Abstract
Campylobacter jejuni is one of the most common bacterial causes of human gastroenteritis, and recent findings suggest that turkeys are an important reservoir for this organism. In this study, 80 C. jejuni isolates from eastern North Carolina were characterized for resistance to nine antimicrobials, and strain types were determined by fla typing, pulsed-field gel electrophoresis (PFGE) with SmaI and KpnI, and (for 41 isolates) multilocus sequence typing (MLST). PFGE analysis suggested that many of the isolates (37/40 [ca. 93%]) in a major genomic cluster had DNA that was partially methylated at SmaI sites. Furthermore, 12/40 (30%) of the isolates in this cluster were completely resistant to digestion by KpnI, suggesting methylation at KpnI sites. MLST of 41 isolates identified 10 sequence types (STs), of which 4 were new. Three STs (ST-1839, ST-2132 and the new ST-2934) were predominant and were detected among isolates from different farms. The majority of the isolates (74%) were resistant to three or more antimicrobials, and resistance to ciprofloxacin was common (64%), whereas resistance to the other drug of choice for treatment of human campylobacteriosis, erythromycin, was never encountered. Most (33/34) of the kanamycin-resistant isolates were also resistant to tetracycline; however, only ca. 50% of the tetracycline-resistant isolates were also kanamycin resistant. Isolates with certain antimicrobial resistance profiles had identical or closely related strain types. Overall, the findings suggest dissemination of certain clonal groups of C. jejuni isolates in the turkey production industry of this region.
Campylobacter is a leading bacterial agent of food poisoning in the United States and other industrialized nations. The majority (>90%) of campylobacteriosis cases are caused by Campylobacter jejuni, with the remainder being caused primarily by Campylobacter coli (12, 14). In addition to acute gastrointestinal symptoms, substantial disease burden due to C. jejuni infection is associated with severe sequelae, including the autoimmune-mediated demyelinating neuropathies Guillain-Barré and Miller Fisher syndromes (28, 32). Human campylobacteriosis is typically self-limited; when antibiotic treatment is suggested, the drugs of choice are ciprofloxacin (or other fluoroquinolones) and erythromycin (or other macrolides) (29, 32).
Transmission to humans mainly involves contaminated food, untreated drinking water, or raw milk (1, 12). Raw or undercooked poultry is a major risk factor for food-borne campylobacteriosis, either directly or through cross-contamination of foods that are ready to eat (3). In the United States and many other nations, significant effort has been invested in determinations of Campylobacter prevalence in broiler carcasses and in assessment of the associated public health risks (for examples, see references 7, 10, and 34).
In comparison with studies involving broilers, relatively few reports are available on C. jejuni isolates from turkeys. Studies in Europe indicated that C. jejuni was frequently isolated from turkeys preharvest or from turkey meat (4, 36, 37). Surveys in the United States have revealed that both C. jejuni and C. coli colonize turkeys, with the relative prevalence of the two species varying among different flocks (33, 39, 40) and among samples from different processing plants (23). In certain flocks, treatments such as feed withdrawal and transport also appeared to influence the C. jejuni-to-C. coli ratio (39). Production systems (organic versus conventional) may also impact this ratio, with the C. jejuni-to-C. coli ratio being higher in samples from conventionally grown turkeys than in those from birds grown organically (24). Previous surveys of five commercial turkey flocks in eastern North Carolina (2001 to 2003) revealed a high prevalence of Campylobacter isolates, primarily C. coli, in turkeys, and a high prevalence of resistance to several antimicrobials (22, 33). Resistance to several antimicrobials was also prevalent among C. coli and C. jejuni isolates from conventionally grown turkeys (24). However, strain subtyping information on C. jejuni isolates from turkeys in the United States is limited. Furthermore, possible correlations between strain types and antimicrobial susceptibility profiles of the organisms remain unidentified.
Eastern North Carolina is a major contributor to turkey production in the United States. Recently, we described the relative prevalence of Campylobacter isolates, and of C. jejuni versus C. coli, in fecal samples from turkeys grown on farms located in close proximity to swine farms in this region. C. jejuni was found to be the predominant species (isolated from 60% of the Campylobacter-positive samples), and its prevalence in samples from different flocks varied from 31% to 86% (40). In this study, 80 C. jejuni isolates from this region were characterized for susceptibility to nine antimicrobials. In addition, the isolates were subtyped using three different strain subtyping tools: fla typing, pulsed-field gel electrophoresis (PFGE), and multilocus sequence typing (MLST). These investigations were pursued to characterize the population structure of C. jejuni isolates from turkeys grown in this region and to elucidate the possible associations between strain types and antimicrobial susceptibility phenotypes of the organisms.
MATERIALS AND METHODS
Campylobacter strains and growth conditions.
The C. jejuni isolates in this study are listed in Table 1. These isolates were from the Campylobacter culture collection in our laboratory and had been isolated between 2003 and 2006 from fecal or cecal samples from turkeys grown conventionally in eastern North Carolina. Information on specific antimicrobials that may have been administered to the flocks therapeutically, or for growth promotion, was not available. The majority of these isolates (n = 63) were chosen from a larger collection of C. jejuni isolates obtained during a survey of Campylobacter colonization of turkeys in North Carolina, from 2003 to 2005 (40). The remaining 17 isolates were obtained during 2006. To maximize diversity, isolates were chosen so as to represent different flocks, farms, and antibiotic susceptibility phenotypes (based on original screens at a single concentration for each respective antimicrobial).
TABLE 1.
