Abstract
Shiga toxin (Stx) genes produce proteins that are pathogenic to humans, leading to severe gastrointestinal illness. This work focuses on examining the abundance and distribution of stx genes in relation to common microbial indicators in beach water and streams in the vicinity of Presque Isle State Park in Erie, PA. By use of quantitative PCR, the relative abundance levels of stx DNA in over 700 samples in the sampling area were determined. The results demonstrate that the abundance and distribution of stx genes are variable and do not correlate with the abundance of Escherichia coli bacteria, enterococci, or viral particles. These results suggest that microbial indicators of water quality are not adequate in predicting the occurrence of organisms that harbor stx genes and highlight the need for standardized pathogen-specific detection protocols for waters utilized for recreational swimming.
Stx genes produce proteins that are pathogenic to humans. Based on the amino acid similarity of the protein product, stx genes are categorically divided into two types, those encoding Shiga toxin 1 (stx1) and Shiga toxin 2 (stx2) (3). Both types produce proteins that have the potential to inhibit translation in certain eukaryotic cells (3). The Stx-dependent cessation of protein synthesis, in conjunction with other cellular disrupting activities, has been linked to damage to human blood vessels in the colon, bloody diarrhea, and hemolytic uremic syndrome, the number one cause of acute renal failure in children (7).
The etiological agents that harbor stx genes are bacteriophages and bacteria (21). Of these, the most epidemiological information is available for Stx-producing Escherichia coli (STEC) serotype O157:H7, which is responsible for approximately 73,000 cases of illnesses annually in the United States (12). In addition to E. coli O157:H7, over 500 other serotypes of STEC (1) and several non-E. coli bacterial species (16, 17) have been reported to be capable of producing Stx. Thus, the actual incidence of Stx-dependent illness is likely to be much higher than the cases reported for E. coli O157:H7 alone.
The presence of Stx-producing organisms has been reported for a number of aquatic ecosystems, including sewer water, swimming pools, rivers, lagoons, lakes, oceans, and drinking water (13). Due to the high rate of usage of some of these water sources, Stx-dependent illness can occur in water that harbors Stx-producing agents. For example, 12 confirmed cases of STEC-dependent illness acquired from bathing beach water were reported for Illinois in 1995 (2). Over the past decade, STEC arising from water sources has been implicated in thousands of infections and hundreds of deaths (13). Hence, recreational waters are a major source of Stx transmission to the general populace.
The recommendations set forth by the Environmental Protection Agency (EPA) for measuring recreational water quality for microbe content rely on the use of the indicator bacterial groups E. coli and enterococcus (14). For freshwater utilized for recreational purposes, a single sample advisory limit is exceeded when the number of E. coli bacteria is above 235 CFU/100 ml of water sampled or when the number of enterococci exceeds 61 CFU/100 ml of sampled water (22). One such beach area particularly impacted by high levels of indicator bacteria is Presque Isle State Park, in Erie, PA. This heavily utilized swimming area, which attracts over 4 million visitors annually, had 37 beach notification actions in 2006 due to levels of E. coli that exceeded the EPA advisory limit (22). While it is clear that high levels of indicator bacteria are commonly present in Presque Isle State Park beach waters, it is unclear whether these indicators accurately reflect levels of microbes that could potentially represent a danger to people swimming in these waters.
The purpose of this study was to screen for the presence of organisms that might have the ability to produce Stx in the heavily utilized beach waters of Presque Isle State Park. Since the production of Stx by such a diverse array of microorganisms is possible, we utilized a DNA-based approach for the identification, enumeration, and comparison of the stx gene to common microbial indicators of water quality in an aquatic environment. A DNA-based approach has proven useful in determining the presence of toxin genes in other aquatic ecosystems (13, 23, 24), including the stx gene, which has been shown to be associated with algal mats in the waters of Lake Michigan (10). Moreover, studies performed on stool, food, soil, and water samples have provided evidence that the concentration of Stx-producing organisms in a sample correlates with the amplification of DNA from those samples (18, 19, 20). A recent study of wastewater samples found that the concentration of stx1 and stx2 extracted from STEC-spiked samples linearly correlated with the number of colonies observed on growth plates, which suggests that a DNA approach is a useful strategy for quick and sensitive screening for Stx-producing organisms in recreational waters (20).
