratory, a novel method allowing the quantification of mucosa-associated bacteria was performed first by gently washing an intestinal sample for luminal-debris elimination, thereby preserving only the bacteria associated with the mucosa, and second by measuring the mucosal surface for bacterial quantification (number of bacteria/mm2).
Studies incorporating fluorescent in situ hybridization technology to investigate microorganisms in human histological material (23, 47, 52) or animal intestines (22) focused on the detection and enumeration of bacteria and on the composition and spatial organization of the intestinal microbiota. In our experiment, the daily gavage of rats with B. pseudolongum strain Patronus for 1 week increased the number of bifidobacteria in their cecal contents and in their cecal mucosa-associated microbiota compared with data for control rats, as expected. The increase in bifidobacteria in cecal contents was similar to that obtained with MTZ treatment for 1 week. Unlike with cecal contents, the bifidobacterial increase in cecal mucosa-associated microbiota was higher in the MTZ group than in the Bif group, and yet, antioxidant effects of MTZ administration were lower than with B. pseudolongum strain Patronus administration. Antioxidant effects could be linked to the bifidobacterial increase but also to other bacterial changes. The stabilization of the rat gut mucosal barrier by the stimulation of mucosal bifidobacteria with fructans could be of benefit in gastrointestinal-disorder prophylaxis (22).
The relative resistance of bifidobacteria to MTZ suggested that these bacteria might survive the antibiotic treatment. Strain Patronus possibly lacks the ferredoxin system responsible for the reduction of the parent compound, which generates an intermediate product responsible for DNA strand breaks. This can lead to resistance to this drug (37).
Other studies with different strains of lactic acid bacteria have shown therapeutic activity in experimental models of IBD (24, 40). The reduction in mucosa-associated bifidobacteria in patients with IBD supports the hypothesis that an imbalance between potentially beneficial and pathogenic bacteria may contribute to its pathogenesis (38).
In conclusion, based on these findings, B. pseudolongum strain Patronus exerted an effect on oxidative stress that protected the intestinal mucosa through a relative predominance of protective species. MTZ treatment also augmented counts of this commensal species. Further investigations need to be conducted to test the effect of MTZ alone or in association with this strain in experimental animal models of IBD. In the future, it would be interesting to study the effect of a human Bifidobacterium strain as a probiotic in IBD patients.
Acknowledgments
We are grateful to Marie-Vincent Dessoy, Irène Mangin, Ester Pereira, and Claire Seigneurgens for their technical assistance.
Footnotes
Published ahead of print on 21 November 2008.
REFERENCES
- 1.Castillo, M., S. M. Martin-Orue, E. G. Manzanilla, I. Badiola, M. Martin, and J. Gasa. 2006. Quantification of total bacteria, enterobacteria and lactobacilli populations in pig digesta by real-time PCR. Vet. Microbiol. 114:165-170. [DOI] [PubMed] [Google Scholar]
- 2.Conte, M. P., S. Schippa, I. Zamboni, M. Penta, F. Chiarini, L. Seganti, J. Osborn, P. Falconieri, O. Borrelli, and S. Cucchiara. 2006. Gut-associated bacterial microbiota in paediatric patients with inflammatory bowel disease. Gut 55:1760-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corfield, A. P., N. Myerscough, R. Longman, P. Sylvester, S. Arul, and M. Pignatelli. 2000. Mucins and mucosal protection in the gastrointestinal tract: new prospects for mucins in the pathology of gastrointestinal disease. Gut 47:589-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.da S. Queiroz-Monici, K., G. E. A. Costa, N. da Silva, S. M. P. M. Reis, and A. C. de Oliveira. 2005. Bifidogenic effect of dietary fiber and resistant starch from leguminous on the intestinal microbiota of rats. Nutrition 21:602-608. [DOI] [PubMed] [Google Scholar]
