Abstract
Proteorhodopsin (PR) genes related to Flavobacteria were found to be highly diverse in the East and South China seas and displayed a distinct geographic pattern, which appeared to reflect cold versus warm adaptation when Global Oceanic Sampling database metagenomic data were included. Flavobacterial PR genes were more abundant offshore than nearshore, implying that inheritance of the PR gene could be important for Flavobacteria living in the oligotrophic environment.
Proteorhodopsin (PR) proteins are bacterial retinal binding membrane pigments that are predicted to have a phototrophic potential (4, 5, 27). Thousands of PR sequences have been retrieved from various marine (4-6, 9, 20-24, 31) and freshwater (3) environments. These PR genes belong to diverse groups of bacteria (4, 9, 13, 19, 21, 23, 27, 31), but are found mainly in three major marine phylum: Alphaproteobacteria, Gammaproteobacteria, and Bacteroidetes. The diversity of PR genes related to Proteobacteria in nature has been studied extensively (21-23, 28). However, little is known about the distribution and diversity of PR genes related to Bacteroidetes in the marine environment.
Flavobacteria, an important class of Bacteroidetes, often make up a significant portion of marine microbial communities (1, 2, 14, 18). They are found both free-living and attached to organic aggregates and are considered as major mineralizers of organic matter (1, 7, 8, 18). A recent study showed that PR genes in Flavobacteria formed a unique cluster divergent from those in Proteobacteria and other taxa (19). So far, PR proteins from five different flavobacterial strains have been sequenced (see the primer design for reference), and the conservation of their PR genes allowed us to design a primer set specifically targeting this group of bacteria. Here, we report the diversity and geographic distribution of flavobacterial PR genes in the East and South China seas.
Surface water samples were collected using Niskin bottles from three stations (W1, W2 and W8) in the East China Sea on 27 to 28 November 2006 and two stations (A1 and A9) in the South China Sea on 19 to 20 January 2007 on board the R/V Dong Fang Hong No. 2 (Fig. 1). Water samples from depths of 30 m, 50 m, and 70 m at station A1 and 60 m, 100 m, and 200 m at station W8 were also collected. Seawater was filtered onto a 0.22-μm-pore-size polycarbonate filter (Millipore Co., Bedford, MA) after prefiltration through a 200-μm-pore-size mesh, except for the surface samples from stations A1 and A9, which were filtered onto a 3-μm-pore-size polycarbonate filter (Millipore Co.) and subsequently onto a 0.22-μm filter after the same prefiltration. Community DNA was extracted following a protocol described elsewhere (11). The primers used to amplify flavobacterial PR genes for both diversity and quantification studies were designed based on the PR sequences from five flavobacteria (see Fig. S1 in the supplemental material), including Dokdonia donghaensis MED134 (accession no. EAQ40507), Polaribacter dokdonensis MED152 (EAQ40925), Polaribacter irgensii 23-P (ZP_01117885), Psychroflexus torquis ATCC 700755 (ZP_01253360), and BAL38 (ZP_01734914). The forward primer (5′-GGCTATGATGGCWGCWT-3′) and reverse primer (5′-CTTCACCAAWRTAWCCAGTWAC-3′) flanked a 310-bp fragment, which contained several key amino acid positions for the functionality of PR, such as Asp97, Met/Leu/Gln105, and Glu108 (SAR86 eBAC31A08 numbering). PCRs were performed in a total volume of 50 μl containing 5 to 10 ng of template DNA, 200 μM deoxynucleoside triphosphates (dNTPs), 2.5 mM MgCl2, 0.5 μM of each primer, and 2.5 U of ExTaq polymerase (TaKaRa, Co., Dalian, China) under the following conditions: 95°C for 2 min, followed by 35 cycles of 95°C for 1 min, 54°C for 30 s, and 72°C for 1 min. PCR products of each sample were used to construct clone libraries, and about 10 positive clones were sequenced in each library. Multiple sequence alignments were performed using the CLUSTALX 1.81 program (30). The neighbor-joining (N-J) phylogenetic tree was inferred using Mega 4.0 (29). The rarefaction analysis and coverage estimation were conducted using DOTUR (25). The fraction of shared operational taxonomic units (OTU [at 5% genetic divergence]) between every two clone libraries was indicated by the classic Sørenson index (S) using the program SONS (26).
FIG. 1.