C. jejuni isolates characterized in this study
| Isolatea | Date | Age (wk) | Company | Farm | Antimicrobial resistanceb | Antimicrobial resistance profile | ST |
|---|---|---|---|---|---|---|---|
| 5966 | 31 Oct 2003 | 5.5 | D | 2 | AQ | 2 | 2934 |
| 5996 | 14 Nov 2003 | 5.5 | D | 13 | ATSQ | 10 | 2936 |
| 6009 | 14 Nov 2003 | 7.5 | D | 2 | ATSKQ | 12 | |
| 6013 | 14 Nov 2003 | 7.5 | D | 2 | ATKQ | 8 | |
| 6040 | 1 Dec 2003 | 8 | D | 13 | ATQ | 6 | |
| 6041 | 1 Dec 2003 | 8 | D | 13 | ATKQ | 8 | |
| 6102 | 15 Dec 2003 | 10 | D | 13 | ATSKQ | 12 | 2132 |
| 6137 | 15 Feb 2004 | 5 | D | 1 | ATSK | 11 | 2132 |
| 6161 | 19 Feb 2004 | 7 | D | 1 | ATSQ | 10 | |
| 6209 | 11 Mar 2004 | 10 | D | 1 | ATSKQ | 12 | 1162 |
| 6447 | 8 Apr 2004 | 8 | D | 8 | ATKQ | 8 | 1796 |
| 6451 | 8 Apr 2004 | 8 | D | 8 | ATSQ | 10 | |
| 6470 | 9 Apr 2004 | 7 | D | 4 | ATSK | 11 | |
| 6471 | 9 Apr 2004 | 7 | D | 4 | AT | 5 | |
| 6532 | 22 Apr 2004 | 10 | D | 8 | ATSQ | 10 | 1839 |
| 6535 | 22 Apr 2004 | 10 | D | 8 | AT | 5 | |
| 6565 | 29 Apr 2004 | 10 | D | 5 | ATS | 9 | 1839 |
| 6608 | 4 May 2004 | 11 | D | 4 | ATSQ | 10 | 1839 |
| 6628 | 6 May 2004 | 12 | D | 8 | ATSQ | 10 | 1839 |
| 6631 | 6 May 2004 | 12 | D | 8 | AT | 5 | 1839 |
| 7287 | 13 July 2004 | 8 | D | 2 | ATSQ | 10 | 1839 |
| 7290 | 13 July 2004 | 8 | D | 2 | ATS | 9 | |
| 7294 | 13 July 2004 | 8 | D | 2 | ATSK | 11 | |
| 7320 | 13 July 2004 | 8 | D | 2 | ATSQ | 10 | |
| 7812 | 5 Aug 2004 | 10 | D | 5 | ATK | 7 | |
| 7813 | 5 Aug 2004 | 10 | D | 5 | AS | 3 | 2132 |
| 7895 | 11 Aug 2004 | 12 | D | 2 | ATKQ | 8 | |
| 8092 | 20 Aug 2004 | 12 | D | 5 | ATSQ | 10 | |
| 8359 | 22 Sep 2004 | 5 | D | 3 | AT | 5 | |
| 8633 | 6 Oct 2004 | 7 | D | 3 | ATKQ | 8 | |
| 8635 | 6 Oct 2004 | 7 | D | 3 | AQ | 2 | 2934 |
| 8647 | 6 Oct 2004 | 7 | D | 3 | ATSKQ | 12 | |
| 8665 | 6 Oct 2004 | 7 | D | 3 | A | 1 | 1838 |
| 8803 | 23 Oct 2004 | 7 | D | 12 | ATSKQ | 12 | |
| 8828 | 23 Oct 2004 | 7 | D | 12 | ATKQ | 8 | |
| 8865 | 27 Oct 2004 | 10 | D | 3 | ATKQ | 8 | |
| 8878 | 27 Oct 2004 | 10 | D | 3 | ATK | 7 | |
| 8904 | 27 Oct 2004 | 10 | D | 3 | A | 1 | 1838 |
| 8950 | 10 Nov 2004 | 10 | D | 12 | ATQ | 6 | |
| 8953 | 10 Nov 2004 | 10 | D | 12 | ATSK | 11 | |
| 8954 | 10 Nov 2004 | 10 | D | 12 | ATSQ | 10 | |
| 8959 | 10 Nov 2004 | 10 | D | 12 | ATS | 9 | |
| 9044 | 17 Nov 2004 | 13 | D | 3 | ATSK | 11 | |
| 9355 | 30 Dec 2004 | 6 | D | 12 | AS | 3 | 2132 |
| 9585 | 1 Feb 2005 | 7 | D | 3 | AT | 5 | |
| 9669 | 2 Feb 2005 | 11 | D | 12 | ATSKQ | 12 | |
| 9778 | 23 Feb 2005 | 14 | D | 12 | ASKQ | 4 | |
| 9838 | 24 Feb 2005 | 10 | D | 3 | ATSQ | 10 | |
| 9850 | 24 Feb 2005 | 10 | D | 3 | AS | 3 | 2132 |
| 10287 | 6 Jul 2005 | 7 | D | 1 | ATSKQ | 12 | |
| 10296 | 6 Jul 2005 | 7 | D | 1 | AQ | 2 | |
| 10297 | 6 Jul 2005 | 7 | D | 1 | AQ | 2 | 2934 |
| 10306 | 6 Jul 2005 | 7 | D | 1 | AS | 3 | 2132 |
| 10328 | 6 Jul 2005 | 7 | D | 1 | ATSK | 11 | |
| 10371 | 26 Jul 2005 | 10 | D | 1 | ATSQ | 10 | 2936 |
| 10882 | 9 Sep 2005 | 8 | D | 4 | ATSKQ | 12 | 1939 |
| 10884 | 9 Sep 2005 | 8 | D | 4 | ATSQ | 10 | |
| 10888 | 9 Sep 2005 | 8 | D | 4 | ATS | 9 | |
| 10977 | 23 Sep 2005 | 10 | D | 4 | AQ | 2 | 2934 |
| 11055 | 23 Sep 2005 | 10 | D | 8 | ATSQ | 10 | |
| 11211 | 17 Oct 2005 | 14 | D | 4 | AS | 3 | 2132 |
| 11222 | 17 Oct 2005 | 14 | D | 8 | ATSKQ | 12 | |
| 11250 | 17 Oct 2005 | 14 | D | 8 | AQ | 2 | |
| 11599 | 26 Sep 2006 | 12 | A | 6 | ATSKQ | 12 | 1839 |
| 11600 | 26 Sep 2006 | 12 | A | 6 | ATSKQ | 12 | 1839 |
| 11603 | 26 Sep 2006 | 11 | A | 17 | ATQ | 6 | 1839 |
| 11607 | 26 Sep 2006 | 13 | A | 7 | ATSK | 11 | 2132 |
| 11618 | 6 Oct 2006 | 8 | D | 10 | T | 13 | 1698 |
| 11619 | 6 Oct 2006 | 8 | D | 10 | ATKQ | 8 | 2934 |
| 11640 | 20 Oct 2006 | 10 | D | 11 | ATKQ | 8 | 2934 |
| 11599MD | 26 Sep 2006 | 12 | A | 6 | ATSK | 11 | 2132 |
| 11600MD | 26 Sep 2006 | 12 | A | 6 | ATSKQ | 12 | 2132 |
| 11601MD | 26 Sep 2006 | 12 | A | 6 | ATKQ | 8 | 2934 |
| 11618MD | 6 Oct 2006 | 8 | D | 10 | ATSQ | 10 | 2937 |
| 11640MD | 20 Oct 2006 | 10 | D | 11 | T | 13 | 1698 |
| SC1453 | 13 Dec 2006 | 3 | A | 9 | A | 1 | 2934 |
| SC1471 | 13 Dec 2006 | 3.5 | A | 15 | ATSKQ | 12 | 1839 |
| SC1486 | 13 Dec 2006 | 5 | B | 18 | ATSQ | 10 | 2935 |
| SC1527 | 14 Dec 2006 | 2.5 | C | 14 | ATQ | 6 | 1839 |
| SC1532 | 14 Dec 2006 | 7.5 | C | 16 | ATSQ | 10 | 2936 |
Isolates in boldface type were a subset of the C. jejuni isolate collection obtained from a previously described survey (40).