MATERIALS AND METHODS
Sample collection.
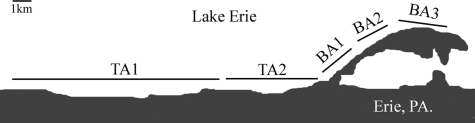
Samples were collected from the beach water and tributary sites indicated in Fig. 1, using the procedure described in reference 4. A complete listing of sample dates and locations for this study is provided in the supplemental material.
FIG. 1.
Schematic of sample site locations for tributary area 1 (TA1), tributary area 2 (TA2), beach area 1 (BA1), beach area 2 (BA2), and beach area 3 (BA3). The exact location of water collection for each site is listed in the figure in the supplemental material.
Bacteriological plating.
Water samples were filtered onto 0.45-μm mixed cellulose ester filters, following outlined procedures (4). E. coli bacteria were enumerated using modified mTEC agar (Difco), and enterococci were enumerated using mEI agar (Difco), following the instructions provided by the manufacturer. Enumeration of bacteria on growth plates was performed using EPA established guideline 1603 for E. coli or EPA established guideline 1600 for enterococci.
DNA isolation.
Water samples were filtered onto 0.45-μm mixed cellulose ester filters as described previously (4). Following filtration, the membranes were placed in 600 μl of a TE (10 mM Tris, pH 7.5, 0.5 mM EDTA) solution, and bacterial DNA was released into solution by using acid-washed glass beads as described by Haugland et al. (8). The glass beads were allowed to settle, and 200 μl of the TE solution was placed into a new Eppendorf tube. RNase A was added to the new tube at a final concentration of 50 μg/ml, and tubes were incubated at 45°C for 15 min. The tubes were allowed to cool, and an equal volume of a 25:24:1 ratio of phenol-chloroform-isoamyl alcohol (pH 6.7) was used to remove proteins. Of the remaining supernatant, 125 μl was placed in a new tube for precipitation with sodium acetate (pH 5.2) and ethanol. The final DNA pellets were resuspended in 50 μl of 10 mM Tris (pH = 7.5).
Quantitative PCR.
For each DNA sample analyzed, 5 μl of the final isolated DNA was diluted to a volume of 20 μl that contained final concentrations of 0.33 μM forward primer (for stx1, GACTGCAAAGACGTATGTAGATTCG; and for stx2, ATTAACCACACCCCACCG), 0.33 μM reverse primer (for stx1, ATCTATCCCTCTGACATCAACTGC; and for stx2, GTCATGGAAACCGTTGTCAC), 0.25 μM fluorescent probe (for stx1, TGAATGTCATTCGCTCTGCAATAGGTACTC; and for stx2, CAGTTATTTTGCTGTGGATATACGAGGGCTTG), 0.34 μM Tris (pH 7.0), and 1× PCR buffer (12 mM Tris, pH 7.0, 50 mM KCl, 5 mM MgCl2, 0.15 g trehalose, 1.6% glycerol, 0.2% Tween, 200 μg/ml nonacetylated ultrapure bovine serum albumin, 500 μM deoxyribonucleotide mixture, and 5 U of AmpliTaq Gold DNA polymerase). The primer sequences were designed from a previous study (9). The reporter dyes for the fluorescent probes were FAM (6-carboxyfluorescein) at the 5′ end of the probe for stx1 and ROX (6-carboxy-x-rhodamine) at the 5′ end of the probe for stx2. The quencher at the 3′ end of the probe was Black Hole Quencher 1 (BHQ-1) for stx1 and BHQ-2 for stx2. All primer probes were ordered through Integrated DNA Technologies.
The DNA was amplified using an Applied Biosystems 7300 real-time PCR system, using the following conditions: initial denaturing at 95°C for 5 min and 45 cycles of 95°C for 15 s and 60°C for 1 min. Threshold cycle (CT) values were determined using the SDS 7300 real-time PCR software program. All samples that tested positive for stx1 or stx2 were confirmed by running two more separate real-time PCRs. A subset of the positive samples from various water sources were sequenced or subjected to a nested PCR, followed by agarose gel electrophoresis using the primers and conditions reported by Dumke et al. for further identification purposes (6). A subset of samples were also seeded with plasmid DNA of a known concentration, processed as described above, and subjected to quantitative PCR for genes on the plasmid not found in aquatic ecosystems. The CT value of this amplified signal was compared to that for plasmid DNA of the same concentration to demonstrate that negative inhibition of PCR signal was not occurring in these water samples. The final concentrations of samples testing positive for the stx2 gene were determined by normalization of CT values to a standard curve of DNA isolated from Global Resource Center bacterial stock (ATCC 43889).