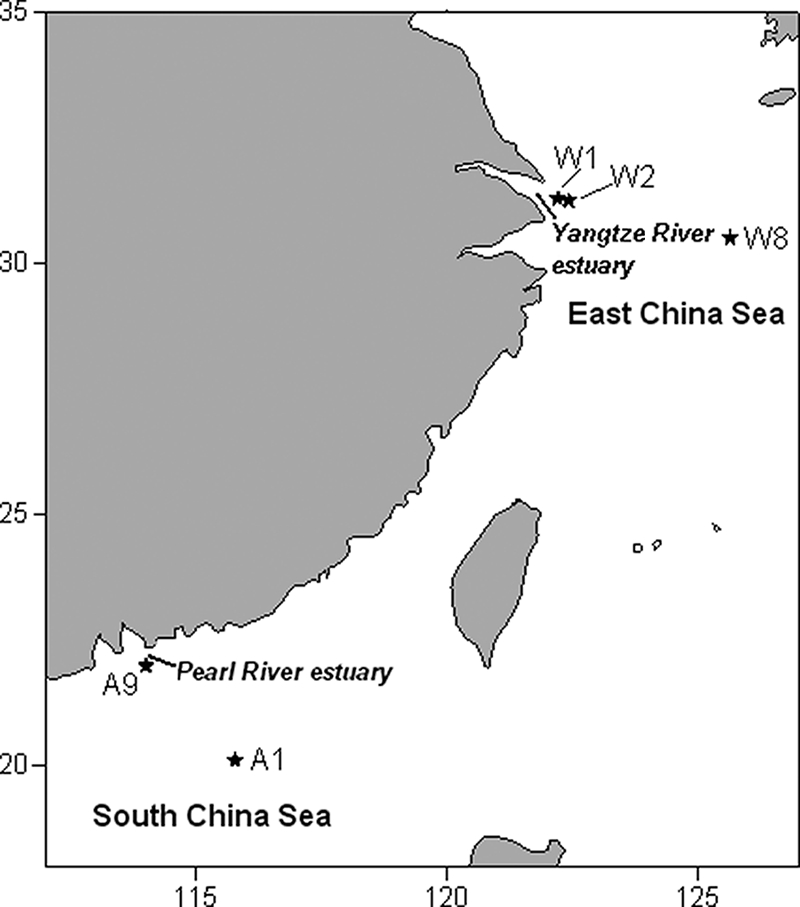
Sampling locations (★) in the East and South China seas.
Real-time quantitative PCR (qPCR) was conducted with the ABI 7500 real-time PCR system (Applied Biosystems, CA) using the Sybr Premix Ex Taq kit (TaKaRa). The mixture solution (25 μl) and amplification conditions followed the manufacturer's protocol. Deionized water without DNA was used as a negative control. The standards were 10-fold serial dilutions of a pMD18-T plasmid (103 to 108 copies) containing a representative PR fragment from our clone library. The melting curve analysis was performed at the end of amplification.
Phylogenetic analysis.
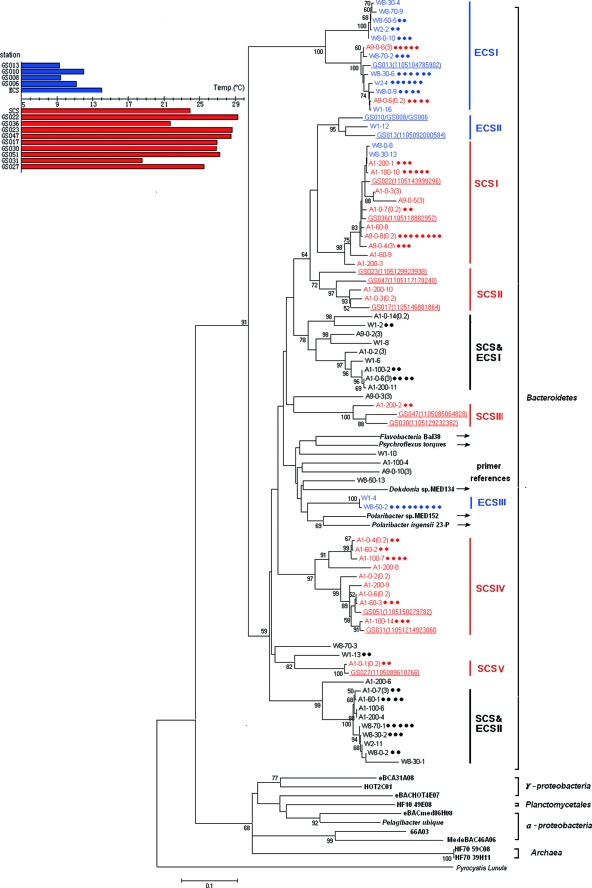
A total of 140 PR sequences were obtained from the East and South China seas. At 5% genetic divergence, the rarefaction curve of these PR sequences tended to reach a maximum (see Fig. S2 in the supplemental material), indicating that most flavobacterial PR genotypes in both China seas have been identified (the coverage by Chao1 richness estimation was 70%). Phylogenetic analysis (Fig. 2) showed that all of the PR sequences recovered from China seas were clustered with the five known flavobacterial PR sequences (nucleotide similarity of 65% to 100%) and distantly related to those from other phyla like Proteobacteria and Archaea (similarity of 48% to 64%), suggesting that our primers were suitable for amplifying the PR genes related to Flavobacteria. Our sequences could be divided into 25 OTU at 10% genetic divergence, indicating a high diversity of flavobacterial PR genes in the China seas. At an e value of 10−19, a total of 83 different PR fragments were retrieved from the Global Ocean Sampling (GOS) metagenomic database using our sequences as queries for BLASTN search. The GOS hits came from a broad range of geographic sites, ranging from the North American coast through the Gulf of Mexico into the equatorial Pacific (20), suggesting a wide distribution of PR genes related to Flavobacteria in the global ocean.
FIG. 2.