A, ampicillin; K, kanamycin; S, streptomycin; T, tetracycline; Q, (fluoro)quinolone (nalidixic acid and ciprofloxacin).
Bacteria were grown as described previously (33) on tryptic soy agar with 5% sheep blood (Remel, Lenexa, KS) at 42°C for 48 h under microaerobic conditions and preserved at −80°C in cryovials containing brain heart infusion medium (Difco, Sparks, MD) with 20% glycerol.
Antibiotic susceptibility and MIC determinations.
MICs were determined for the following nine antimicrobial agents: ampicillin, chloramphenicol, ciprofloxacin, erythromycin, gentamicin, kanamycin, nalidixic acid, streptomycin, and tetracycline. Antimicrobials were purchased from Sigma Chemical Co. (St. Louis, MO), except for gentamicin, kanamycin, nalidixic acid, and ciprofloxacin, which were purchased from Fisher Biotech (Fair Lawn, NJ). The dilution range used for all tested antimicrobial agents was from 0.5 to 256 μg/ml (Table 2). MICs were determined using the agar dilution method as described previously (24). C. jejuni ATCC 33560 (purchased from the American Type Culture Collection) was used as the quality control strain. The plates were incubated at 42°C for 48 h under microaerobic conditions, and the MIC was defined as the lowest concentration of an antimicrobial agent that inhibited growth on the plates. The quality control strain and all tested isolates were also grown on Mueller-Hinton agar plates (Becton Dickinson, Sparks, MD) without antimicrobials to ensure viability. The MIC results of C. jejuni ATCC 33560 were within a 3-dilution range for ampicillin, kanamycin, nalidixic acid, and chloramphenicol, for which quality control ranges are not currently available. The resistance breakpoints for all antimicrobials (except for chloramphenicol and streptomycin) were those described previously (24). For chloramphenicol, the resistance breakpoint was that used for Enterobacteriaceae, as recommended by the Clinical and Laboratory Standards Institute (8). The resistance breakpoint used for streptomycin was ≥64 μg/ml, as described previously (17) (Table 2).
TABLE 2.
MIC distributions and resistance rates of C. jejuni isolates from turkey farms
| Antimicrobial | No. of isolates inhibited at indicated concn (μg/ml)a
|
MIC50/MIC90(μg/ml) | No. (%) of resistant isolates | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | >256 | |||
| Ampicillin | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 76 | >256/>256 | 78 (97.5) |
| Chloramphenicol | 2 | 5 | 32 | 36 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 4/4 | 0 |
| Ciprofloxacin | 27 | 1 | 1 | 0 | 1 | 8 | 29 | 11 | 2 | 0 | 0 | 32/64 | 51 (63.75) |
| Erythromycin | 30 | 31 | 18 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1/2 | 0 |
| Gentamicin | 32 | 45 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1/1 | 0 |
| Kanamycin | 0 | 0 | 1 | 8 | 29 | 7 | 0 | 1 | 0 | 1 | 33 | 16/>256 | 34 (42.5) |
| Nalidixic acid | 0 | 0 | 2 | 12 | 13 | 2 | 0 | 15 | 25 | 7 | 4 | 64/256 | 51 (63.75) |
| Streptomycin | 1 | 8 | 11 | 2 | 0 | 2 | 1 | 7 | 9 | 3 | 36 | 128/>256 | 48 (60) |
| Tetracycline | 2 | 3 | 8 | 1 | 1 | 0 | 2 | 5 | 28 | 21 | 9 | 128/>256 | 65 (81.25) |
Boldface numbers of isolates indicate that the corresponding MIC was the resistance breakpoint.
DNA extractions, species confirmation, fla typing, PFGE, and MLST.
Genomic DNA was extracted using the Qiagen DNeasy kit (Qiagen Inc., Valencia, CA) according to the manufacturer's protocol. To confirm that the isolates were C. jejuni, hip primers were used in PCR, as described previously (33). PFGE was done using SmaI and KpnI (New England Biolabs) following the PulseNet protocol (http://www.cdc.gov/PULSENET/protocols.htm), with a few minor modifications as described by D'lima et al. (9), and fla typing was performed as described previously (33). Tagged image file format images of banding patterns resulting from fla typing and PFGE were analyzed by BioNumerics (version 4.6; Applied Maths, Saint-Marten-Latem, Belgium). Band position tolerance using the Dice coefficient was set at 1%, and clustering was performed using the unweighted-pair group method with arithmetic averages, as described previously (9). For MLST, we employed previously described primer sets for seven housekeeping genes (aspA, atpA, glnA, gltA, glyA, pgm, and tkt), and PCR conditions were as described previously (27) with minor modifications (95°C for 45 s, 53°C for 45 s, and 72°C for 2 min, over 32 cycles). Amplicons were sequenced at the Genome Research Laboratory of North Carolina State University, and at Davis Sequencing (Davis, CA). Sequence type (ST) identifications were done using the C. jejuni/C. coli MLST database (http://pubmlst.org/campylobacter), and new sequences were assigned new allele numbers and STs as described previously (26). Putative phylogenetic relationships among the STs were presented using a minimum spanning tree constructed with BioNumerics (version 4.6), as described previously (9). The numerical index of discrimination (D) was determined as described previously (18).