- 5.Delroisse, J. M., A. L. Boulvin, I. Parmentier, R. D. Dauphin, M. Vandenbol, and D. Portetelle. 2008. Quantification of Bifidobacterium spp. and Lactobacillus spp. in rat fecal samples by real-time PCR. Microbiol. Res. 163:663-670. [DOI] [PubMed] [Google Scholar]
- 6.Duncan, S. H., G. E. Lobley, G. Holtrop, J. Ince, A. M. Johnstone, P. Louis, and H. J. Flint. 2008. Human colonic microbiota associated with diet, obesity and weight loss. Int. J. Obes. (London) 32:1720-1724. [DOI] [PubMed] [Google Scholar]
- 7.Farhadi, A., A. Keshavarzian, L. R. Fitzpatrick, E. Mutlu, Y. Zhang, and A. Banan. 2002. Modulatory effects of plasma and colonic milieu of patients with ulcerative colitis on neutrophil reactive oxygen species production in presence of a novel antioxidant, rebamipide. Dig. Dis. Sci. 47:1342-1348. [DOI] [PubMed] [Google Scholar]
- 8.Freeman, C. D., N. E. Klutman, and K. C. Lamp. 1997. Metronidazole. A therapeutic review and update. Drugs 54:679-708. [DOI] [PubMed] [Google Scholar]
- 9.Gavini, F., A. M. Pourcher, C. Neut, D. Monget, C. Romond, C. Oger, and D. Izard. 1991. Phenotypic differentiation of bifidobacteria of human and animal origins. Int. J. Syst. Bacteriol. 41:548-557. [DOI] [PubMed] [Google Scholar]
- 10.Gibson, G. R., and X. Wang. 1994. Regulatory effects of bifidobacteria on the growth of other colonic bacteria. J. Appl. Bacteriol. 77:412-420. [DOI] [PubMed] [Google Scholar]
- 11.Gionchetti, P., F. Rizzello, K. M. Lammers, C. Morselli, L. Sollazzi, S. Davies, R. Tambasco, C. Calabrese, and M. Campieri. 2006. Antibiotics and probiotics in treatment of inflammatory bowel disease. World J. Gastroenterol. 12:3306-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grisham, M. B. 1994. Oxidants and free radicals in inflammatory bowel disease. Lancet 344:859-861. [DOI] [PubMed] [Google Scholar]
- 13.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 14.Hicks, R. E., R. I. Amann, and D. A. Stahl. 1992. Dual staining of natural bacterioplankton with 4′,6-diamidino-2-phenylindole and fluorescent oligonucleotide probes targeting kingdom-level 16S rRNA sequences. Appl. Environ. Microbiol. 58:2158-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoensch, H., I. Morgenstern, G. Petereit, M. Siepmann, W. H. Peters, H. M. Roelofs, and W. Kirch. 2002. Influence of clinical factors, diet, and drugs on the human upper gastrointestinal glutathione system. Gut 50:235-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hooper, L. V., and J. I. Gordon. 2001. Commensal host-bacterial relationships in the gut. Science 292:1115-1118. [DOI] [PubMed] [Google Scholar]
- 17.Ito, M., K. Ohishi, Y. Yoshida, W. Yokoi, and H. Sawada. 2003. Antioxidative effects of lactic acid bacteria on the colonic mucosa of iron-overloaded mice. J. Agric. Food Chem. 51:4456-4460. [DOI] [PubMed] [Google Scholar]
- 18.Ito, M., H. Sawada, K. Ohishi, Y. Yoshida, W. Yokoi, T. Watanabe, and T. Yokokura. 2001. Suppressive effects of bifidobacteria on lipid peroxidation in the colonic mucosa of iron-overloaded mice. J. Dairy Sci. 84:1583-1589. [DOI] [PubMed] [Google Scholar]
- 19.Kalliomaki, M., M. C. Collado, S. Salminen, and E. Isolauri. 2008. Early differences in fecal microbiota composition in children may predict overweight. Am. J. Clin. Nutr. 87:534-538. [DOI] [PubMed] [Google Scholar]
- 20.Kane, M. D., L. K. Poulsen, and D. A. Stahl. 1993. Monitoring the enrichment and isolation of sulfate-reducing bacteria by using oligonucleotide hybridization probes designed from environmentally derived 16S rRNA sequences. Appl. Environ. Microbiol. 59:682-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keshavarzian, A., A. Banan, A. Farhadi, S. Komanduri, E. Mutlu, Y. Zhang, and J. Z. Fields. 2003. Increases in free radicals and cytoskeletal protein oxidation and nitration in the colon of patients with inflammatory bowel disease. Gut 52:720-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kleessen, B., L. Hartmann, and M. Blaut. 2003. Fructans in the diet cause alterations of intestinal mucosal architecture, released mucins and mucosa-associated bifidobacteria in gnotobiotic rats. Br. J. Nutr. 89:597-606. [DOI] [PubMed] [Google Scholar]