Phylogenetic tree of flavobacterial PR genes from the East China Sea (ECS) and South China Sea (SCS) along with their homologs in the GOS database (underlined) relative to PR genes found in other major groups (bold). The five flavobacterial strains used for the primer design are marked by arrows. The N-J tree was constructed based on 310 nucleotide positions. Bootstrap values (1,000 resamplings) over 50% are indicated. The sequences obtained in this study are named by station (W1, W2, W8 in the ECS and A1 and A9 in the SCS), followed by the sampling depth (only for A1 and W8) and the clone number. Sequences postfixed with “(3)” or “(0.2)” represent PR from the 3- to 200-μm-size fraction or 0.22- to 3-μm-size fraction (only for A1 and A9 surface samples). Dots represent each clone with >95% DNA sequence identity to the representative sequence within one sample. Sampling temperatures in each GOS site and the two China seas (average temperature) are indicated in the left panel, and the sequences from the temperate and warm water are differentiated in blue and red. The omitted open reading frame accession no. of GS010/GS008/GS006 is (1105088900124)/(1105145485660)/(1105102499548).
Geographic pattern.
Most of the flavobacterial PR genotypes detected in both China seas appeared to be region specific. For examples, five major SCS clusters (SCSI to SCSV) were basically comprised of PR sequences from the South China Sea and three major ECS clusters (ECSI to ECSIII) were basically comprised of PR sequences from the East China Sea (Fig. 2). A few PR genotypes, such as those in clusters ECS & SCSI and ECS & SCSII (Fig. 2), were found in both China seas. The region-specific PR sequences accounted for about 2/3 of all the sequences recovered from each sea, respectively, reflecting a genetic divergence of PR between the two sea areas. Fifteen of 83 PR sequences retrieved from the GOS database were included in the phylogenetic analysis as they shared >90% similarity with our sequences. Interestingly, five GOS sequences from North Atlantic coastal water (GS006, -008, -010, and -013) were clustered with the PR sequences from the East China Sea in ECSI and ECSII (Fig. 2, clusters in blue) with sampling temperatures ranging from 9°C to 14°C (Fig. 2, left panel, bars in blue), while others (GS017, -022, -023, -027, -030, -031, -036, -047, and -051) from the tropical oceans were all clustered with the PR sequences from the South China Sea in SCSI to SCSV (clusters in red), with sampling temperatures ranging from 18°C to 29°C (bars in red). It is intriguing that common flavobacterial PR genotypes were found in both Pacific and Atlantic oceans where water temperatures are similar.