RESULTS
Antimicrobial susceptibility phenotypes and MICs.
MIC determinations revealed that all 80 C. jejuni isolates were susceptible to erythromycin, chloramphenicol, and gentamicin. There was always agreement between nalidixic acid and ciprofloxacin resistances, and since they have the same mechanism of action, the results for these two antimicrobials are reported here as a single class. No pansensitive isolates were identified. Only five isolates were resistant to a single antimicrobial; SC1453, 8665, and 8904 were resistant only to ampicillin, whereas 11618 and 11640MD were resistant only to tetracycline (Table 1). Resistance to four or five antimicrobials was encountered among 36 (45%) and 13 (16.3%) isolates, respectively (Tables 1 and 3). Thus, even though a total of 13 different antimicrobial resistance profiles were identified among the 80 C. jejuni isolates, multidrug resistance profiles (resistance to three or more antimicrobials, with nalidixic acid and ciprofloxacin counted as one) were found in the majority (ca. 74%) of the isolates (Tables 1 and 3).
TABLE 3.
Strain subtypes of isolates with specific antimicrobial resistance profilesd
| Antimicrobial resistance profile no. | Antimicrobial resistance patterna | No. of isolates | ST(s)b | Subtype cluster (no. of isolates)c |
|---|---|---|---|---|
| 1 | A | 3 | 1828 (2/3), 2934 (1/3) | A (3) |
| 2 | AQ | 6 | 2934 (4/4) | A (6) |
| 3 | AS | 5 | 2132 (5/5) | C (5) |
| 4 | ASKQ | 1 | ND | B (1) |
| 5 | AT | 5 | 1839 (1/1) | A (1), B (4) |
| 6 | ATQ | 4 | 1839 (2/2) | A (1), B (3) |
| 7 | ATK | 2 | ND | A (1), B (1) |
| 8 | ATKQ | 10 | 2934 (3/4), 1796 (1/4) | A (9), B (1) |
| 9 | ATS | 4 | 1839 (1/1) | B (4) |
| 10 | ATSQ | 17 | 1839 (4/9), 2935 (1/9), 2936 (3/9), 2937 (1/9) | A (1), B (15), C (1) |
| 11 | ATSK | 8 | 2132 (3/3) | B (2), C (6) |
| 12 | ATSKQ | 13 | 1839 (4/7), 2132 (2/7), 1162 (1/7) | B (9), C (4) |
| 13 | T | 2 | 1698 (2/2) | C (2) |
A, ampicillin; K, kanamycin; S, streptomycin; T, tetracycline; Q, (fluoro)quinolone (nalidixic acid and ciprofloxacin).
Data in parentheses represent the number of isolates with the ST out of the total number of isolates. ND, not determined.
Based on fla typing and PFGE profiles (Fig. 1).
For the 13 antimicrobial resistance profiles, there were totals of 13 patterns, 80 isolates, 10 STs, 22 subtype cluster A isolates, 40 subtype cluster B isolates, and 18 subtype cluster C isolates.
When the prevalence of resistance to individual antimicrobials was assessed, we noted high prevalence (60% to ca. 98%) of resistance to ampicillin, tetracycline, streptomycin, and nalidixic acid/ciprofloxacin (Table 2). There was a wide range of MICs for all of the antimicrobials, especially tetracycline and streptomycin, for which the widest range (32 to >256 μg/ml) was observed (Table 2). MICs exceeding the maximum of the tested range (>256 μg/ml) were noted in the majority of the isolates resistant to ampicillin (76/78 [97.4%]), kanamycin (33/34 [97.1%]), or streptomycin (36/48 [75%]) (Table 2).
Association between resistance to kanamycin and resistance to tetracycline.
A strong correlation was found between kanamycin resistance and resistance to tetracycline. Of the 34 kanamycin-resistant isolates, all but one (strain 9778) were also resistant to tetracycline. The reverse, however, was not true. Only ca. 50% of the tetracycline-resistant isolates were also kanamycin resistant (Tables 1 and 3).
Strain subtyping reveals three predominant clusters.
All 80 isolates were subtyped by fla typing and PFGE using SmaI and KpnI (Fig. 1). In addition, a subset of 41 of the 80 isolates was chosen for analysis by MLST. These 41 isolates were chosen to represent different subtypes based on fla typing and PFGE profiles, as well as diverse antimicrobial susceptibility profiles. The resulting numerical indices of discrimination (D) for fla typing, PFGE with SmaI, PFGE with KpnI, and MLST, each used separately, were 0.97, 0.969, 0.958, and 0.824, respectively. Higher D values were obtained when the subtyping schemes were combined; D for the combination of fla typing and PFGE (with both SmaI and KpnI) was 0.996 and was not appreciably changed (D = 0.995, for the subset of 41 isolates that was typed by MLST) when MLST subtyping data were also included.
FIG. 1.
Dendrogram depicting fla and PFGE (SmaI and KpnI) profiles of the 80 isolates investigated in this study. Cluster analysis of the patterns was performed by BioNumerics (version 4.6; Applied Maths). The arrow indicates a large undigested DNA band observed in isolates incompletely digested with SmaI. A, ampicillin; T, tetracycline; S, streptomycin; E, erythromycin; K, kanamycin; Q, (fluoro)quinolones (nalidixic acid and ciprofloxacin).
PFGE analysis revealed that 14 isolates (18%) failed to cut with KpnI, in spite of repeated efforts, and therefore, band-based subtyping analysis of these isolates was based on fla typing and PFGE using SmaI (Fig. 1). Bionumerics-based cluster analysis of the compiled fla types and PFGE profiles identified 69 distinct strain subtypes and yielded three major clusters of isolates (with overall identity within each cluster being >55%), designated A (22 isolates), B (40 isolates), and C (18 isolates). The isolates in the major clusters were derived from several different farms and time points during the 2003 to 2006 period (Fig. 1 and Table 1). Only a few were identical with both fla typing and PFGE. Eight pairs of isolates (three in cluster A and five in cluster B) exhibited identical fla and PFGE subtypes, with five of these eight subtypes involving organisms isolated at different times from the same farm (Fig. 1). The only subtype with more than two isolates consisted of four isolates (11600, 11603, 8092, and 9585) from four different farms and three different times (Fig. 1 and Table 1).
