Abstract
Shiga toxin 2 (stx2) gene-carrying bacteriophages have been shown to convert Escherichia coli strains to Shiga toxin-producing Escherichia coli (STEC). In this study, 79 E. coli strains belonging to 35 serotypes isolated from wastewaters of both human and animal origin were examined for the presence of stx2-carrying bacteriophages in their genomes. The lytic cycle of the bacteriophages was induced by mitomycin, and the bacteriophage fraction was isolated and used for morphological and genetic characterization. The induced bacteriophages showed morphological diversity, as well as restriction fragment length polymorphism variation, in the different strains belonging to different serotypes. The ability to infect new hosts was highly variable, although most of the induced phages infected Shigella sonnei host strain 866. In summary, in spite of carrying either the same or different stx2 variants and in spite of the fact that they were isolated from strains belonging to the same or different serotypes, the induced bacteriophages were highly variable. The high level of diversity and the great infectious capacity of these phages could enhance the spread of the stx2 gene and variants of this gene among different bacterial populations in environments to which humans may be exposed.
Horizontal transmission of pathogenic factors through mobile elements, such as bacteriophages, plasmids, or transposable elements, is important in the emergence of new pathogenic bacteria (25). The pathogenic factors that are encoded in bacteriophages include toxins, such as the Streptococcus pyogenes toxin (48), the A, D, and E enterotoxins and toxic shock syndrome 1 toxin of Staphylococccus aureus (4), the C, D, and E neurotoxins of Clostridium botulinum (44), the diphtheria toxin of Corynebacterium diphtheriae (15), the enterotoxin of Vibrio cholerae (48), and the Shiga toxins (Stxs) of Shigella dysenteriae and Escherichia coli (5, 24).
Shiga toxin-producing E. coli (STEC) emerged in 1982 as a human pathogen that causes hemorrhagic colitis and the hemolytic-uremic syndrome (30, 49). STEC strains have been shown to produce bacteriophage-encoded toxins belonging to two main groups, Stx1 and Stx2, and some variants of these main groups have been described (Stx1, Stx1c, Stx1d, Stx2, Stx2c, Stx2d, Stx2e, Stx2f, and Stx2g) (10, 20, 24, 37, 43, 45). Although the presence of stx genes is not sufficient to cause infection, the fact that these genes were first detected in association with increased disease in humans suggests that they may be linked to the emergence of STEC. However, little information is available about the epidemiological associations of stx2 genes before the association with STEC was found (23). Thus, establishing a link to evolutionary trends in E. coli is difficult without further study of the dispersal of stx2 genes in the environment and in other genetically related bacteria, such as Shigella.
stx2 gene-carrying bacteriophages (Stx2 phages) have been shown to occur naturally in the environment (18, 22) and have been shown to spread stx2 genes among bacterial populations (1, 36, 41). It has been suggested that these phages play a direct role in STEC pathogenesis since Stx production is linked to induction of the lysis cycle of the phages (17, 47). Several mechanisms induce lysis, and mitomycin treatment and UV irradiation are the methods most commonly used for research purposes (26). These treatments induce the SOS system, leading to an increase in phage gene expression and consequently an increase in Stx production (14, 50). The environment is a potential reservoir for the phages since they show high resistance in the environment (19). The ability of these phages to transduce stx genes has been demonstrated in vivo and in vitro (2, 35).
The genetic structure of Stx2 phages is similar to that of the lambda bacteriophage, and thus they are frequently referred to as lambdoid bacteriophages (33). Some of these phages have been totally or partially sequenced (13, 20, 28, 34, 46). Rietra et al. concluded that induced bacteriophages from E. coli O157:H7 isolates of human origin exhibited similar restriction fragment length polymorphism (RFLP) patterns and high levels of DNA-DNA cross-hybridization, suggesting that these bacteriophages were genetically closely related (29). However, other authors have shown that there is heterogeneity in Stx2 phages isolated from strains with different pulsed-field gel electrophoresis profiles (26, 31). The DNA phage heterogeneity of different isolates derived from the same strain may be due to different rearrangements of the Stx phages with other temperate bacteriophage genes present in the genomes of these strains (11, 27). In fact, in two previously sequenced E. coli O157 strains, different prophages with a structure similar to the lambda structure were detected, suggesting that these phages have an important role in the evolution of pathogenic strains since they have not been found in E. coli K-12.
The genetic heterogeneity reported for the Stx phages is consistent with some diversity in virion morphology. Thus, some bacteriophages are morphologically similar to the lambda bacteriophage and have icosahedral capsids with hexagonal symmetry and long tails, while other bacteriophages have long capsids with hexagonal symmetry and noncontractile tails (21). Moreover, there have been several descriptions of strains carrying two different bacteriophages (1, 3, 21).
The majority of the studies addressing the diversity among Stx2 phages have focused on phages isolated from E. coli O157:H7 strains which have been involved in STEC outbreaks (29). Other studies have examined free Stx2 phages in the environment (22). In this study, the presence of inducible bacteriophages in a collection of 79 STEC strains belonging to 35 serotypes isolated from wastewaters from different animal slaughterhouses and human sewage was evaluated for the first time. This study included detection of Stx2 phages by PCR, localization of the stx2 gene in the genomes of these phages by RFLP analysis and specific hybridization of RFLP products with an stx2 probe, and morphological analysis of the phages. Moreover, in vitro host infection assays were performed to determine the abilities of the phages to transduce the stx2 gene in different hosts. Toxin production in new lysogenic strains generated upon infection by some of the Stx2 phages was also assessed. As far as we know, this is the first analysis of the prevalence of these phages in nonclinical strains having different origins in which the real prevalence of the phages in environmental strains was assessed.