Viral epifluorescence.
Water (1.0 ml) from each sample site was filtered through a 0.45-μm mixed cellulose ester filter to remove bacteria and larger particulates. The filtrate was recovered and then filtered through a 0.2-μm Anodisc 25 filter (Whatman) to trap viral particles. The filters were then stained with 80 μl of a 25× solution of the intercalating dye Sybr green I (Cambrex). The filters were then mounted on microscope slides that contained 20 μl of mounting solution (1× phosphate-buffered saline, 50% glycerol). Viral particles on each slide were visualized with a Leica DML fluorescence microscope. For each slide, viral particles from 12 regions of equal and known dimensions from three separate pictures at random locations on a filter were counted to obtain final viral particle concentrations as previously described (15).
RESULTS
We employed the use of quantitative PCR to test for the presence of stx genes in the beach waters and tributaries west of Presque Isle State Park (Fig. 1). The primers recognize stx1 or stx2 (9). In total, two tributary areas and three beach areas (Fig. 1; also see the supplemental material) were monitored over a course of 1 year. A summary of our findings is presented in Table 1. Of the 716 samples that were probed for the presence of the stx1 gene, only 5, or 0.7%, were found to be above the limit of detection of 0.1 pg/μl of amplified signal when the results were standardized to those for control DNA samples. The presence of the stx2 gene at these locations was detected with an approximately fivefold-higher frequency than that for to stx1, present in 37, or 5.17%, of the 716 samples that were examined.
TABLE 1.
Beach and tributary water samples that tested positive for the stx1 or stx2 gene
| Locationa |
stx1 samples
|
stx2 samples
|
||
|---|---|---|---|---|
| No. | Total (%) positive | No. | Total (%) positive | |
| Beach water | 329 | 2 (0.60) | 329 | 34 (10.33) |
| Tributary water | 387 | 3 (0.78) | 387 | 3 (0.78) |
Beach water and tributary water locations include all tested samples from the beach (BA1, BA2, and BA3) and tributary (TA1 and TA2) areas presented in Fig. 1.
In addition to the difference in detection rates between the stx gene types, we also found spatial variability in detection frequency for a particular stx type. This is most evident upon comparison of the detection frequency of the stx2 gene in tributaries to that in beach water samples. While only 3, or 0.78%, of the 387 tributary samples tested positive for stx2 DNA, 34, or 10.33%, of the 329 beach water samples were above the same detection limit for stx2 gene presence, a >10-fold difference in these water sources.
To refine our analysis of stx2 gene presence in beach waters, we examined the spatial distribution of samples that tested positive for stx2 DNA, which comprised the majority of samples that tested positive in this study (Tables 1 and 2). Despite being separated from one another by only approximately 3 km in total length (Fig. 1; also see the supplemental material), the different beach sites showed variability in the presence of the stx2 gene. This is most pronounced upon consideration of the >3-fold-higher detection frequency of stx2 DNA in beach area 2 than in beach area 1 water samples, despite that the water samples collected at these two sites were separated by at most 1 km (Fig. 1).
TABLE 2.
Spatial distribution of beach water samples that tested positive for the stx2 gene
| Locationa | No. of samples | Total no. (%) positive |
|---|---|---|
| BA1 | 77 | 4 (5.19) |
| BA2 | 82 | 13 (15.85) |
| BA3 | 170 | 17 (10.0) |
Beach area (BA) locations are described in the legend to Fig. 1.