MLST analysis of 41 of the isolates identified 10 STs, of which four (2934 to 2937) were new. Three of the 10 STs were predominant, detected in 73% of the isolates, as follows: ST-2934 (n = 8), ST-2132 (n = 10), and ST-1839 (n = 12). The apparent dissemination of these three STs was further evidenced by the fact that each was found in isolates from different farms and from birds of different ages (Table 1). The remaining seven STs were represented by one to three isolates each (Fig. 2).
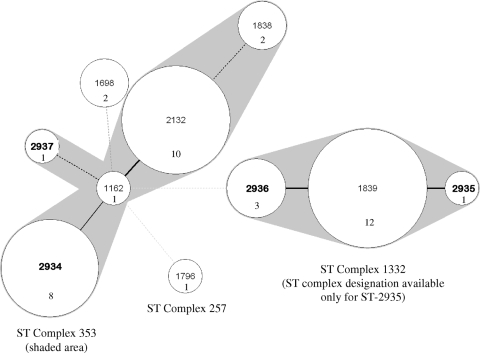
FIG. 2.
Minimum spanning tree depicting the clustering of 10 STs identified among 41 C. jejuni isolates. The tree was created using BioNumerics (version 4.6; Applied Maths). Each ST is represented by a circle, and the size of each circle is proportional to the number of isolates that comprise that ST. Thick, short lines indicate a single allele difference; thin, longer lines indicate two allele differences; and dashed lines represent three (black) or more (gray) allele differences. New STs are in boldface type. The shaded area depicts clusters of highly related STs. ST complex designations were obtained from http://pubmlst.org/campylobacter. ST complex designations were not available for STs 1698, 2936, and 1839.
Construction of a minimum spanning tree to evaluate the relatedness among the 10 STs revealed two clusters, one including two predominant STs (ST-2132 and ST-2934) and the other including predominant ST-1839 and closely related ST-2935 and ST-2936 (Fig. 2). ST-2935 was the only ST in this cluster identified as belonging to a greater ST complex, specifically 1332. The second cluster was primarily comprised of STs belonging to ST complex 353 (STs 2934, 1162, 2132, 1838, and 2937) whereas distantly related ST-1796 was included in ST complex 257 (Fig. 2). An ST complex designation was not available for ST-1698 (http://pubmlst.org/campylobacter).
Isolates with the same STs were grouped into the same fla typing- and PFGE-based clusters (Fig. 1), with one exception; isolate 6102 (ST-2132) was placed in cluster B, whereas all other isolates with ST-2132 were in cluster C. Furthermore, isolates with closely related STs (e.g., 1839, 2935, and 2936) had related fla and PFGE fingerprints. Interestingly, even though STs 1162, 2132, and 2934 were closely related based on MLST (Fig. 2), they were placed into two different clusters based on fla typing and PFGE (Fig. 1).
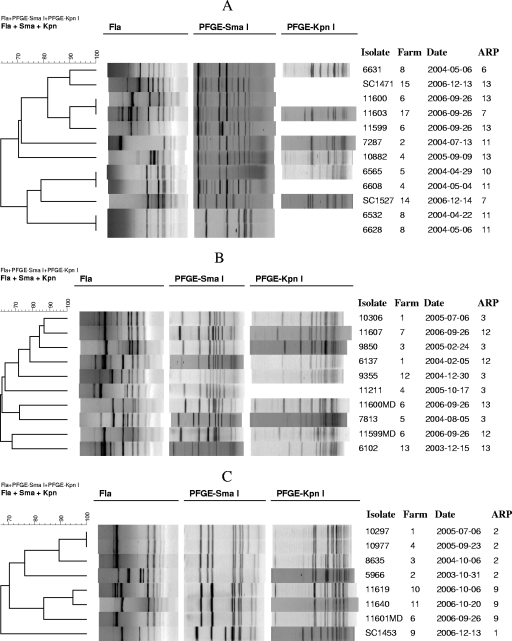
Within each of the three predominant STs (ST-1839, ST-2132, ST-2934), several related strain subtypes could be identified with the combination of fla typing and PFGE (Fig. 3A to C). In several instances, isolates with the same ST and with identical PFGE patterns were differentiated by distinct fla types (e.g., 7287 and 10882 in Fig. 3A, 5966 and 8635 in Fig. 3C). Different antimicrobial profiles were noted for strains with the same ST, especially in the case of ST-1839 (five profiles) (Fig. 3A).
FIG. 3.
fla and PFGE (SmaI and KpnI) diversity among isolates with the same ST. fla-PFGE subtypes of isolates with ST-1839 (A), ST-2132 (B), and ST-2934 (C). ARP refers to antimicrobial resistance patterns (Table 3).
PFGE data suggest DNA methylation among many isolates, mostly in cluster B.
In repeated trials, PFGE with SmaI suggested incomplete digestion of genomic DNA of 39 isolates, characterized by the appearance of multiple weak bands in the background between the major bands and a prominent high-molecular-weight band (ca. 1 Mb) at the top of each lane (Fig. 1). Furthermore, 14 isolates were completely resistant to digestion by KpnI and thus untypeable with this enzyme (Fig. 1). Digestion of these isolates with KpnI yielded only a large band of presumably undigested DNA, and the corresponding wells in the PFGE gels were unusually bright, also suggesting undigested DNA (see Fig. S1 in the supplemental material).
The majority (37/39 [ca. 95%]) of the isolates with DNA partially digested with SmaI were in cluster B, and conversely, most (37/40 [ca. 93%]) of the cluster B isolates had such partially digested DNA. When known, the STs of such isolates were 1839 (12 isolates), 2936 (two isolates), and 2935 (one isolate). Three cluster B isolates (6532, 6628, and 6451) had DNA that could be completely digested with SmaI; all three of these isolates had the same fla and SmaI PFGE profiles, and two were of ST-1839 (ST of the third isolate was not determined) (Fig. 1). The majority (12/14 [86%]) of the KpnI-untypeable isolates were also in cluster B, and several (9/14) had DNA that was incompletely digested with SmaI (Fig. 1).