MATERIALS AND METHODS
Bacterial strains, bacteriophages, and culture media.
A total of 79 STEC strains carrying the stx2 gene isolated from municipal human sewage (sewage mostly from humans) and animal sewage were screened for the presence of inducible Stx2 phages. Sixty-five of the strains were isolated by using no specific selection method other than selection for the presence of the stx2 gene in the coliform population; the phenotypic and genotypic characteristics of these strains, as well as their serotypes, have been described previously (8). The 14 remaining strains belonged to the O157:H7 serotype and were isolated by using specific recovery on CT-SMAC medium as previously described, and their phenotypic and genotypic characteristics have been described previously (9). The E. coli C600 (933W) strain (17), which carries the 933W bacteriophage integrated as a prophage in the genome, was used as a positive control in the bacteriophage induction experiments, while the E. coli C600 laboratory strain, which does not carry any Stx2 phage, was used as a negative control.
Bacteriophage induction.
The lytic cycle of temperate bacteriophages was induced by mitomycin C treatment as described previously (17). Briefly, 200-ml cultures of each STEC strain were prepared from single colonies in Luria-Bertani (LB) medium supplemented with 5 mM CaCl2. The cultures were grown at 37°C to an optical density at 600 nm (OD600) of 0.3, and then each culture was divided and placed into two flasks. Mitomycin C was added to one of the flasks at a final concentration of 0.5 mg ml−1, and both cultures were then incubated overnight in the dark at 37°C with agitation. Phage-mediated lysis was monitored by determining the decrease in OD600.
After overnight incubation, the culture was centrifuged at 14,300 × g for 30 min, and the supernatant was filtered through 0.22-μm-pore-size, low-protein-binding polyether sulfone membranes (Millex-GP; Millipore, Mosheim, France) to recover the bacteriophage fraction.
Phage DNA isolation.
Phage DNA was isolated from the 200-ml mitomycin C-induced cultures after centrifugation and filtration as described above by using polyethylene glycol precipitation, DNase treatment, and phenol-chloroform extraction performed as previously described (22).
PCR.
The presence of the stx2 gene in the isolated phage DNA fraction was determined by PCR with the specific primers used to detect the different stx2 variants (stx2, stx2c, stx2d, stx2e, and stx2g) in the original STEC strains, as previously described (9).
Bacteriophage RFLP analysis and detection of stx2 gene.
In order to obtain insight into the genetic diversity of the lysogenic phages found in the environmental STEC strains, isolated phage DNA was subjected to RFLP analysis by digestion with the EcoRI restriction enzyme, analysis by electrophoresis in a 0.8% Tris-borate-EDTA 1× agarose gel, and subsequent staining with ethidium bromide. Alternatively, in some cases the ClaI enzyme was used when an EcoRI pattern was not clearly visible.
Localization of the stx2 gene in the different RFLP patterns obtained for the different DNA phage fractions was performed by Southern blotting using the stx2 gene probe used previously for isolation of the STEC strains by colony blotting, which codes for a 378-bp region of the A subunit (8).
Evaluation of the infectious capacity of Stx2 phages.
The infectious capacity of Stx2 phages was evaluated using the following host strains in the infection assays: E. coli O157:H7 strain ATCC 43888, three E. coli O157 environmental isolates, the E. coli DH5α and C600 laboratory strains, Enterobacter aerogenes ATTC 13048, a Citrobacter freundii environmental isolate, and several clinical isolates, including Shigella dysenteriae strain 500H, Shigella boydii strains 316 and 351, Shigella sonnei strains 635 and 866, Shigella flexneri strains 668 and 805, E. coli O26 strains 216 and 224, and E. coli O111 strain 209. None of these strains carry the stx2 gene. Briefly, 10 μl of induced and filtered phage supernatant was spotted onto LB agar overlaid with LB soft agar supplemented with 5 mM CaCl2 containing the corresponding host strain at an OD600 of 0.3. After overnight incubation at 37°C, the formation of plaques was monitored. Then the plaques were transferred onto nylon membranes using standard procedures (32) and were hybridized using the stx2 probe, as described above. Only the tests showing lysis and a positive hybridization signal were considered positive.
Construction of lysogens.
The induced phages were transduced into S. sonnei strain 866 and E. coli DH5α by spotting 0.5 ml of the phage lysate as described above. After overnight incubation at 37°C, the LB soft agar overlay was removed and resuspended in 2 ml SM buffer (32). The mixture was incubated for 90 min at 37°C, and then 0.1-ml portions of 10-fold dilutions in phosphate-buffered saline were plated onto LB agar. The plates were incubated overnight at 37°C. Lysogens carrying the stx2 gene were selected by a colony hybridization method as described previously (6). The presence of stx2 genes in the new lysogens was confirmed by PCR, as indicated above.
Phage integration site.
PCR was used to assess whether the bacteriophages studied were integrated in the same locus as the 933W phage (the wrbA locus) or the VT1-Sakai phage (the yehV locus), before and after the transduction of the bacteriophages into the new host strains. Briefly, the bacterial DNA with the inserted prophages was extracted, and a PCR was carried out using the primers described previously (39).
Evaluation of Stx production.
The production of Stxs (both Stx1 and Stx2, including Stx2 variants) by the constructed lysogens was tested using a commercial Duopath VT detection kit (Merck, Darmstadt, Germany) according to the manufacturer's instructions.
Electron microscopy imaging.
Twenty-milliliter portions of phage-induced cultures were filtered as described above and were concentrated 1,000-fold using Amicon Ultra-4 filters according to the manufacturer's instructions (Millipore). Phages were adsorbed onto Formvar-coated grids and negatively stained with 2% KOH, phosphotungstic acid (pH 7.2) for 2.5 min. Negatively stained samples were observed with an Hitachi 800 STEM microscope (Portland, OR) at 80 kV.