- 23.Kleessen, B., A. J. Kroesen, H. J. Buhr, and M. Blaut. 2002. Mucosal and invading bacteria in patients with inflammatory bowel disease compared with controls. Scand. J. Gastroenterol. 9:1034-1041. [DOI] [PubMed] [Google Scholar]
- 24.Lee, H. S., S. Y. Han, E. A. Bae, C. S. Huh, Y. T. Ahn, J. H. Lee, and D. H. Kim. 2008. Lactic acid bacteria inhibit proinflammatory cytokine expression and bacterial glycosaminoglycan degradation activity in dextran sulfate sodium-induced colitic mice. Int. Immunopharmacol. 8:574-580. [DOI] [PubMed] [Google Scholar]
- 25.Levine, R. L., D. Garland, C. N. Oliver, A. Amici, I. Climent, A. G. Lenz, B. W. Ahn, S. Shaltiel, and E. R. Stadtman. 1990. Determination of carbonyl content in oxidatively modified proteins. Methods Enzymol. 186:464-478. [DOI] [PubMed] [Google Scholar]
- 26.Lih-Brody, L., S. R. Powell, K. P. Collier, G. M. Reddy, R. Cerchia, E. Kahn, G. S. Weissman, S. Katz, R. A. Floyd, M. J. McKinley, S. E. Fisher, and G. E. Mullin. 1996. Increased oxidative stress and decreased antioxidant defenses in mucosa of inflammatory bowel disease. Dig. Dis. Sci. 41:2078-2086. [DOI] [PubMed] [Google Scholar]
- 27.Lin, M. Y., and F. J. Chang. 2000. Antioxidative effect of intestinal bacteria Bifidobacterium longum ATCC 15708 and Lactobacillus acidophilus ATCC 4356. Dig. Dis. Sci. 45:1617-1622. [DOI] [PubMed] [Google Scholar]
- 28.Lin, M. Y., and C. L. Yen. 1999. Inhibition of lipid peroxidation by Lactobacillus acidophilus and Bifidobacterium longum. J. Agric. Food Chem. 47:3661-3664. [DOI] [PubMed] [Google Scholar]
- 29.Lin, M. Y., and C. L. Yen. 1999. Reactive oxygen species and lipid peroxidation product-scavenging ability of yogurt organisms. J. Dairy Sci. 82:1629-1634. [DOI] [PubMed] [Google Scholar]
- 30.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 31.Magne, F., M. Abely, F. Boyer, P. Morville, P. Pochart, and A. Suau. 2006. Low species diversity and high interindividual variability in faeces of preterm infants as revealed by sequences of 16S rRNA genes and PCR-temporal temperature gradient gel electrophoresis profiles. FEMS Microbiol. Ecol. 57:128-138. [DOI] [PubMed] [Google Scholar]
- 32.Magne, F., W. Hachelaf, A. Suau, G. Boudraa, I. Mangin, M. Touhami, K. Bouziane-Nedjadi, and P. Pochart. 2006. A longitudinal study of infant faecal microbiota during weaning. FEMS Microbiol. Ecol. 58:563-571. [DOI] [PubMed] [Google Scholar]
- 33.Maidak, B. L., J. R. Cole, T. G. Lilburn, Jr., C. T. Parker, P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mangin, I., A. Suau, F. Magne, D. Garrido, M. Gotteland, C. Neut, and P. Pochart. 2006. Characterization of human intestinal bifidobacteria using competitive PCR and PCR-TTGE. FEMS Microbiol. Ecol. 55:28-37. [DOI] [PubMed] [Google Scholar]
- 35.Matsuda, H., Y. Fujiyama, A. Andoh, T. Ushijima, T. Kajinami, and T. Bamba. 2000. Characterization of antibody responses against rectal mucosa-associated bacterial flora in patients with ulcerative colitis. J. Gastroenterol. Hepatol. 15:61-68. [DOI] [PubMed] [Google Scholar]
- 36.Matsuki, T., K. Watanabe, J. Fujimoto, T. Takada, and R. Tanaka. 2004. Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in human feces. Appl. Environ. Microbiol. 70:7220-7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moubareck, C., F. Gavini, L. Vaugien, M. J. Butel, and F. Doucet-Populaire. 2005. Antimicrobial susceptibility of bifidobacteria. J. Antimicrob. Chemother. 55:38-44. [DOI] [PubMed] [Google Scholar]