Antimicrobial susceptibility profiles and strain subtypes.
Strains with some of the predominant antimicrobial resistance profiles tended to be found in the same cluster on the dendrogram constructed using fla typing and PFGE data. For instance, 15 of the 17 isolates with resistance profile 10 (ATSQ), and 9 of the 13 isolates with resistance profile 12 (ATSKQ), were placed in cluster B; isolates with less-predominant antimicrobial resistance profiles also tended to be in the same fla-PFGE cluster (Table 3). However, a bidirectional association between specific fla-PFGE fingerprints and antimicrobial resistance profiles was not observed except in the case of the two isolates with profile 13 (resistance to only tetracycline) and the six isolates with profile 2 (AQ), which had related fingerprints (≥70% similarity) (Fig. 1).
Similar findings were obtained with MLST. For instance, most (8/9) of the MLST-typed isolates with antimicrobial resistance profile 10 (ATSQ) had closely related STs: ST-1839 (n = 4), ST-2935 (n = 1), and ST-2936 (n = 3). As mentioned earlier, ST-2935 and ST-2936 were closely related to ST-1839 (one allele difference each) (Fig. 2). All tested isolates with resistance profile 2 (AQ) had ST-2934, and all (of those tested) with profile 3 (AS) had ST-2132 (Table 3). However, bidirectional agreement between ST and resistance profile was detected only in the case of ST-1698, which was exclusively found in the two isolates with relatively rare resistance profile 13 (resistance to tetracycline only) (Table 3). These two isolates also harbored identical fla-PFGE fingerprints (Fig. 1). However, the availability of only two isolates with this resistance profile does not permit conclusions as to possible associations between ST-1698 and this resistance profile.
DISCUSSION
In the current study, we analyzed population structure and resistance to several antimicrobials among 80 isolates of C. jejuni from eastern North Carolina. None of the isolates were found to be resistant to chloramphenicol, gentamicin, or erythromycin. In contrast, we observed a high prevalence of resistance to ampicillin, tetracycline, streptomycin, kanamycin, nalidixic acid and ciprofloxacin, and most isolates were resistant to multiple (three or more) antimicrobials. Prevalence of resistance to ampicillin was noticeably higher in the current study than previously reported in conventionally grown turkeys (97.5% versus 12.5%) (24). Furthermore, none of the 80 isolates that we investigated were resistant to erythromycin, in contrast to the erythromycin resistance prevalence of 19.2% reported previously (24). MIC distributions in the current study also showed several differences from those previously reported (24), especially in regard to ampicillin and tetracycline, for which MICs were noticeably higher among our isolates. Such differences could be caused by a variety of factors, including region, time period, and farm practices related to antimicrobial use.
Investigation of resistance profiles showed that most isolates resistant to kanamycin were also resistant to tetracycline, although the reverse was not the case. In C. jejuni, tetracycline resistance is typically conferred by plasmid-associated tet(O) (13, 35), and genetic determinants for resistance to kanamycin and tetracycline can be cotransferred via conjugation (19, 21). Isolates that were resistant both to kanamycin and to tetracycline had diverse STs and fla-PFGE types, as did those in which resistance to tetracycline was not accompanied by kanamycin resistance. Such findings suggest that plasmids harboring a kanamycin resistance determinant as well as tet(O) may have disseminated among C. jejuni strains of different genetic backgrounds. It will be of interest to determine whether C. coli from turkeys may also exhibit the observed correlation between resistance to kanamycin and resistance to tetracycline.
The combination of fla typing and PFGE identified distinct subtypes among isolates of the same ST, which is consistent with findings reported by Bae et al. (5). These findings indicate the usefulness of MLST for detection of specific clonal groups, the population structure of which can then be characterized at higher resolution with fla typing and PFGE. Similar findings were described with multidrug-resistant clonal groups in C. coli isolates from turkeys (9).
In the case of C. jejuni isolates from turkeys, MLST may be especially valuable, because numerous isolates exhibited partial digestion with SmaI, and several also had DNA that could not be digested at all by KpnI, suggesting the presence of methylases. Interestingly, the putative methylases were largely a characteristic of one clonal group (fla-PFGE cluster B). The identification of a substantial number of isolates that were completely refractory to digestion with KpnI points to limitations concerning the use of this enzyme for PFGE of C. jejuni isolates from turkeys. The biological significance of DNA methylation such as that suggested by our data (partial methylation at SmaI sites and complete methylation at KpnI sites) remains to be investigated.
Of the 10 STs identified among the isolates that were subtyped by MLST in this study, three (ST-1839, ST-1162, and ST-1838) were previously reported only in turkey isolates. On the other hand, ST-1698 was encountered in an isolate from a chicken, and STs 1796 and 2132 were detected in isolates from human cases of illness (http://www.pubmlst.org/campylobacter). Further studies are needed to assess possible host associations of some of the STs (e.g., ST-1839 with turkeys).
We identified several isolates that were derived from different farms and had identical STs and either identical or closely related fla-PFGE patterns. Such findings suggest the dissemination of specific clonal groups among different farms in the turkey industry of the region. Possible vehicles for strain dissemination may include insects, other animals, groundwater, or human traffic (6, 15, 30, 31). Clonal groups were also identified among multidrug-resistant C. coli isolates from turkeys in this region (9, 22), as well as among C. coli and C. jejuni isolates from cattle and other meat animals (2, 5, 11, 20).
Several isolates with the same ST and identical or closely related PFGE patterns (with SmaI and KpnI) were found to have different fla types. Diversity in fla types among isolates with identical PFGE patterns was also described earlier in a study of C. coli isolates from successive turkey flocks at the same farm (22). The observed diversity in fla types among apparently related isolates likely reflects genetic instability involving the flagellin locus, described in several studies (16, 25, 38), and may contribute to antigenic diversity and host evasion strategies of the bacteria.