RESULTS
Induction and detection of Stx2 phages from environmental STEC.
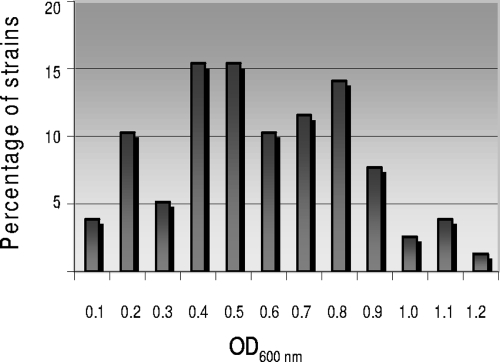
The levels of induction of the different prophages present in the STEC strains showed great variability. As shown in Fig. 1, for some of the prophages the induction levels were high, and there was a >1-OD600 unit difference compared with their noninduced counterparts after mitomycin treatment, while other prophages did not respond to this treatment and there was no significant difference. In summary, around 60% of the prophages present in STEC strains showed high levels of induction, and the values ranged from 0.1 to 0.6 OD600 unit. For the corresponding noninduced cultures the values were 1.6 ± 0.2 OD600 units after 24 h of incubation.
FIG. 1.
Levels of induction of STEC integrated prophages, expressed in OD600 units, after mitomycin treatment. A low OD600 correlates with lysis of bacterial cells and induction. The bars indicate the percentages of the strains having specific OD600 values after mitomycin treatment. The average values for noninduced cultures of the same strains were 1.6 ± 0.2 OD600 units.
The amounts of phage DNA extracted from the mitomycin-induced cultures correlated with the levels of induction of the prophages determined by measuring the absorbance, and phage DNA was more abundant in the cultures exhibiting higher levels of induction. A minimum of 0.5 μg/μl of phage DNA was observed for the strains showing a good level of induction.
Of the 79 STEC strains examined, 70 (89%) contained inducible bacteriophages; 11 of 14 (79%) STEC strains from human sewage contained inducible bacteriophages, 47 of 50 (94%) and 5 of 6 (83%) STEC strains from cattle and pig abattoirs, respectively, contained inducible bacteriophages, and 7 of 9 (78%) STEC strains from mixed sewage contained inducible bacteriophages.
Genetic diversity among Stx2 phages as determined by RFLP analysis.
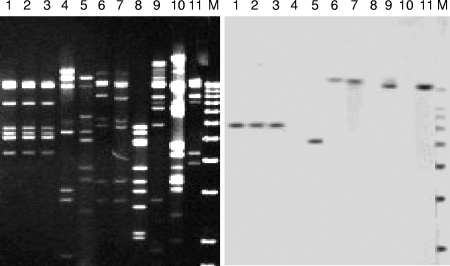
RFLP analysis of phage DNA revealed diversity among induced phages from different STEC strains. However, some prophages isolated from different strains had the same profile, suggesting that the same prophage could be integrated in the different STEC strains. Since any temperate phage present in an STEC strain could have been induced in the assay, detection of the stx2 gene was necessary to confirm the presence of DNA from Stx2 phages. stx2 was detected by Southern blotting and hybridization with the stx2 probe described previously within the RFLP pattern obtained. This probe has neither EcoRI nor ClaI restriction sites, and therefore only one hybridization signal was expected for each stx2 gene copy present in the extracted phage DNA. The stx2 gene was detected by hybridization of the RFLP products with the stx2 probe in 43% of the induced STEC prophages. The RFLP patterns observed were diverse, although some induced prophages from STEC strains having different origins had the same pattern. The molecular weight of the RFLP fragment in which the stx2 gene was located (according to the positive hybridization signal) was highly conserved. The stx2 gene was located in the same RFLP fragment for all phages having the same RFLP pattern. Moreover, for some of the phages with different RFLP patterns, the sizes of the RFLP fragments in which the stx2 gene was located were similar (Fig. 2, lanes 9 and 11). As shown in Table 1, the sizes of the positive RFLP fragments from the induced Stx2 phages from different serotypes ranged from 4.9 to 18 kb. Two bands were detected in the RFLP analysis of the induced prophages from strain O91:H21, which is consistent with previous results showing that this strain carried two stx2 genes (stx2 and the stx2c variant). It was not possible to detect the stx2 gene after RFLP analysis in some of the induced prophage cultures (Fig. 2, lanes 4, 8, and 10), although it was noticed that a prophage was inside the strain.
FIG. 2.
EcoRI-digested phage DNA from some of the induced phages present in the STEC strains (left panel) and hybridization signals with the stx2 probe after Southern blotting (right panel). Lanes 1 to 11 contained strains belonging to serotypes O157:H7, O157:H7, O157:H7, O157:H7, O146:H−, O171:H2, O171:H2, O171:H2, O181:H20, O90:H−, and O90:H−, respectively. Lane M contained a 1-kb DNA ladder.
TABLE 1.
Serotypes carrying inducible phages showing positive hybridization signals with the stx2 gene probe after RFLP analysis and Southern blotting
| Serotype | No. of phages/ no. of strains | Size(s) of EcoRI fragment(s) (kb) |
|---|---|---|
| O146:H− | 1/1 | 4.9 |
| O91:H21 | 1/1 | 6, 4.9 |
| O113:H21 | 1/1 | 6 |
| O157:H7 | 14/14 | 6 |
| O157:H− | 1/1 | 6 |
| O100:H− | 1/1 | 11 |
| O2:H25 | 3/5 | 12 |
| O26:H− | 1/2 | 12 |
| O90:H− | 1/4 | 12 |
| O171:H2 | 6/15 | 12 (5), 15 (1)a |
| O181:H20 | 2/2 | 12 |
| O98:HNT | 1/1 | 15 |
| O1:H20 | 2/2 | 18 |
The numbers in parentheses are numbers of phages.