PR sequences from the five known flavobacteria were deeply branched within the Bacteroidetes group, probably due to their unique ecological adaptation. Strains Med134 and Med152 were isolated from the Mediterranean Sea (15), 23-P (16) and Psychroflexus torquis were isolated from Antarctica, and BAL38 was isolated from the Baltic Sea. We speculate that the two psychrophilic strains living in Antarctica might represent PR genotypes adapted to a lower temperature. It has been reported that temperature could affect the photocycle kinetics of PR (10). We hypothesize that temperature could be an important factor affecting the geographic distribution of different PR genotypes in the ocean. Further investigations are needed to fully understand the environmental impact on the global distribution of PR.
Free-living versus attached bacteria.
Distinct PR genotypes were found between particle-attached (3- to 200-μm-size fraction) and free-living Flavobacteria (<0.22-μm-size fraction) at the surface water of station A1, as evidenced by the classic Sørenson index (S = 0.2) between the two clone libraries. However, no major difference (S = 0.8) between these two fractions was seen at station A9. This could be related to the rapid clogging on the 3-μm-filter as station A9 usually contains a large quantity of detritus from the Pearl River. As a result, all of the PR genotypes from the 0.22-μm filter could be found on the 3-μm filter at this station.
Abundance of flavobacterial PR genes in the sea.
The abundance of flavobacterial PR genes in the two China seas ranged from 54 to 4,354 copies/ml based on the qPCR estimation. The loss of DNA by the extraction procedure (DNA recovery rate of 25 to 50%) (11) was not counted in this estimation. The maximum value occurred at the surface water of station A1, the most oligotrophic site (chlorophyll a, 0.2 μg/liter) among the sampling areas, while the minimum value occurred at the 200-m depth of this station, the deepest of all of the samples. In the surface waters, flavobacterial PR genes at the offshore stations were more abundant than those at the nearshore stations. The quantity of PR genes increased from 1,822 copies/ml at a nearshore station (A9) to 4,354 copies/ml at an offshore station (A1) in the South China Sea and from ca. 900 copies/ml at nearshore stations (W1 and W2) to 2,136 copies/ml at an offshore station (W8) in the East China Sea. The nearshore stations in both seas were affected by a large amount of land-source organic matters and nutrients from either the Yangtze River (East China Sea) (32) or the Pearl River (South China Sea) (17) (chlorophyll a, 2.0 μg/liter at station W1 and 2.8 μg/liter at station A9). We speculate that the increase of PR abundance from inshore to offshore waters might be related to the trophic conditions. It has been demonstrated in the laboratory that a flavobacterium (MED134) with PR exhibited a light-stimulated growth under a carbon-limited condition (15). In the natural marine environment, it seems logical that a poor nutrient condition would favor more abundant PR genes to complement bacterial chemotrophic life style by phototrophy. In addition, certain PR types in Alphaproteobacteria were also found more abundant in the low-chlorophyll environments (6). Meanwhile, other studies suggested that the light benefit by PR in oligotrophic oceans might not be true for all individual taxa (12), with an explanation that PR might have other physiological role besides phototrophy.
The abundance of flavobacterial PR genes appeared to be low in the ocean. PR-containing Flavobacteria were roughly estimated to account for 0 to 1.6% of total bacteria in our sampling regions, similar to those reported in the North Atlantic Ocean (0 to 3%) using the same qPCR method but differently designed primers (6). The qPCR method applied here has been well calibrated. The standard curve for the quantification was linear (R2 > 0.999) over a 6-log dynamic range with a PCR efficiency close to 100%. It is possible that the PCR primers may not cover all of the PR genotypes in Flavobacteria. However, given that highly diverse PR sequences were retrieved using this primer set, the quantification of flavobacterial PR genes in the China seas is probably close to the actual abundance.
This study provides a detailed analysis of the diversity and distribution of PR genes related to Flavobacteria in the natural marine environment. A temperature-related geographic pattern was noticed in this PR type. Whether such a pattern exists in other bacterial PR proteins is still not known and deserves further investigation.
Nucleotide sequence accession numbers.
The PR sequences obtained in this study have been deposited in GenBank under accession no. EU683472 to EU683610.
Supplementary Material
Acknowledgments
We thank Bo Wei, Yonghui Zeng, and Anyi Hu for advice on experimental design and data analysis and Ning Hong and Wei Shen for assistance with sampling.
This work was supported by an NSFC project (40632013), MOST projects (2007CB815904 and 2006BAC11B04), a COMRA project (DYXM-115-02-4-3), and an SOA project (2008418068).
