Abstract
Sinorhizobium meliloti can form a nitrogen-fixing symbiotic relationship with alfalfa after bacteria in the soil infect emerging root hairs of the growing plant. To be successful at this, the bacteria must be able to survive in the soil between periods of active plant growth, including when conditions are dry. The ability of S. meliloti to withstand desiccation has been known for years, but genes that contribute to this phenotype have not been identified. Transposon mutagenesis was used in combination with novel screening techniques to identify four desiccation-sensitive mutants of S. meliloti Rm1021. DNA sequencing of the transposon insertion sites identified three genes with regulatory functions (relA, rpoE2, and hpr) and a DNA repair gene (uvrC). Various phenotypes of the mutants were determined, including their behavior on several indicator media and in symbiosis. All of the mutants formed an effective symbiosis with alfalfa. To test the hypothesis that UvrC-related excision repair was important in desiccation resistance, uvrA, uvrB, and uvrC deletion mutants were also constructed. These strains were sensitive to DNA damage induced by UV light and 4-NQO and were also desiccation sensitive. These data indicate that uvr gene-mediated DNA repair and the regulation of stress-induced pathways are important for desiccation resistance.
Sinorhizobium meliloti is a gram-negative soil bacterium that forms a nitrogen-fixing symbiotic relationship with alfalfa by inducing the formation of root nodules. Within these nodules, S. meliloti converts dinitrogen gas from the atmosphere into ammonia, a form of nitrogen that can be used by the plant; the plant supplies the bacteria with carbon sources that are catabolized to provide the energy needed for nitrogen fixation (14). The formation of a nodule occurs after soil rhizobia infect the emerging root hairs of the plant. As a soil bacterium, S. meliloti must be able to cope with the conditions found in the soil or rhizosphere, including suboptimal pH and desiccation. Resistance to desiccation is an issue for “wild” rhizobia living in the soil, but it is also important in agricultural applications of these bacteria. The desiccation susceptibility of rhizobia has led to the use of carrier materials, such as peat and mineral soils, to increase the viability of commercial rhizobium inocula (5, 24). Spore or cyst formation by rhizobia has not been observed, suggesting that they have alternate adaptive mechanisms for surviving harsh environmental conditions.
A recent study by Vriezen et al. (38) examined the effects of various physiological and physical conditions on the survival of S. meliloti during desiccation. They found that cells from a stationary culture resulted in 2.9-fold-greater cell survival during desiccation than exponentially growing cells. The drying medium also affected the survival of S. meliloti cells. More viable cells were recovered from inoculated sand or nitrocellulose filters than from alfalfa seeds. Drying S. meliloti at temperatures above 37°C also decreased the survival rate. Finally, drying S. meliloti in the presence of NaCl increased the survival rate by three- to fivefold depending on the culture medium and NaCl concentration used. The results of this study demonstrated that there are many factors that contribute to the survival of S. meliloti during desiccation.
Rhizobium strains that are more or less tolerant to desiccation have been characterized (19, 20), but the genes involved in this process are only now being identified. Recently, a microarray-based study of Bradyrhizobium japonicum USDA 110 measured gene induction under desiccation stress (6). In B. japonicum, 225 genes were upregulated during all desiccation incubations, with an additional 446 genes being upregulated during longer periods of desiccation. The genes that were upregulated during desiccation were diverse, but included genes involved in exopolysaccharide formation, transcriptional regulators, sigma factors, oxidative and heat stress response systems, and DNA repair and modification (6). A ctpA mutant of Rhizobium leguminosarum 3841 has been found to be about 21 times more sensitive to desiccation than the wild type (9). CtpA is a periplasmic protease, and the increased desiccation sensitivity of the R. leguminosarum ctpA mutant is believed to be due to defects in the structure of the cell envelope. Trehalose accumulation has also been implicated in desiccation resistance. An otsA (trehalose-6-phosphate synthase) and treY (maltooligosyltrehalose synthase) double mutant of R. leguminosarum bv. trifolii, which did not accumulate trehalose, exhibited an increased sensitivity to drying (21). Taken together, the results of these studies indicate that the genes involved in desiccation resistance are varied and many cellular processes are involved.