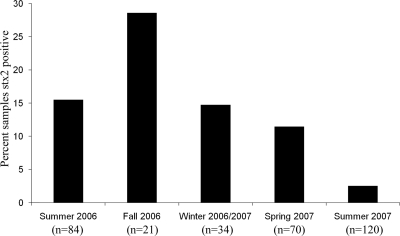
The beach area that is the most heavily utilized for recreational use is beach area 3, and this use occurs predominantly during the summer months. Because of the relatively high detection rate of the stx2 gene in this beach area, the temporal distribution of the stx2 gene was examined in greater detail. The results are displayed in Fig. 2 and demonstrate that there is seasonal variability in stx2 gene detection frequency in these beach waters. For example, nearly 30% of the samples tested positive for the presence of stx2 DNA in the fall of 2006, while only 3% tested positive in the summer of 2007 (Fig. 2). It is noteworthy that the summer of 2006 had a >6-fold-higher percentage of samples that tested positive for the stx2 gene than the summer of 2007, suggesting that these seasonal variations might also fluctuate on a larger time scale.
FIG. 2.
Percentages of beach water (BA1, BA2, and BA3) (Fig. 1) samples that tested positive for stx2 within each season. The total number of samples tested within each season is shown (n) for summer 2006 (July 2006-August 2006), fall 2006 (September 2006 to November 2006), winter 2006/2007 (December 2006 to March 2007), spring 2007 (April 2007-May 2007), and summer 2007 (June 2007 to August 2007).
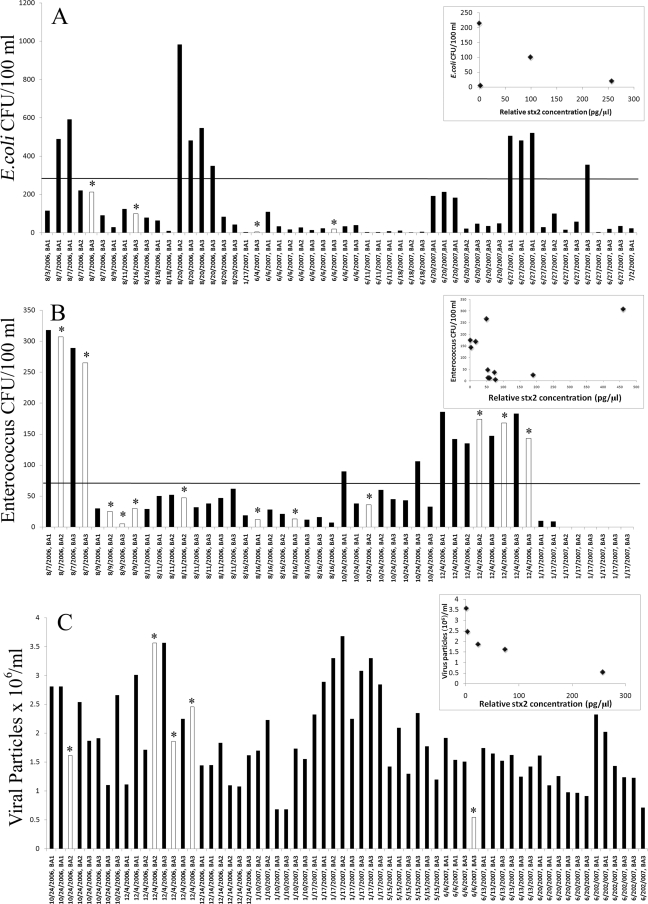
To directly test how the presence of stx2 DNA in a water sample relates to commonly used microbial parameters of water quality, the levels of E. coli or enterococcus in a water sample were compared to the relative concentration of stx2 DNA in those samples. The real-time PCR assay detected the presence of stx2 DNA in 4 of the 52 samples in which E. coli colonies were enumerated (Fig. 3A) and 12 of the 48 samples in which enterococcus colonies were determined (Fig. 3B). The data demonstrate that the majority of samples above the advisory threshold for enterococcus or the advisory threshold for E. coli did not test positive for the presence of the stx2 gene. In total, only 5 of the total 24 samples above the advisory threshold limit for either bacterial indicator tested positive for stx2 DNA (Fig. 3A and B). Instead, the majority of samples testing positive for the stx2 gene were below the advisory threshold limit for either E. coli or enterococcus. For example, 7 of the 12 samples that were found to test positive for stx2 DNA when enterococcus colonies were counted were below the advisory threshold limit for enterococcus (Fig. 3B), while all four of the stx2 gene-positive samples were found to be below the detection limit for E. coli when colonies of E. coli were measured (Fig. 3A). Moreover, there was not a clear correlation between the number of enterococcus colonies or E. coli colonies and the relative concentration of stx2 DNA in the water samples analyzed (Fig. 3A and B, insets). Hence, neither enterococcus nor E. coli colony growth appears to be a reliable indicator for detecting the presence or relative abundance of the stx2 gene in these waters.