The size of the total phage DNA was estimated by adding the sizes of the different RFLP fragments, and in most cases it was around 50 kb (data not shown), which was consistent with the assumption that a single phage had been induced.
It should be noted that this study could be performed only with prophages that exhibited sufficient levels of induction, which was possible for only 65 of the 79 STEC strains, even though larger culture volumes were prepared for the strains in which no RFLP of phage DNA was detected.
Evaluation of the infection capacity of the induced Stx2 phages.
The ability of induced Stx2 phages to infect different hosts was analyzed both by a plaque-forming assay and by hybridization of the generated plaques with the 378-bp stx2 gene probe described previously. Infection was induced in 50 of the 79 STEC strains (63%) (Table 2). The Stx2 phages infected mainly S. sonnei strain 866 (51%), E. coli DH5α (46%), S. sonnei strain 635 (44%), and E. coli C600 (40%). Some STEC strains yielded lytic phages (probably non-Stx2 phages), which may have accounted for the lack of detection of the stx2 gene in the phage plaques generated. Six induced Stx2 phages from non-O157 STEC strains were able to infect the E. coli O157:H7 host strain used in this study. It is noteworthy that all induced Stx2 phages from O157 strains were able to infect the O157 host strains, as well as the most infected host strains described above, including C. freundii and E. aerogenes.
TABLE 2.
Detection of bacteriophage infection by plaque blotting and hybridization with the stx2 probe in different host strains analyzed
| Serotypea | No. of phages | E. coli C600 | E. coli DH5a | E. coli O111 | E. coli O26 strain 216 | E. coli O26 strain 224 | E. coli O157:H7 strain ATCC 43888 | E. coli O157 strain 11 | E. coli O157 strain 44 | E. coli O157 strain 151 | S. sonnei 866 | S. sonnei 635 | S. dysenteriae 500 | S. boydii 316 | S. boydii 351 | S. flexneri 805 | S. flexneri 668 | E. aerogenes | C. freundii |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| O1:H20 | 2 | + (2)b | + (2) | + (2) | + (2) | + (2) | + (2) | + (2) | + (2) | + (2) | + (2) | + (2) | + (2) | + (2) | + (2) | + (2) | + (2) | + (2) | + (2) |
| O2:H2 | 1 | + (1) | + (1) | + (1) | + (1) | ||||||||||||||
| O2:H21 | 1 | ||||||||||||||||||
| O2:H25 | 5 | + (3) | + (2) | + (1) | + (1) | + (5) | + (5) | + (1) | + (1) | ||||||||||
| O8:H− | 1 | ||||||||||||||||||
| O8:H31 | 1 | ||||||||||||||||||
| O8:H9 | 3 | ||||||||||||||||||
| O22:H8 | 2 | ||||||||||||||||||
| O26:H− | 2 | + (1) | + (1) | + (1) | + (1) | ||||||||||||||
| O54:H21 | 1 | ||||||||||||||||||
| O70:H− | 1 | ||||||||||||||||||
| O76:H2 | 1 | ||||||||||||||||||
| O89:H19 | 1 | ||||||||||||||||||
| O90:H− | 4 | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | ||||||||||
| O91:H21 | 1 | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | ||||||||||
| O98:HNT | 1 | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | ||||||
| O100:H− | 1 | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + 1 | ||||||||
| O113:H21 | 1 | + (1) | |||||||||||||||||
| O127:H− | 1 | ||||||||||||||||||
| O136:H1 | 1 | + (1) | |||||||||||||||||
| O146:H− | 1 | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) |
| O146:H21 | 1 | ||||||||||||||||||
| O157:H− | 1 | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | ||||||
| O157:H7 | 14 | + (13) | + (13) | + (13) | + (13) | + (13) | + (13) | + (13) | + (13) | + (13) | + (13) | + (13) | + (2) | + (13) | + 2 | + (13) | + (13) | ||
| O159:H− | 1 | + (1) | |||||||||||||||||
| O162:H7 | 1 | + (1) | + (1) | + (1) | |||||||||||||||
| O166:H21 | 1 | + (1) | |||||||||||||||||
| O171:H2 | 15 | + (5) | + (9) | + (2) | + (2) | + (4) | + (7) | + (9) | + (5) | + (5) | + (6) | + (3) | + (4) | + (4) | |||||
| O175:H16 | 1 | ||||||||||||||||||
| O177:H− | 1 | ||||||||||||||||||
| O181:H20 | 2 | + (2) | + (2) | + (1) | + (1) | + (2) | + (1) | + (1) | + (1) | ||||||||||
| O181:H49 | 1 | + (1) | + (1) | ||||||||||||||||
| ONT:H− | 4 | ||||||||||||||||||
| ONT:H21 | 1 | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | + (1) | |||||||||
| ONT:H9 | 1 | + (1) | |||||||||||||||||
| ONT:HNT | 1 |
E. coli serotypes from which the bacteriophages were induced.
The numbers in parentheses are the numbers of induced prophages from all of the strains belonging to different E. coli serotypes showing a positive signal.
The presence of infectious Stx2 phages varied according to the isolation source, and such phages were detected mainly in STEC obtained from animal wastewaters. Thus, 80% of the STEC strains isolated from cattle abattoirs and 50% of the STEC strains isolated from pig abattoirs, but only 28 and 33% of the STEC strains isolated from human and mixed sewage, respectively, contained detectable Stx2 infectious prophages.