In this work, four desiccation-sensitive mutants of Sinorhizobium meliloti Rm1021 were identified. Three regulatory genes (relA, rpoE2, and hpr) and one DNA repair gene (uvrC) were shown to contribute to normal S. meliloti desiccation resistance. Deletion mutants of uvrA, uvrB, and uvrC were also constructed and tested in order to further investigate the role of nucleotide excision repair (NER) during desiccation. A novel desiccation screen for quantifying the sensitivity of S. meliloti to desiccation was also developed in the process of the research.
MATERIALS AND METHODS
Strains, culture conditions, and reagents.
Escherichia coli strains (described below as needed) were grown in LB medium at 37°C. All Sinorhizobium meliloti strains, including Rm1021 (22), the mutants described below, and the previously described relA mutants (DW strains) (41, 42), were cultured at 30°C in LB (25), YMB (33), or minimal ammonium medium (33) with either 1% mannitol (MMNH4) or 0.2% succinate (MNH4Succ) as the carbon source. Antibiotics were used at the following final concentrations: neomycin, 200 μg/ml, and tetracycline, 10 μg/ml. For phenotypic assays and growth curves, the following media and reagents were used. Calcofluor (17) was added to LB medium at a final concentration of 0.02%. Congo red was added to YMB as described previously (15). Nadi tests were performed on colonies grown on MNH4Succ as described previously (18). YMB-grown colonies were stained with iodine (8) to determine glycogen accumulation. TY medium (3) supplemented with either NaCl or polyethylene glycol (PEG) 200 at final concentrations of 0.2 M and 0.4 M was used for growth curve experiments. M9 medium supplemented with 3-amino-1,2,4-triazole (AT) was used to test for relA mutant sensitivity as previously described (41).
Transposon mutagenesis.
In order to identify mutants that were more susceptible to desiccation, S. meliloti Rm1021 was randomly mutagenized with either Tn5 or Tn5-B50. The Tn5 suicide plasmids pSUP5011 and pSUPTn5-B50 (31), which are maintained in E. coli S17-1 (30), were mated into Rm1021 via biparental matings. Mating mixtures were placed on LB agar plates, incubated overnight at 30°C, and then removed from the LB plates and plated on MMNH4 plates supplemented with either neomycin or tetracycline to select for Tn5 or Tn5-B50 integration, respectively. The cells that contained transposon insertions were picked onto MMNH4 agar plates in a grid pattern (36 transposon mutants/plate) and incubated for 3 days at 30°C. The colonies were then replicated via stamping onto three plates: a MMNH4 plate for the screen (see below), a MNH4Succ plate to check the ability of the mutants to grow on MNH4Succ, and another MMNH4 plate to generate a new library copy. The stamped plates were incubated at 30°C until the colony growth was uniform.
Initial screen for desiccation sensitivity.
To test the ability of the transposon mutants to resist desiccation, the colonies were lifted off of a MMNH4 plate by placing a sterile 6-mm circle of Whatman #1 filter paper on top of the colonies until the filter paper was moist and then removing it and placing it into a sterile petri dish. The petri dishes containing the filters were placed into a plastic box with a loosely fitting lid and incubated at 30°C for 1 week. After incubation, the filters were rehydrated by inverting them onto fresh MMNH4 plates until the filters were completely moist. The filters were then removed and placed back in the sterile petri dishes they were in and set aside. The MMNH4 plates were incubated at 30°C until bacterial growth started to be visible. About 3,000 transposon mutants were screened in this manner. Forty-four mutants that were delayed in growth, presumably due to lower cell survival, were identified as putative desiccation-sensitive mutants.
Quantitative screen for desiccation sensitivity.