FIG. 3.
Comparison of samples that tested positive (white bars with *) or negative (dark bars) for the stx gene when the number of E. coli (A) or enterococcus (B) CFU or the number of viral particles (C) in the sample was known. The advisory limit for E. coli or enterococcus is indicted by a solid horizontal line. The normalized concentration of stx2, expressed in pg/μl, is compared to the number of E. coli (A) or enterococcus (B) CFU or the number of viral particles (C) in the inset of each panel. The data point for 9 August 2006 BA3 that tested positive for stx2 is not shown in the inset of the graph for comparison to number of enterococcus CFU, because the determined stx2 concentration of 7,200 pg/μl was too large to fit the scale of the graph.
In addition to enterococcus and E. coli colony numbers, we also compared stx2 gene presence and concentration in a water sample to virus concentration, which is an important part of the microbial fauna in aquatic ecosystems that have been monitored in a number of studies (25). Of the 64 viral samples taken, only 5, or 7.8%, tested positive for the presence of the stx2 gene (Fig. 3C). Of the samples that did test positive for the stx2 gene, there was a trend toward increasing concentration of the stx2 gene compared to decreasing viral concentration (Fig. 3C, inset). However, there were many samples in the lower end of the viral concentration spectrum that did not test positive for stx2 DNA presence, demonstrating that viral particles are not reliable indicators for detection of the stx2 gene in these waters.
DISCUSSION
This study represents, to our knowledge, the first long-term quantitative characterization of stx gene distribution in waters commonly used for recreational purposes. The results of this characterization indicate a low overall presence of STEC in the waters analyzed. In these studies, the lower limits of detection for both stx1 and stx2 were approximately 200 cells/ml, as determined by comparison of standard curves of extracted DNA in control samples to colony counts of those samples on modified mTEC agar. Based on these limits of detection, this assay would be capable of detecting approximately 3,200 cells that harbor the stx gene for the 16 ml of water that adults are estimated to ingest during swimming activities (5). Since we used a DNA-based assay, it is impossible to know whether the DNA detected came from individual cells or from fewer cells with multiple copies. Moreover, these experiments did not determine whether the genes detected in this assay are capable of producing infectious toxins. However, given the extremely low dose of 10 to 100 STEC cells needed to cause human illness (21), it is likely that these results are within a range that is important for detection of infectious dosages of Stx-producing bacteria. Thus, while the overall frequency of stx gene detection was low, those samples that did test positive for Stx could be within levels that are harmful to humans.
It is surprising that the majority of samples that tested positive for stx DNA were biased to the stx2 type and were present in the beach waters and not the tributaries. This result has important implications for recreational water use for many reasons. First, since the tributaries tend to be more polluted with E. coli and other bacteria than the beach waters (11), it would appear that stx2 gene distribution is not directly related to E. coli colony counts. The data presented in Fig. 3 provide support for this idea and point to the need for Stx-specific assays for determining the presence of bacteria which are capable of causing Stx-dependent illness. Second, the uneven distribution of recreational water samples that tested positive for stx1 or stx2 genes indicates that the detection of Stx-producing bacteria by PCR will be dependent on which type of the stx gene is being probed for. Hence, these results highlight the need for the development of multiple assays or detection systems that probe for the presence of Stx-producing organisms in a water sample.
In addition to the variability in detection frequency of the different types of stx genes, our results also find variability in the spatial and temporal distributions of the stx2 gene in these water sources. While a large proportion of samples testing positive for the stx2 gene were found during seasons when the beach water is not utilized for recreational swimming (fall, winter, and spring months), there were also several instances where the stx2 gene was found in water obtained from populated swimming beaches during the summer season. Overall, the beaches that are furthest from one another are separated by approximately 3 km of beach front. While the stx2 gene was often detected in multiple beach water samples on the same day (see the figure in the supplemental material), the uneven distribution of stx2 gene detection in these beach water samples demonstrates the need for testing beach water on an individual basis instead of using one beach as a model for the other in a continuous water system.