Construction of new lysogens.
The abilities of the Stx2 phages to infect and to establish lysogeny in two previously described hosts (E. coli DH5α and S. sonnei strain 866) were assessed. Nine bacteriophages carrying different stx2 genes (stx2, stx2c, stx2g, and stx2e) were successfully transduced (Table 3). The infection and subsequent stabilization of the stx2 gene within the different hosts generated new lysogenic strains, which in some cases changed some phenotypic characteristics. The new lysogens showed levels of induction different than those obtained with the original strains. In general, the ability to produce Stxs was maintained, but there were some exceptions. Thus, in the case of strain S8 (serotype O146:H−, carrying an Stx2 prophage), after transduction of the Stx2 phage to E. coli DH5α, the prophage was poorly induced and no Stx2 production was detected. The opposite was found to be the case for strain S150 (serotype O26:H−, carrying an Stx2e prophage), in which a higher level of prophage induction was observed and the Stx2 protein was detected after transduction to S. sonnei. The seven other transduced prophages retained the same phenotypic characteristics in the new lysogenic strains. The RFLP prophage patterns obtained were the same as the patterns obtained for the induced prophages from the original strains.
TABLE 3.
Genetic and morphological characteristics of the transduced bacteriophages
| Transduced bacteriophagea | Host | stx2 gene | Size of EcoRI fragment (kb) | Morphology |
|---|---|---|---|---|
| S8 | E. coli DH5α | stx2 | NDb | ND |
| S48 | E. coli DH5α | stx2e | ND | ND |
| S62 | S. sonnei 866 | stx2c | 12 | H19-likec |
| S63 | S. sonnei 866 | stx2c | 12 | H19-likec |
| S67 | E. coli DH5α | stx2c | 12 | H19-likec |
| S68 | E. coli DH5α | stx2c | ND | H19-likec |
| S86 | S. sonnei 866 | stx2g | 12 | 933W-liked |
| S134 | S. sonnei 866 | stx2g | 12 | 933W-liked |
| S150 | S. sonnei 866 | stx2e | ND | 933W-liked |
Induced prophages from the original STEC strains.
ND, not detected.
Bacteriophage morphology similar to that of phage H19.
Bacteriophage morphology similar to that of phage 933W.
Phage integration site.
One strain belonging to serotype O146:H− was shown to amplify the wrbA phage-bacterium attachment site, suggesting that the phage was integrated in this locus. Unexpectedly, 3 strains amplified only one of the wrbA locus junctions (1 serotype O146:H21 strain, 1 serotype O162:H7 strain, and 1 serotype O90:H− strain), while 22 strains amplified only one of the yehV locus junctions (3 serotype O171:H2 strains, 1 serotype O113:H21 strain, 15 serotype O157:H7 strains, 1 serotype O91:H21 strain, and 2 serotype O22:H8 strains). Additionally, two other strains belonging to serotypes O127:H− and O90:H− amplified one junction from both the wrbA and yehV loci. It should be noted that in the newly constructed lysogens most of the prophages showed the same integration profile; the only exception was a prophage that became integrated in the yehV locus after transduction.
Morphological diversity of STEC prophages.
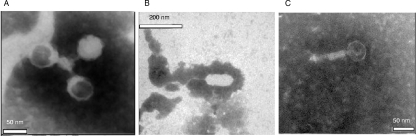
In most cases the morphology of the induced bacteriophages resembled that of the previously described Stx2 phages 933W and H19, which have icosahedral capsids and a short tail and long capsids and a long tail, respectively. In a few cases other phages were detected (Fig. 3), but they could not be transduced, and therefore the presence of the stx2 gene could not be determined. No relationship between the presence of a particular stx variant and the morphology of the observed phage was found.
FIG. 3.
Electron micrographs showing the Stx2 phages isolated most frequently from the newly constructed 933W-like and H19-like lysogens (A and B, respectively) and one non-Stx2 phage isolated from the original STEC (C), which may have hindered the detection of Stx2 phages.
DISCUSSION
Phages can be considered important reservoirs of genes that can be mobilized in the bacterial population, which is especially relevant for the emergence of new pathogenic strains. Bacteriophage-mediated transmission of the stx2 gene has been examined in various studies since the emergence of STEC strains. However, most authors have focused on Stx2 phages isolated from clinical STEC strains or on the O157:H7 serotype. In this study, the presence of inducible bacteriophages in a collection of 79 STEC strains belonging to 35 serotypes isolated from different animal wastewaters and human sewage was evaluated in order to assess the real prevalence of these phages in environmental strains.
Eighty-nine percent of the strains contained mitomycin-inducible bacteriophages, as measured by stx2 gene PCR amplification of phage DNA, and a high proportion (63%) of the bacteriophages were capable of infecting the laboratory strains used in this study. However, it should be noted that only 28% of the strains isolated from human sewage carried infectious Stx2 prophages. It could be hypothesized that loss of functional stx2 genes occurs as STEC emerges in human populations. The lack of infectious and inducible Stx2 phages in human sewage isolates was supported by the absence of Stx production, a feature that is highly related to the pathogenicity of the STEC strains, although this was not the subject of this study. This finding suggests that inactivation of the phages may have occurred after the phage integration process. Moreover, only a small proportion of the phages (9 of 79) were maintained after several passages in the host strain to generate new lysogens, which implies that although the phages are infectious in different hosts, their insertion and persistence as prophages in the host genome are dependent on the genetic background of the strains (38), including the presence of other bacteriophages. This effect was also obvious after transduction of some of the phages whose lysogenic strains lost or gained the ability to produce Stx.