Footnotes
Published ahead of print on 17 October 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abell, G. C. J., and J. P. Bowman. 2005. Ecological and biogeographic relationships of class Flavobacteria in the Southern Ocean. FEMS Microbiol. Ecol. 51:265-277. [DOI] [PubMed] [Google Scholar]
- 2.Alonso, C., F. Warnecke, R. Amann, and J. Pernthaler. 2007. High local and global diversity of Flavobacteria in marine plankton. Environ. Microbiol. 9:1253-1266. [DOI] [PubMed] [Google Scholar]
- 3.Atamna-Ismaeel, N., G. Sabehi, I. Sharon, K.-P. Witzel, M. Labrenz, K. Jurgens, T. Barkay, M. Stomp, J. Huisman, and O. Béjà. 2008. Widespread distribution of proteorhodopsins in freshwater and brackish ecosystems. ISME J. 2:656-662. [DOI] [PubMed] [Google Scholar]
- 4.Béjà, O., E. N. Spudich, J. L. Spudich, M. Leclerc, and E. F. DeLong. 2001. Proteorhodopsin phototrophy in the ocean. Nature 411:786-789. [DOI] [PubMed] [Google Scholar]
- 5.Béjà, O., L. Aravind, E. V. Koonin, M. T. Suzuki, A. Hadd, L. P. Nguyen, S. B. Jovanovich, C. M. Gates, R. A. Feldman, J. L. Spudich, E. N. Spudich, and E. F. DeLong. 2000. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289:1902-1906. [DOI] [PubMed] [Google Scholar]
- 6.Campbell, B. J., L. A. Waidner, M. T. Cottrell, and D. L. Kirchman. 2008. Abundant proteorhodopsin genes in the North Atlantic Ocean. Environ. Microbiol. 10:99-109. [DOI] [PubMed] [Google Scholar]
- 7.Cottrell, M. T., and D. L. Kirchman. 2000. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66:1692-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crump, B. C., E. V. Armbrust, and J. A. Baross. 1999. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia River, its estuary, and the adjacent coastal ocean. Appl. Environ. Microbiol. 65:3192-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de la Torre, J. R., L. M. Christianson, O. Béjà, M. T. Suzuki, D. M. Karl, J. Heidelberg, and E. F. DeLong. 2003. Proteorhodopsin genes are distributed among divergent bacterial taxa. Proc. Natl. Acad. Sci. USA 100:12830-12835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedrich, T., S. Geibel, R. Kalmbach, I. Chizhov, K. Ataka, J. Heberle, M. Engelhard, and E. Bamberg. 2002. Proteorhodopsin is a light-driven proton pump with variable vectoriality. J. Mol. Biol. 321:821-838. [DOI] [PubMed] [Google Scholar]
- 11.Fuhrman, J. A., D. E. Comeau, Å. Hagström, and A. M. Chan. 1988. Extraction from natural planktonic microorganisms of DNA suitable for molecular biological studies. Appl. Environ. Microbiol. 54:1426-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuhrman, J. A., M. S. Schwalbach, and U. Stingl. 2008. Proteorhodopsins: an array of physiological roles? Nature 6:488-494. [DOI] [PubMed] [Google Scholar]
- 13.Giovannoni, S. J., L. Bibbs, J. C. Cho, M. D. Stapels, R. Desiderio, K. L. Vergin, M. S. Rappe, S. Laney, L. J. Wilhelm, H. J. Tripp, E. J. Mathur, and D. F. Barofsky. 2005. Proteorhodopsin in the ubiquitous marine bacterium SAR11. Nature 438:82-85. [DOI] [PubMed] [Google Scholar]
- 14.Glockner, F. O., B. M. Fuchs, and R. Amann. 1999. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 65:3721-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gómez-Consarnau, L., J. M. González, M. Coll-Lladó, P. Gourdon, T. Pascher, R. Neutze, C. Pedrós-Alió, and J. Pinhassi. 2007. Light stimulates growth of proteorhodopsin-containing marine Flavobacteria. Nature 445:210-213. [DOI] [PubMed] [Google Scholar]
- 16.Gosink, J. J., C. R. Woese, and J. T. Staley. 1998. Polaribacter gen. nov., with three new species, P. irgensii sp. nov., P. franzmannii sp. nov. and P. filamentus sp. nov., gas vacuolate polar marine bacteria of the Cytophaga-Flavobacterium-Bacteroides group and reclassification of ‘Flectobacillus glomeratus’ as Polaribacter glomeratus comb. nov. Int. J. Syst. Bacteriol. 48:223-235. [DOI] [PubMed] [Google Scholar]