While these data provide important information on the presence of stx genes in water, they do not provide an understanding on how these genes might relate to the ability of bacteria to produce Stx or lead to increased incidences of water-related illness. In the duration of this study, there were no reports of an outbreak of Stx-dependent illness related to the ingestion of beach water that tested positive for stx genes. However, epidemiological studies were not conducted during the time of this study, so it is difficult to assess whether individual cases of illness occurred but were not reported. Regardless, by demonstrating that there is not a stable genetic reservoir for the stx2 gene in these recreational waters, these results provide important insight into how Stx-dependent outbreaks can arise in aquatic habitats. Specifically, the results of this study suggest that aquatic outbreaks of Stx-dependent illness do not arise from the activation of a stable population of Stx-producing bacteria in a water source but instead result from the appearance of infectious Stx-producing bacteria transiently inhabiting the water.
Since quantitative PCR can be completed in a few hours, these results also demonstrate the utility of this procedure for screening water samples over a prolonged period of time for the purpose of alerting health officials when the presence of a genetic reservoir for Stx arises in a body of water. Given the limited understanding that we currently have on conditions that favor outbreaks of Stx-dependent illness, such information will allow for increased precautionary measures to be put into practice when there is a detection of Stx in aquatic ecosystems. The low infectious-dose rate required for Stx-dependent illness, the occurrence of Stx-dependent illness arising from water sources, the correlation of stx gene concentration with concentration of STEC, and the finding in this study that there is not a correlation between stx2 gene presence and commonly used microbial indicators of water quality all warrant such a monitoring program.
Supplementary Material
Acknowledgments
We acknowledge the Pennsylvania Water Resources Research Center, Coastal Zone Management, the Department of Environmental Protection, the EPA, and Mercyhurst College for funding parts of this project.
We also thank the Erie County Department of Health, the Regional Science Consortium, Pennsylvania Sea Grant, and members of the laboratory for valuable input and assistance in the study.
Footnotes
Published ahead of print on 14 November 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Blanco, M. J., J. E. Blanco, A. Mora, G. Dahbi, M. P. Alonso, A. Gonzalez, M. I. Bernardez, and J. Blanco. 2004. Serotypes, virulence genes, and intimin types of Shiga toxin (verotoxin)-producing Escherichia coli isolates from cattle in Spain and identification of a new intimin variant gene (eae-ξ). J. Clin. Microbiol. 42:645-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 1996. Lake-associated outbreak of Escherichia coli O157:H7—Illinois, 1995. MMWR Morb. Mortal. Wkly. Rep. 45:437-439. [PubMed] [Google Scholar]
- 3.Cherla, R. P., S. Y. Lee, and V. L. Tesh. 2003. Shiga toxins and apoptosis. FEMS Microbiol. Lett. 228:159-166. [DOI] [PubMed] [Google Scholar]
- 4.Clescerl, L. S., A. E. Greenberg, and A. D. Eaton (ed.). 1985. Standard methods for examination of water and wastewater, 16th ed., p. 886-901, 977. American Public Health Association, Washington, DC.
- 5.Dufour, A. P., O. Evans, T. D. Behymer, and R. Cantu. 2006. Water ingestion during swimming activities in a pool: a pilot study. J. Water Health 4:425-430. [PubMed] [Google Scholar]
- 6.Dumke, R., U. Schroter-Bobsin, E. Jacobs, and I. Roske. 2006. Detection of phages carrying the Shiga toxin 1 and 2 genes in waste water and river water samples. Lett. Appl. Microbiol. 42:48-53. [DOI] [PubMed] [Google Scholar]
- 7.Gyles, C. L. 2007. Shiga toxin-producing Escherichia coli: an overview. J. Anim. Sci. 85:E45-E62. [DOI] [PubMed] [Google Scholar]
- 8.Haugland, R. A., S. C. Siefring, L. J. Wymer, K. P. Brenner, and A. P. Dufour. 2005. Comparison of Enterococcus measurements in freshwater at two recreational beaches by quantitative polymerase chain reaction and membrane filter culture analysis. Water Res. 39:559-568. [DOI] [PubMed] [Google Scholar]
- 9.Ibekwe, M. A., P. M. Watt, C. M. Grieve, V. K. Sharma, and S. R. Lyons. 2002. Multiplex fluorogenic real-time PCR for detection and quantification of Escherichia coli O157:H7 in dairy wastewater wetlands. Appl. Environ. Microbiol. 68:4853-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishii, S., T. Yan, D. A. Shively, M. N. Byappanahalli, R. L. Whitman, and M. J. Sadowsky. 2006. Cladophora (Chlorophyta) spp. harbor human bacterial pathogens in nearshore water of Lake Michigan. Appl. Environ. Microbiol. 72:4545-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lybrook, R. P. 2006. Operation Creek Sweep—surface water E. coli assessment of major tributaries to the western portion of the Lake Erie watershed, Erie County. Pennsylvania Department of Environmental Protection, Northwest Regional Office, Meadville, PA.