Phages carrying different stx2 gene variants, including the recently reported stx2g variant, were detected in this study (16). The differences between stx genes from S. dysenteriae, which traditionally appear to be chromosomally encoded, and stx genes from E. coli strains, which are bacteriophage encoded, are unknown. However, an stx gene-carrying bacteriophage was isolated from the related species S. sonnei (5). This bacteriophage was adapted to different Shigella species (42). Adaptation of the induced Stx2 phages from STEC strains to S. sonnei could be inferred from the results obtained in this study since the majority of the prophages were able to infect S. sonnei 866. This is not surprising since similar results were obtained with free Stx2 phages isolated from sewage (22). These observations support the hypothesis that Shigella could act as a reservoir of the stx2 gene. To our knowledge, no studies have examined the prevalence of Stx2 phages in the phages in nonclinical isolates of Shigella species, which could provide new insight into the horizontal transmission of stx2 genes in the environment. In any case, it is important to point out that the Stx2 phages that are present mostly in potentially nonpathogenic STEC strains are similar to the phages that have previously been linked to human outbreaks (8, 9).
Ten STEC strains contained two stx2 genes, although only one phage was induced from the majority of these strains. Similar results have been obtained in other studies, showing that simultaneous induction of two prophages results in an induction rate for one of the prophages that is lower than the induction rate when a single prophage is present in the strain (21).
A low proportion of the induced Stx2 phages from non-O157 STEC strains were capable of infecting the E. coli O157 host strains used in this study. It has been reported previously that short-tailed Stx phages share a common receptor, YaeT, which is an outer membrane protein commonly found in members of the Enterobacteriaceae (40). Although the possibility of the presence of another receptor in O157 that is preferentially used by induced phages from O157 strains cannot be ruled out, this information suggests that some other mechanisms in the genetic background of the O157 strains can also hinder infection with Stx phages isolated from non-O157 strains. Accordingly, 14 of 15 induced phages from O157 STEC were able to infect the O157 serotype host strains, as well as the majority of the host strains tested.
The RFLP studies revealed a high degree of heterogeneity among the different induced STEC prophages, which could be attributed to the presence of more than one phage (probably non-Stx2) in the genomes of the strains. However, the degree of heterogeneity of Stx2 phages was supported by the finding that the stx2 gene was located in fragments with different molecular weights in the RFLP patterns. The differences were attributed to variations in the genetic sequences of the phages. Recombination of Stx2 phages with other prophages or phage genes in intergenic regions that do not disrupt functional modules is a plausible explanation for reorganization events leading to an increase in such genetic heterogeneity (7). Genes that are present in some phage genomes may also increase the fitness of the phage (12). It is noteworthy that some of the induced Stx2 phages from different STEC strains having different origins had similar RFLP patterns, suggesting that the horizontal transfer of these phages could have been a recent event or that the phages are subjected to high selective pressure. In addition, two induced phages were from a strain belonging to E. coli serotype O91:H21, which is consistent with a previous study that found two stx2 genes in a genome (8). The heterogeneity of the Stx2 prophages present in the environmental STEC strains is consistent with the heterogeneity reported in previous studies performed with free environmental Stx2 phages (22).
Numerous Stx phages have been shown to be integrated in the wrbA and yehV loci (39). In this study only 1 strain showed PCR amplification of both wrbA bacteriophage-bacterium junctions, 2 strains showed simultaneous PCR amplification of a single wrbA-yehV junction, while 3 and 22 strains showed amplification of the wrbA and yehV junctions, respectively. The two strains that showed PCR amplification of both loci had previously been shown to carry two stx2 gene variants (8). The change of integration site that occurred in one of the transduced bacteriophages is not surprising since it has been reported that an Stx phage has a preferred integration site when it is available but is also able to integrate at different sites when this site is occupied (38). The high amplification rate for only one of the locus junctions may be related to slight variability in one of the junctions, although this has not been reported to date.
Some instability of free Stx2 phages after propagation has been reported previously (22), which suggests that these phages could have evolved so that they were integrated in an STEC bacterial host as a prophage. A prophage can be induced when the environmental conditions activate its lytic cycle. Accordingly, in this study all the strains studied allowed induction of their Stx phages at moderate or high levels, ensuring that free viral particles remained free in the environment, which ensured the survival necessary to allow new infection of a suitable host once environmental conditions were optimal again. The cycle is thus closed and allows the stx genes to be prevalent in the environment, which may be their real reservoir.
Acknowledgments
We thank Guillem Prats for providing the clinical isolates of Shigella and E. coli.
This study was supported by the Generalitat de Catalunya (grant 2005SGR00592), the Ministerio de Educación y Ciencia (grant AGL200601566/ALI), and the Xarxa de Referència en Biotecnologia (XeRBa).
Footnotes
Published ahead of print on 14 November 2008.