The 44 putative mutants were then screened with a quantitative drying assay. Fifty microliters of YMB agar was aliquoted into wells of flat-bottomed 96-well microtiter plates and allowed to solidify. The mutant strains were cultured in triplicate for 2 days in YMB broth at 30°C, and then 4 μl of each sample was aliquoted into one row of wells (seven mutants/plate) in a 96-well plate with the YMB agar. Rm1021 was also put on each plate in one row as a control. The plates were then covered with an AirPore tape sheet (Qiagen), and the plate lid was also put on the plate. The plates were then kept at 30°C for 2 days. After incubation, the plate lids were removed and a time zero colony count was performed. The AirPore sheet was peeled back, exposing only the first column of wells, and 100 μl of minimal ammonium buffer (MNH4), which is minimal medium without a carbon source, was added to each well to rehydrate the agar plug. The liquid was removed from the wells and transferred to microcentrifuge tubes. The agar plugs were also removed from the wells by using sterile toothpicks and placed into the tubes with the corresponding liquid. An additional 100 μl of MNH4 was added, and the tubes were agitated with a vortex mixer to dislodge cells from the agar plug. The suspended cells were then diluted 1:1,000, and 50 μl of each dilution was plated in triplicate on YMB plates by using a Spiral Biotech Autoplate model 3000 spiral plater. The plates were incubated for 3 to 4 days at 30°C and counted according to the manufacturer's directions. After the time zero agar plugs were harvested, the AirPore sheet was folded back down and the plates were placed in a plastic box with a loosely fitting lid but without the plate covers. The plastic box was placed in a 30°C incubator. The AirPore sheet acts as a physical barrier to prevent contamination and allows the samples to dehydrate, since the sheet is permeable to air. After a few days, the agar plugs dry down to very thin, adherent films on the bottoms of the wells. Additional samples were rehydrated as described above at 2, 4, 6, and 8 weeks, and a 1:1,000 dilution was plated. To obtain sufficient viable colonies of the desiccation-sensitive strains, lower dilutions (1:100, 1:10, and 1:2) were also evaluated.
Location of transposon inserts.
In order to identify the location of the transposon insertions, total cell DNA was isolated from mutants by using a DNeasy tissue kit (Qiagen). Arbitrary PCR (16) was used to amplify fragments containing transposon junctions, and DNA sequences were determined. Arbitrary PCR primers ARB1-A and ARB2 are described in Griffitts and Long (10). The transposon-specific primers used with the arbitrary PCR primers (Tn5-4 and Tn5-2) and the sequencing primer (TZTn5) are described in Yurgel et al. (43). DNA sequencing was performed on an Applied Biosystems 373 DNA sequencer at the Washington State University Laboratory for Bioanalysis and Biotechnology. The DNA sequences were located in the mutant genomes by using nucleotide BLAST (http://www.ncbi.nlm.nih.gov/blast).
Transposon transduction.
Transducing-phage ΦM12 lysates grown on mutants AL5, AL15, B15-4, and T2 (Table 1) were prepared as described previously (7) except that the phage lysate was separated from the lysed cell debris by filtering through a 0.2-μm syringe filter (Nalgene). Transductants of Rm1021 containing the Tn5 mutations in AL5, AL15, or B15-4 were selected on LB medium containing neomycin, and transductants containing the Tn5-B50 mutation in T2 were selected on LB medium with tetracycline. The positions of the transposons in the transductants were confirmed by arbitrary PCR as described above.
TABLE 1.