Isolates with certain antimicrobial resistance profiles were found to have the same ST and belonged to the same fla-PFGE cluster. On the other hand, isolates with identical or closely related strain types often differed in antimicrobial resistance profiles. The latter types of isolates may have shared a common ancestor and acquired (or lost) determinants related to antimicrobial resistance, possibly in response to selection pressures related to exposure to relevant antimicrobial agents. Isolates with ST-2934 and antimicrobial resistance profiles 2 (AQ) and 8 (ATKQ) may represent an example of such a scenario. Such isolates had identical or closely related fla-PFGE patterns, and it is conceivable that the differences in their antimicrobial resistance profiles could reflect the presence or absence of a plasmid conferring resistance to both tetracycline and kanamycin and not detectable by PFGE subtyping. As discussed earlier, resistance to both kanamycin and tetracycline has been reported to be plasmid mediated in Campylobacter (19, 21).
In conclusion, the combined use of different subtyping schemes (fla typing, PFGE, MLST) has revealed distinct clonal groups of C. jejuni colonizing conventionally grown turkeys in eastern North Carolina, a major turkey production region in the United States. Although isolates of certain antimicrobial resistance profiles had identical or closely related strain types, strain subtype data generally did not allow the prediction of antimicrobial resistance profiles. Further studies are needed to determine whether genotypes and clonal groups of the isolates analyzed in this study are representative of those in other turkey-growing regions and to assess the extent to which C. jejuni strains and clonal groups predominant in turkeys may also be found in other animal hosts.
Supplementary Material
Acknowledgments
The project was supported in part by the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service, grant number 2003-0299.
This publication made use of the Campylobacter jejuni MLST website (http://pubmlst.org/campylobacter/) developed by Keith Jolley and Man-Suen Chan and sited at the University of Oxford. The development of this site has been funded by the Wellcome Trust.
Footnotes
Published ahead of print on 21 November 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Adak, G. K., J. M. Cowden, S. Nicholas, and H. S. Evans. 1995. The Public Health Laboratory Service national case-control study of primary indigenous sporadic cases of campylobacter infection. Epidemiol. Infect. 115:15-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adhikari, B., J. H. Connolly, P. Madie, and P. R. Davies. 2004. Prevalence and clonal diversity of Campylobacter jejuni from dairy farms and urban sources. N. Z. Vet. J. 52:378-383. [DOI] [PubMed] [Google Scholar]
- 3.Altekruse, S. F., and L. K. Tollefson. 2003. Human campylobacteriosis: a challenge to the veterinary profession. J. Am. Vet. Med. Assoc. 223:445-451. [DOI] [PubMed] [Google Scholar]
- 4.Atanassova, V., F. Reich, L. Beckmann, and G. Klein. 2007. Prevalence of Campylobacter spp. in turkey meat from a slaughterhouse and in turkey meat retail products. FEMS Immunol. Med. Microbiol. 49:141-145. [DOI] [PubMed] [Google Scholar]
- 5.Bae, W., D. D. Hancock, D. R. Call, Y. H. Park, A. C. Berge, R. M. Finger, W. M. Sischo, and T. E. Besser. 2007. Dissemination of antimicrobial resistant strains of Campylobacter coli and Campylobacter jejuni among cattle in Washington State and California. Vet. Microbiol. 122:306-315. [DOI] [PubMed] [Google Scholar]
- 6.Bates, C., K. L. Hiett, and N. J. Stern. 2004. Relationship of Campylobacter isolated from poultry and from darkling beetles in New Zealand. Avian Dis. 48:138-147. [DOI] [PubMed] [Google Scholar]
- 7.Bull, S. A., V. M. Allen, G. Domingue, F. Jorgensen, J. A. Frost, R. Ure, R. Whyte, D. Tinker, J. E. Corry, J. Gillard-King, and T. J. Humphrey. 2006. Sources of Campylobacter spp. colonizing housed broiler flocks during rearing. Appl. Environ. Microbiol. 72:645-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2007. Performance standards for antimicrobial susceptibility testing; 17th informational supplement. CLSI document M100-S17. Clinical and Laboratory Standards Institute, Wayne, PA.
- 9.D'lima, C. B., W. G. Miller, R. E. Mandrell, S. L. Wright, R. M. Siletzky, D. K. Carver, and S. Kathariou. 2007. Clonal population structure and specific genotypes of multidrug-resistant Campylobacter coli from turkeys. App. Environ. Microbiol. 73:2156-2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans, S. J., and A. R. Sayers. 2000. A longitudinal study of campylobacter infection of broiler flocks in Great Britain. Prev. Vet. Med. 46:209-223. [DOI] [PubMed] [Google Scholar]
- 11.Fitzgerald, C., K. Stanley, S. Andrew, and K. Jones. 2001. Use of pulsed-field gel electrophoresis and flagellin gene typing in identifying clonal groups of Campylobacter jejuni and Campylobacter coli in farm and clinical environments. Appl. Environ. Microbiol. 67:1429-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedman, C. R., J. Neimann, H. C. Wegener, and R. V. Tauxe. 2000. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations, p. 121-138. In I. Nachamkin and M. Blaster (ed.), Campylobacter, 2nd ed. American Society for Microbiology, Washington, DC.