REFERENCES
- 1.Acheson, D. W. K., and G. T. Keusch. 1996. Which Shiga toxin-producing types of Escherichia coli are important? ASM News 6:302-306. [Google Scholar]
- 2.Acheson, D. W. K., J. Reidl, X. Zhang, G. T. Keusch, J. J. Mekalanos, and M. K. Waldor. 1998. In vivo transduction with Shiga toxin 1-encoding phage. Infect. Immun. 66:4496-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allison, H. E., M. J. Sergeant, C. E. James, J. R. Saunders, D. L. Smith, R. J. Sharp, T. S. Marks, and A. J. McCarthy. 2003. Immunity profiles of wild-type and recombinant Shiga-like toxin-encoding bacteriophages and characterization of novel double lysogens. Infect. Immun. 71:3409-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Betley, M., and J. Mekalanos. 1985. Staphylococcal enterotoxin A is encoded by a phage. Science 229:185-187. [DOI] [PubMed] [Google Scholar]
- 5.Beutin, L., E. Strauch, and I. Fischer. 1999. Isolation of Shigella sonnei lysogenic for a bacteriophage encoding gene for production of Shiga toxin. Lancet 353:1498. [DOI] [PubMed] [Google Scholar]
- 6.Blanch, A. R., C. García-Aljaro, M. Muniesa, and J. Jofre. 2003. Detection, enumeration and isolation of strains carrying the stx2 gene from urban sewage. Water Sci. Technol. 47:109-116. [PubMed] [Google Scholar]
- 7.Brüssow, H., C. Canchaya, and W. D. Hardt. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68:560-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Aljaro, C., M. Muniesa, J. E. Blanco, M. Blanco, J. Blanco, J. Jofre, and A. R. Blanch. 2005. Characterization of Shiga toxin producing Escherichia coli isolated from aquatic environments. FEMS Microbiol. Let. 246:55-65. [DOI] [PubMed] [Google Scholar]
- 9.García-Aljaro, C., X. Bonjoch, and A. R. Blanch. 2005. Combined use of an immunomagnetic separation method and immunoblotting for the enumeration and isolation of E. coli O157 in wastewaters. J. Appl. Microbiol. 98:589-597. [DOI] [PubMed] [Google Scholar]
- 10.García-Aljaro, C., M. Muniesa, J. Jofre, and A. R. Blanch. 2006. Newly identified bacteriophages carrying the stx2g gene isolated from Escherichia coli strains in polluted waters. FEMS Microbiol. Lett. 258:127-135. [DOI] [PubMed] [Google Scholar]
- 11.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 12.Hendrix, R. W., J. G. Lawrence, G. F. Hatfull, and S. Casjens. 2000. The origins and ongoing evolution of viruses. Trends Microbiol. 8:504-508. [DOI] [PubMed] [Google Scholar]
- 13.Johansen, B. K., Y. Wasteson, P. E. Granum, and S. Brynestad. 2001. Mosaic structure of Shiga-toxin-2-encoding phages isolated from Escherichia coli O157:H7 indicates frequent gene exchange between lambdoid phage genomes. Microbiology 147:1929-1936. [DOI] [PubMed] [Google Scholar]
- 14.Kimmitt, P. T., C. R. Harwood, and M. R. Barer. 2000. Toxin gene expression by Shiga toxin-producing Escherichia coli: the role of antibiotics and the bacterial SOS response. Emerg. Infect. Dis. 6:458-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laird, W., and N. Groman. 1976. Orientation of the tox gene in the prophage of corynebacteriophage beta. J. Virol. 19:228-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leung, P. H., J. S. Peiris, W. W. Ng, R. M. Robins-Browne, K. A. Bettelheim, and W. C. Yam. 2003. A newly discovered verotoxin variant, VT2g, produced by bovine verocytotoxigenic Escherichia coli. Appl. Environ. Microbiol. 69:7549-7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mühldorfer, I., J. Hacker, G. T. Keusch, D. W. Acheson, H. Tschäpe, A. V. Kane, A. Ritter, T. Olschlager, and A. Donohue-Rolfe. 1996. Regulation of the Shiga-like toxin II operon in Escherichia coli. Infect. Immun. 64:495-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muniesa, M., and J. Jofre. 1998. Abundance in sewage of bacteriophages that infect Escherichia coli O157:H7 and that carry the Shiga toxin 2 gene. Appl. Environ. Microbiol. 64:2443-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muniesa, M., F. Lucena, and J. Jofre. 1999. Comparative survival of free Shiga toxin 2-encoding phages and Escherichia coli strains outside the gut. Appl. Environ. Microbiol. 65:5615-5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muniesa, M., J. Recktenwald, M. Bielaszewska, H. Karch, and H. Schmidt. 2000. Characterization of a Shiga toxin 2e-converting bacteriophage from an Escherichia coli strain of human origin. Infect. Immun. 68:4850-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muniesa, M., M. de Simon, G. Prats, D. Ferrer, H. Panella, and J. Jofre. 2003. Shiga toxin 2-converting bacteriophages associated with clonal variability in Escherichia coli O157:H7 strains of human origin isolated from a single outbreak. Infect. Immun. 71:4554-4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muniesa, M., R. Serra-Moreno, and J. Jofre. 2004. Free Shiga toxin bacteriophages isolated from sewage showed diversity although the stx genes appeared conserved. Environ. Microbiol. 6:716-725. [DOI] [PubMed] [Google Scholar]
- 23.O'Brien, A. D., M. K. Gentry, M. R. Thompson, B. P. Doctor, P. Gemski, and S. B. Formal. 1979. Shigellosis and Escherichia coli diarrhea: relative importance of invasive and toxigenic mechanisms. Am. J. Clin. Nutr. 32:229-233. [DOI] [PubMed] [Google Scholar]
- 24.O'Brien, A. D., J. W. Newland, S. F. Miller, R. K. Holmes, H. W. Smith, and S. B. Formal. 1984. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 226:694-696. [DOI] [PubMed] [Google Scholar]
- 25.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 26.Osawa, R., S. Iyoda, S. I. Nakayama, A. Wada, S. Yamai, and H. Watanabe. 2000. Genotypic variations of Shiga toxin-converting phages from enterohaemorrhagic Escherichia coli O157:H7 isolates. J. Med. Microbiol. 49:565-574. [DOI] [PubMed] [Google Scholar]