Location of transposon insertion and phenotypes of desiccation mutants
| Strain | ORF | Gene | No. of aa encoded by ORF | Position of Tn in ORF (no. of aa)a | Results for indicated assayb
|
Symbiotic effectivenessc | Plant dry mass ± SD (mg)d | Description of nodules | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Congo red | Calcofluor | Iodine | Nadi | ||||||||
| None | NAe | NA | NA | NA | NA | NA | NA | NA | − | 4.9 ± 0.8 | None |
| Rm1021 | NA | NA | NA | NA | R | F | B | P | + | 8.8 ± 1.2 | Pink (wt) |
| AL5 | SMc02659 | relA | 741 | 128 | R+ | F+ | B+ | P+ | +/− | 6.9 ± 0.9 | wt, large white |
| AL15 | SMc01506 | rpoE2 | 184 | 114 | R | F | B | P | + | 7.6 ± 1.3 | wt |
| B15-4 | SMc02754 | hpr | 96 | 55 | R+ | F+ | B+ | P+ | + | 7.7 ± 1.9 | wt |
| T2 | SMc00602 | uvrC | 674 | 175 | R | F | B | P | + | 7.5 ± 1.5 | wt |
ORF, open reading frame; aa, amino acids.
R, red colonies; R+, dark red colonies; F, fluorescent colonies; F+, brightly fluorescent colonies; B, brown colonies; B+, dark brown colonies; P, purple colonies; P+, dark purple colonies.
−, not effective (yellow plants); +/−, partially effective (green plants with yellow tint); +, fully effective (green plants).
Bold numbers indicate that the dry mass of the shoots and leaves of the inoculated alfalfa plants was significantly lower than that of plants inoculated wild-type Rm1021 (Student's t test; P < 0.005). Eight magenta boxes with six plants each were inoculated for each strain. Some plants did not survive the assay due to damage at planting, so the average dry mass per plant was calculated for each magenta box. These eight averages were used to calculate the numbers in the table.
NA, not applicable.
Growth curves.
Growth curves were performed to measure the sensitivities of the desiccation-sensitive mutants to salt and osmotic stress. Strains were cultured in TY broth for 48 h at 30°C, and then 2 μl of these cultures was used to inoculate microtiter plate wells containing 200 μl of TY or TY with either NaCl or PEG 200 (see above). The microtiter plate was incubated at 30°C in a Molecular Devices SpectraMax 250 plate reader using SoftMax Pro version 1.2.0 software, and readings of the optical density at 600 nm were taken every 30 min; the plate was mixed for 10 s before each reading.
Plant tests.
All of the mutants were tested on alfalfa (Medicago sativa cv. Champ) for the ability to form effective symbiotic root nodules. The plant tests were performed as previously described (44).
uvr gene deletion mutants.
Deletion mutants of uvrA, uvrB, and uvrC in Rm1021 were constructed by using homologous recombination. “Shoulders,” regions flanking the genes, were amplified by PCR from Rm1021 genomic DNA. The primers (Table 2) are designated as follows for each gene: F1 and R1 primer pairs amplify the 5′ shoulder and F2 and R2 primer pairs amplify the 3′ shoulder. The shoulders were designed to give one shoulder of about 400 bp and another of about 600 bp in order to provide about 1,000 bp for homologous recombination. After amplification, the shoulders were cut with BamHI, ligated together, and then reamplified using the outside primers (F1 and R2) to produce a DNA fragment that resulted in an in-frame deletion of the gene. Only 246 bp and 72 bp of the 3′ end of the uvrA and uvrB genes remained, respectively; 342 bp of the 5′ end of the uvrC gene was still present. The resulting PCR fragments were then cut with EcoRI, ligated to EcoRI-cut pK19-mob-sacB (27), and transformed into E. coli DH5α (11). Positive clones were confirmed by PCR and DNA sequencing. The plasmids that contained the correct shoulder regions were then transformed into E. coli S17-1 (30), and the plasmids were then conjugated into Rm1021. Neomycin-resistant isolates containing the plasmid integrated by homologous recombination were selected. After purification, these were streaked onto YMB agar supplemented with 5% sucrose to select for plasmid loss. If the second recombination leading to plasmid loss occurs in the shoulder not used for the first recombination, the isolate will lose the sequence in the gene that was between the shoulder regions (44). Isolates that were Neos and sucrose resistant were tested by PCR, using the external shoulder primers (F1 and R2). After the PCR screen, isolates missing the full-length gene were sequenced by using arbitrary PCR. The ARB1-A and ARB2 primers and PCR conditions were as previously described (10, 16). The primers used with ARB1-A were uvrA-R2, uvrBseq-R, and uvrC-F1 for the uvrA, uvrB, and uvrC deletions, respectively (Table 2). The primers used in conjunction with ARB2 were SMc01235R for uvrA, SMc04231R for uvrB, and SMc00602F for uvrC and are described in Schroeder et al. (28). Sequences for these primers can also be found at http://www.bioinformatics.wsu.edu/kahn. Primers SMc01235R, SMc04231R, and SMc00602F were also used in sequencing reactions as described above.