- 38.Mylonaki, M., N. B. Rayment, D. S. Rampton, B. N. Hudspith, and J. Brostoff. 2005. Molecular characterization of rectal mucosa-associated bacterial flora in inflammatory bowel disease. Inflamm. Bowel Dis. 11:481-487. [DOI] [PubMed] [Google Scholar]
- 39.Ohkawa, H., N. Ohishi, and K. Yagi. 1979. Assay for lipid peroxides in animal tissues by thiobarbituric acid reaction. Anal. Biochem. 95:351-358. [DOI] [PubMed] [Google Scholar]
- 40.Osman, N., D. Adawi, S. Ahrne, B. Jeppsson, and G. Molin. 2004. Modulation of the effect of dextran sulfate sodium-induced acute colitis by the administration of different probiotic strains of Lactobacillus and Bifidobacterium. Dig. Dis. Sci. 49:320-327. [DOI] [PubMed] [Google Scholar]
- 41.Pavlick, K. P., F. S. Laroux, J. Fuseler, R. E. Wolf, L. Gray, J. Hoffman, and M. B. Grisham. 2002. Role of reactive metabolites of oxygen and nitrogen in inflammatory bowel disease. Free Radic. Biol. Med. 33:311-322. [DOI] [PubMed] [Google Scholar]
- 42.Pelissier, M. A., P. Marteau, and P. Pochart. 2007. Antioxidant effects of metronidazole in colonic tissue. Dig. Dis. Sci. 52:40-44. [DOI] [PubMed] [Google Scholar]
- 43.Rezaie, A., R. D. Parker, and M. Abdollahi. 2007. Oxidative stress and pathogenesis of inflammatory bowel disease: an epiphenomenon or the cause? Dig. Dis. Sci. 52:2015-2021. [DOI] [PubMed] [Google Scholar]
- 44.Rutgeerts, P., M. Hiele, K. Geboes, M. Peeters, F. Penninckx, R. Aerts, and R. Kerremans. 1995. Controlled trial of metronidazole treatment for prevention of Crohn's recurrence after ileal resection. Gastroenterology 108:1617-1621. [DOI] [PubMed] [Google Scholar]
- 44a.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 45.Sartor, R. B. 2004. Therapeutic manipulation of the enteric microflora in inflammatory bowel diseases: antibiotics, probiotics, and prebiotics. Gastroenterology 126:1620-1633. [DOI] [PubMed] [Google Scholar]
- 46.Satokari, R. M., E. E. Vaughan, A. D. Akkermans, M. Saarela, and W. M. De Vos. 2001. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:504-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schultsz, C., F. M. Van Den Berg, F. W. Ten Kate, G. N. Tytgat, and J. Dankert. 1999. The intestinal mucus layer from patients with inflammatory bowel disease harbors high numbers of bacteria compared with controls. Gastroenterology 117:1089-1097. [DOI] [PubMed] [Google Scholar]
- 48.Sedlak, J., and R. H. Lindsay. 1968. Estimation of total, protein-bound, and nonprotein sulfhydryl groups in tissue with Ellman's reagent. Anal. Biochem. 25:192-205. [DOI] [PubMed] [Google Scholar]
- 49.Sido, B., V. Hack, A. Hochlehnert, H. Lipps, C. Herfarth, and W. Droge. 1998. Impairment of intestinal glutathione synthesis in patients with inflammatory bowel disease. Gut 42:485-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Siems, W. G., H. Zollner, T. Grune, and H. Esterbauer. 1997. Metabolic fate of 4-hydroxynonenal in hepatocytes: 1,4-dihydroxynonene is not the main product. J. Lipid Res. 38:612-622. [PubMed] [Google Scholar]
- 51.Suzuki, K., K. Sugimura, K. Hasegawa, K. Yoshida, A. Suzuki, K. Ishizuka, K. Ohtsuka, T. Honma, R. Narisawa, and H. Asakura. 2001. Activated platelets in ulcerative colitis enhance the production of reactive oxygen species by polymorphonuclear leukocytes. Scand. J. Gastroenterol. 36:1301-1306. [DOI] [PubMed] [Google Scholar]
- 52.Swidsinski, A., A. Ladhoff, A. Pernthaler, S. Swidsinski, V. Loening-Baucke, M. Ortner, J. Weber, U. Hoffmann, S. Schreiber, M. Dietel, and H. Lochs. 2002. Mucosal flora in inflammatory bowel disease. Gastroenterology 122:44-54. [DOI] [PubMed] [Google Scholar]
- 53.Tanabe, S., Y. Kinuta, and Y. Saito. 2008. Bifidobacterium infantis suppresses proinflammatory interleukin-17 production in murine splenocytes and dextran sodium sulfate-induced intestinal inflammation. Int. J. Mol. Med. 22:181-185. [PubMed] [Google Scholar]
- 54.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]