- 17.Huang, X. P., L. M. Huang, and W. Z. Yue. 2003. The characteristics of nutrients and eutrophication in the Pearl River estuary, South China. Mar. Pollut. Bull. 47:30-36. [DOI] [PubMed] [Google Scholar]
- 18.Kirchman, D. L. 2002. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol. Ecol. 39:91-100. [DOI] [PubMed] [Google Scholar]
- 19.McCarren, J., and E. F. DeLong. 2007. Proteorhodopsin photosystem gene clusters exhibit co-evolutionary trends and shared ancestry among diverse marine microbial phyla. Environ. Microbiol. 9:846-858. [DOI] [PubMed] [Google Scholar]
- 20.Rusch, D. B., A. L. Halpern, G. Sutton, K. B. Heidelberg, S. Williamson, S. Yooseph, D. Wu, J. A. Eisen, J. M. Hoffman, K. Remington, K. Beeson, B. Tran, H. Smith, H. Baden-Tillson, C. Stewart, J. Thorpe, J. Freeman, C. Andrew-Pfannkoch, J. E. Venter, K. Li, S. Kravitz, J. F. Heidelberg, T. Utterback, Y.-H. Rogers, L. I. Falcón, V. Souza, G. Bonilla-Rosso, L. E. Eguiarte, D. M. Karl, S. Sathyendranath, T. Platt, E. Bermingham, V. Gallardo, G. Tamayo-Castillo, M. R. Ferrari, R. L. Strausberg, K. Nealson, R. Friedman, M. Frazier, and J. C. Venter. 2007. The Sorcerer II global ocean sampling expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol. 5:398-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sabehi, G., A. Loy, K. H. Jung, R. Partha, J. L. Spudich, T. Isaacson, J. Hirschberg, M. Wagner, and O. Béjà. 2005. New insights into metabolic properties of marine bacteria encoding proteorhodopsins. PLoS Biol. 3:e273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sabehi, G., B. C. Kirkup, M. Rozenberg, N. Stambler, M. F. Polz, and O. Béjà. 2007. Adaptation and spectral tuning in divergent marine proteorhodopsins from the eastern Mediterranean and the Sargasso Seas. ISME J. 1:48-55. [DOI] [PubMed] [Google Scholar]
- 23.Sabehi, G., O. Béjà, M. T. Suzuki, C. M. Preston, and E. F. DeLong. 2004. Different SAR86 subgroups harbour divergent proteorhodopsins. Environ. Microbiol. 6:903-910. [DOI] [PubMed] [Google Scholar]
- 24.Sabehi, G., R. Massana, J. P. Bielawski, M. Rosenberg, E. F. DeLong, and O. Béjà. 2003. Novel proteorhodopsin variants from the Mediterranean and Red Seas. Environ. Microbiol. 5:842-849. [DOI] [PubMed] [Google Scholar]
- 25.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schloss, P. D., and J. Handelsman. 2006. Introducing SONS, a tool for operational taxonomic unit-based comparisons of microbial community memberships and structures. Appl. Environ. Microbiol. 72:6773-6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spudich, J. L., C. S. Yang, K. H. Jung, and E. N. Spudich. 2000. Retinylidene proteins: structures and functions from archaea to humans. Annu. Rev. Cell Dev. Biol. 16:365-392. [DOI] [PubMed] [Google Scholar]
- 28.Stingl, U., R. A. Desiderio, J.-C. Cho, K. L. Vergin, and S. J. Giovannoni. 2007. The SAR92 clade: an abundant coastal clade of culturable marine bacteria possessing proteorhodopsin. Appl. Environ. Microbiol. 73:2290-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 30.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y. H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 32.Zhang, J., S. M. Liu, J. L. Ren, Y. Wu, and G. L. Zhang. 2007. Nutrient gradients from the eutrophic Changjiang (Yangtze River) estuary to the oligotrophic Kuroshio waters and re-evaluation of budgets for the East China Sea shelf. Prog. Oceanogr. 74:449-478. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.