- 12.Mead, P. S., L. Slutsker, V. Deitz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muniesa, M., J. Jofre, C. Garciaa-Aljaro, and A. R. Blanch. 2006. Occurrence of Escherichia coli O157:H7 and other enterohemorrhagic Escherichia coli in the environment. Environ. Sci. Technol. 40:7141-7149. [DOI] [PubMed] [Google Scholar]
- 14.Office of Science and Technology, Standards and Applied Science Division. 2000. Bacterial water quality standards for recreational waters (fresh and marine). Status report. U.S. Environmental Protection Agency, Washington, DC.
- 15.Patel, A., R. T. Noble, J. A. Steele, M. S. Schwalbach, I. Hewson, and J. A. Fuhrman. 2007. Virus and prokaryote enumeration from planktonic aquatic environments by epifluorescence microscopy with SYBR Green I. Nat. Protoc. 2:269-276. [DOI] [PubMed] [Google Scholar]
- 16.Paton, A. W., and J. C. Paton. 1996. Enterobacter cloacae producing a Shiga-like toxin II-related cytotoxin associated with a case of hemolytic-uremic syndrome. J. Clin. Microbiol. 34:463-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt, H., M. Montag, J. Bockemuhl, J. Heesemann, and H. Karch. 1993. Shiga-like toxin II-related cytotoxins in Citrobacter freundii strains from humans and beef samples. Infect. Immun. 61:534-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sekse, C., A. Solberg, A. Peterson, K. Rudi, and Y. Wasteson. 2005. Detection and quantification of Shiga toxin-encoding genes in sheep faeces by real-time PCR. Mol. Cell. Probes 19:363-370. [DOI] [PubMed] [Google Scholar]
- 19.Spano, G., L. Beneduce, V. Terzi, A. M. Stanca, and S. Massa. 2005. Real-time PCR for the detection of Escherichia coli O157:H7 in dairy and cattle wastewater. Lett. Appl. Microbiol. 40:164-171. [DOI] [PubMed] [Google Scholar]
- 20.Stefan, A., S. Scaramagli, R. Bergami, C. Mazzini, M. Baranera, S. Perelle, and P. Fach. 2007. Real-time PCR and enzyme-linked fluorescent assay methods for detecting Shiga-toxin-producing Escherichia coli in mincemeat samples. Can. J. Microbiol. 53:337-342. [DOI] [PubMed] [Google Scholar]
- 21.Tyler, J. S., J. Livny, and D. I. Friedman. 2005. Lambdoid phages and Shiga toxin, p. 131-164. In M. K. Waldor, D. I. Friedman, and S. L. Adhya (ed.), Phages: their role in bacterial pathogenesis and biotechnology. ASM Press, Washington, DC.
- 22.U.S. Environmental Protection Agency. 2006. BEACH report: Pennsylvania 2006 swimming season. U.S. Environmental Protection Agency, Washington, DC. http://pubweb.epa.gov/waterscience/beaches/seasons/2006/pdf/pa.pdf.
- 23.Whitman, R. L., T. G. Horvath, M. L. Goodrich, M. B. Nevers, M. J. Wolcott, and S. K. Haack. 2001. Characterization of E. coli levels at 63rd Street Beach. Report to the City of Chicago Department of the Environment and the Chicago Park District. U.S. Geological Survey, Porter, IN. http://www.glsc.usgs.gov/_files/reports/63rdStreetBeachReport.pdf.
- 24.Winfield, M. D., and E. A. Groisman. 2003. Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl. Environ. Microbiol. 69:3687-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wommack, E. K., and R. R. Colwell. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.