- 13.Gibreel, A., D. M. Tracz, L. Nonaka, T. M. Ngo, S. R. Connell, and D. E. Taylor. 2004. Incidence of antibiotic resistance in Campylobacter jejuni isolated in Alberta, Canada, from 1999 to 2002, with special reference to tet(O)-mediated tetracycline resistance. Antimicrob. Agents Chemother. 48:3442-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillespie, I. A., S. J. O'Brien, J. A. Frost, G. K. Adak, P. Horby, A. V. Swan, M. J. Painter, and K. R. Neal. 2002. A case-case comparison of Campylobacter coli and Campylobacter jejuni infection: a tool for generating hypotheses. Emerg. Infect. Dis. 8:937-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hänninen, M. L., M. Niskanen, and L. Korhonen. 1998. Water as a reservoir for Campylobacter jejuni infection in cows studied by serotyping and pulsed-field gel electrophoresis (PFGE). Zentralbl. Veterinaermed. B 45:37-42. [DOI] [PubMed] [Google Scholar]
- 16.Harrington, C. S., F. M. Thomson-Carter, and P. E. Carter. 1997. Evidence for recombination in the flagellin locus of Campylobacter jejuni: implications for the flagellin gene typing scheme. J. Clin. Microbiol. 35:2386-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heuer, O. E., K. Pedersen, J. S. Andersen, and M. Madsen. 2001. Prevalence and antimicrobial susceptibility of thermophilic Campylobacter in organic and conventional broiler flocks. Lett. Appl. Microbiol. 33:269-274. [DOI] [PubMed] [Google Scholar]
- 18.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kotarski, S. F., T. L. Merriwether, G. T. Tkalcevic, and P. Gemski. 1986. Genetic studies of kanamycin resistance in Campylobacter jejuni. Antimicrob. Agents Chemother. 30:225-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwan, P. S., A. Birtles, F. J. Bolton, N. P. French, S. E. Robinson, L. S. Newbold, M. Upton, and A. J. Fox. 2008. Longitudinal study of the molecular epidemiology of Campylobacter jejuni in cattle on dairy farms. Appl. Environ. Microbiol. 74:3626-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lambert, T., G. Gerbaud, P. Trieu-Cuot, and P. Courvalin. 1985. Structural relationship between the genes encoding 3′-aminoglycoside phosphotransferase in Campylobacter and in gram-positive cocci. Ann. Inst. Pasteur Microbiol. 136:135-150. [DOI] [PubMed] [Google Scholar]
- 22.Lee, B. C., N. Reimers, H. J. Barnes, C. D'lima, D. Carver, and S. Kathariou. 2005. Strain persistence and fluctuation of multiple-antibiotic resistant Campylobacter coli colonizing turkeys over successive production cycles. Foodborne Pathog. Dis. 2:103-110. [DOI] [PubMed] [Google Scholar]
- 23.Logue, C. M., J. S. Sherwood, L. M. Elijah, P. A. Olah, and M. R. Dockter. 2003. The incidence of Campylobacter spp. on processed turkey from processing plants in the midwestern United States. J. Appl. Microbiol. 95:234-241. [DOI] [PubMed] [Google Scholar]
- 24.Luangtongkum, T., T. Y. Morishita, A. J. Ison, S. Huang, P. E. McDermott, and Q. Zhang. 2006. Effect of conventional and organic production practices on the prevalence and antimicrobial resistance of Campylobacter spp. in poultry. Appl. Environ. Microbiol. 72:3600-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meinersmann, R. J., R. W. Phillips, K. L. Hiett, and P. Fedorka-Cray. 2005. Differentiation of Campylobacter populations as demonstrated by flagellin short variable region sequences. Appl. Environ. Microbiol. 71:6368-6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, W. G., M. D. Englen, S. Kathariou, I. V. Wesley, G. Wang, L. Pittenger-Alley, R. M. Siletzky, W. Muraoka, P. J. Fedorka-Cray, and R. E. Mandrell. 2006. Identification of host-associated alleles by multilocus sequence typing of Campylobacter coli strains from food animals. Microbiology 152:245-255. [DOI] [PubMed] [Google Scholar]
- 27.Miller, W. G., S. L. W. On, G. Wang, S. Fontanoz, A. J. Lastovica, and R. E. Mandrell. 2005a. Extended multilocus sequence typing system for Campylobacter coli, C. lari, C. upsaliensis, and C. helveticus. J. Clin. Microbiol. 43:2315-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nachamkin, I., B. M. Allos, and T. Ho. 1998. Campylobacter species and Guillain-Barré syndrome. Clin. Microbiol. Rev. 11:555-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nachamkin, I., J. Engberg, and F. M. Aarestrup. 2000. Diagnosis and antimicrobial susceptibility of Campylobacter species, p. 45-66. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. American Society for Microbiology, Washington, DC.
- 30.Newell, D. G., and C. Fearnley. 2003. Sources of Campylobacter colonization in broiler chickens. Appl. Environ. Microbiol. 69:4343-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pacha, R. E., G. W. Clark, E. A. Williams, A. W. Carter, J. J. Scheffelmaier, and P. Debusschere. 1987. Small rodents and other mammals associated with mountain meadows as reservoirs of Giardia spp. and Campylobacter spp. Appl. Environ. Microbiol. 53:1574-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skirrow, M. B., and M. J. Blaser. 2000. Clinical aspects of Campylobacter infection, p. 69-88. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. American Society for Microbiology, Washington, DC.
- 33.Smith, K., N. Reimers, H. J. Barnes, B. C. Lee, R. Siletzky, and S. Kathariou. 2004. Campylobacter colonization of sibling turkey flocks reared under different management conditions. J. Food Prot. 67:1463-1468. [DOI] [PubMed] [Google Scholar]
- 34.Stern, N. J., and S. Pretanik. 2006. Counts of Campylobacter spp. on U. S. broiler carcasses. J. Food Prot. 69:1034-1039. [DOI] [PubMed] [Google Scholar]
- 35.Taylor, D. E., and P. Courvalin. 1988. Mechanisms of antibiotic resistance in Campylobacter species. Antimicrob. Agents Chemother. 32:1107-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Looveren, M., G. Daube, L. De Zutter, J.-M. Dumnon, C. Lammens, M. Wijdooghe, P. Vandamme, M. Jouret, M. Cornelis, and H. Goossens. 2001. Antimicrobial susceptibilities of Campylobacter strains isolated from food animals in Belgium. J. Antimicrob. Chemother. 48:235-240. [DOI] [PubMed] [Google Scholar]
- 37.Wallace, J. S., K. N. Stanley, and K. Jones. 1998. The colonization of turkeys by thermophilic campylobacters. J. Appl. Microbiol. 85:224-230. [DOI] [PubMed] [Google Scholar]
- 38.Wassenaar, T. M., B. N. Fry, and B. A. van der Zeijst. 1995. Variation of the flagellin gene locus of Campylobacter jejuni by recombination and horizontal gene transfer. Microbiology 141:95-101. [DOI] [PubMed] [Google Scholar]
- 39.Wesley, I. V., W. T. Muraoka, D. W. Trampel, and H. S. Hurd. 2005. Effect of preslaughter events on prevalence of Campylobacter jejuni and Campylobacter coli in market-weight turkeys. Appl. Environ. Microbiol. 71:2824-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wright, S. L., D. K. Carver, R. M. Siletzky, S. Romine, W. E. M. Morrow, and S. Kathariou. 2008. Longitudinal study of prevalence of Campylobacter jejuni and Campylobacter coli from turkeys and swine grown in close proximity. J. Food Prot. 71:1791-1796. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.