- 27.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 28.Plunkett, G., III, D. J. Rose, T. J. Durfee, and F. R. Blattner. 1999. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J. Bacteriol. 181:1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rietra, P. J. G. M., G. A. Willshaw, H. R. Smith, A. M. Field, S. M. Scotland, and B. Rowe. 1989. Comparison of vero-cytotoxin-encoding phages from Escherichia coli of human and bovine origin. J. Gen. Microbiol. 135:2307-2318. [DOI] [PubMed] [Google Scholar]
- 30.Riley, L. W., R. S. Remis, S. D. Helgerson, H. B. McGee, J. G. Wells, B. R. Davis, R. J. Hebert, E. S. Olcott, L. M. Johnson, N. T. Hargrett, P. A. Blake, and M. L. Cohen. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308:681-685. [DOI] [PubMed] [Google Scholar]
- 31.Rüssmann, H., H. Schmidt, J. Heesemann, A. Caprioli, and H. Karch. 1994. Variants of Shiga-like toxin II constitute a major toxin component in Escherichia coli O157 strains from patients with haemolytic uraemic syndrome. J. Med. Microbiol. 40:338-343. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 33.Sanger, F., A. R. Coulson, G. F. Hong, D. F. Hill, and G. B. Petersen. 1982. Nucleotide sequence of bacteriophage λ DNA. J. Mol. Biol. 162:729-773. [DOI] [PubMed] [Google Scholar]
- 34.Sato, T., T. Shimizu, M. Watarai, M. Kobayashi, S. Kano, T. Hamabata, Y. Takeda, and S. Yamasaki. 2003. Distinctiveness of the genomic sequence of Shiga toxin 2-converting phage isolated from Escherichia coli O157:H7 Okayama strain compared to other Shiga toxin 2-converting phages. Gene 309:35-48. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt, H., M. Bielaszewska, and H. Karch. 1999. Transduction of enteric Escherichia coli isolates with a derivative of Shiga toxin 2-encoding bacteriophage φ3538 isolated from Escherichia coli O157:H7. Appl. Environ. Microbiol. 65:3855-3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt, H., J. Scheef, H. I. Huppertz, M. Frosch, and H. Karch. 1999. Escherichia coli O157:H7 and O157:H− strains that do not produce Shiga toxin: phenotypic and genetic characterization of isolates associated with diarrhea and hemolytic-uremic syndrome. J. Clin. Microbiol. 37:3491-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scotland, S. M., H. R. Smith, G. A. Willshaw, and B. Rowe. 1983. Vero cytotoxin production in a strain of Escherichia coli determined by genes carried on bacteriophage. Lancet ii:216. [DOI] [PubMed] [Google Scholar]
- 38.Serra-Moreno, R., J. Jofre, and M. Muniesa. 2007. Insertion site occupancy by stx2 bacteriophages depends on the locus availability of the host strain chromosome. J. Bacteriol. 189:6645-6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaikh, N., and P. I. Tarr. 2003. Escherichia coli O157:H7 Shiga toxin-encoding bacteriophages: integrations, excisions, truncations, and evolutionary implications. J. Bacteriol. 185:3596-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith, D. L., C. E. James, M. J. Sergeant, Y. Yaxian, J. R. Saunders, A. J. McCarthy, and H. E. Allison. 2007. Short-tailed stx phages exploit the conserved YaeT protein to disseminate Shiga toxin genes among enterobacteria. J. Bacteriol. 189:7223-7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, H. W., P. Green, and Z. Parsell. 1983. Vero cell toxins in Escherichia coli and related bacteria: transfer by phage and conjugation and toxic action in laboratory animals, chickens, and pigs. J. Gen. Microbiol. 129:3121-3137. [DOI] [PubMed] [Google Scholar]
- 42.Strauch, E., R. Lurz, and L. Beutin. 2001. Characterization of a Shiga toxin-encoding temperate bacteriophage of Shigella sonnei. Infect. Immun. 69:7588-7595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strockbine, N. A., L. R. M. Marques, J. W. Newland, H. W. Smith, R. K. Holmes, and A. D. O’ Brien. 1986. Two toxin-converting phages from Escherichia coli O157:H7 strain 933 encode antigenically distinct toxins with similar biological activities. Infect. Immun. 53:135-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sunagawa, H., and K. Inoue. 1992. Biological and biophysical characteristics of phages isolated from Clostridium botulinum type C and D strains, and physicochemical properties of the phage DNAs. J. Vet. Med. Sci. 54:675-684. [DOI] [PubMed] [Google Scholar]
- 45.Teel, L. D., A. R. Melton-Celsa, C. K. Schmitt, and A. D. O'Brien. 2002. One of two copies of the gene for the activatable Shiga toxin type 2d in Escherichia coli O91:H21 strain B2F1 is associated with an inducible bacteriophage. Infect. Immun. 70:4282-4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Unkmeir, A., and H. Schmidt. 2000. Structural analysis of phage-borne stx genes and their flanking sequences in Shiga toxin-producing Escherichia coli and Shigella dysenteriae type 1 strains. Infect. Immun. 68:4856-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wagner, P. L., D. W. Acheson, and M. K. Waldor. 1999. Isogenic lysogens of diverse Shiga toxin 2-encoding bacteriophages produce markedly different amounts of Shiga toxin. Infect. Immun. 67:6710-6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]
- 49.Wells, J. G., B. R. Davis, I. K. Wachsmuth, L. W. Riley, R. S. Remis, R. Sokolow, and G. K. Morris. 1983. Laboratory investigation of hemorrhagic colitis outbreaks associated with a rare Escherichia coli serotype. J. Clin. Microbiol. 18:512-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoh, M., and T. Honda. 1997. The stimulating effect of fosfomycin, an antibiotic in common use in Japan, on the production/release of verotoxin-1 from enterohaemorrhagic Escherichia coli O157:H7 in vitro. Epidemiol. Infect. 119:101-103. [DOI] [PMC free article] [PubMed] [Google Scholar]