TABLE 2.
Primers used to construct uvr gene deletions
| Primer | Sequence (5′ to 3′)a |
|---|---|
| uvrA-F1 | GCATGAATTCTGAAGGACGACCTCGGTCGAATAG |
| uvrA-R1 | GCATGGATCCCGGAAGGCCAGCTTTGTATGT |
| uvrA-F2 | GCATGGATCCGGCTTGCATTTCCACGACGTA |
| uvrA-R2 | CGGCGAATTCGAGCTGCCGCTTTTTCGGAA |
| uvrB-F1 | GCATGAATTCGGTTCCTTCAGCCAGAAAATCGAG |
| uvrB-R1 | GCATGGATCCGGGAATATGGACAGAGTCAGGTCG |
| uvrB-F2 | TACGAGGATCCGGCGGAGGAAAAGCGGCGCAAG |
| uvrB-R2 | GCATGAATTCTCATCGCCGTTCAGGTTGCGA |
| uvrC-F1 | CGAGGAATTCGGACGTGCGATCCGCTTCCTTTTCGA |
| uvrC-R1 | GCATGGATCCCATATGCGCAGTCTCGCGGAC |
| uvrC-F2 | GCATGGATCCGCGTTCAGGCGAAATCCGCCAAATATG |
| uvrC-R2 | GCATGAATTCGATCGAGGCGACCAGCAGCTT |
| uvrBseq-R | TCTTACGGCCGCCTCTTGCAGCG |
Underlined sequences are EcoRI restriction sites, and bold sequences are BamHI restriction sites.
UV killing and zone of inhibition assays.
Uvr proteins repair DNA damage caused by agents such as UV light and 4-nitroquinoline N-oxide (4-NQO) (12, 34). UV light treatments were performed by using a cell dilution stamped onto YMB agar in rectangular OmniTray plates (Nalge Nunc). For each strain tested, 10 μl of 2-day YMB cultures was added to 100 μl of sterile water in wells of a 96-well microtiter plate. At least one row for each strain (12 wells) was done per replicate experiment. Approximately 10 μl of the diluted cells was then transferred onto a YMB agar plate by using a sterile 96-well prong tool (V & P Scientific, Inc.), in duplicate. The drops were allowed to dry, and then the OmniTray plates were divided into six sections (two strips of eight spots each), with each section receiving a different UV exposure (see Results). The UV light source (254-nm wavelength) provided 25 mW/cm2 of intensity to the samples. After UV exposure, the plates were incubated at 30°C for 4 to 5 days, and then the growth in each section was recorded.
Zone-of-inhibition assays were performed to measure the sensitivities of strains to 4-NQO. The strains tested were grown for 2 days in YMB broth prior to the assay. To make the lawns of bacteria for the assay, 100 μl of cells was added to 3.5 ml of YMB top agar (0.7% agar, cooled to 55°C), mixed, and then poured on top of a YMB plate. Ten plates were made for each strain. Whatman #1 filter paper disks, 6 mm in diameter, were prepared by using a hole punch and sterilized. A 0.5 mg/ml solution of 4-NQO was prepared in acetone and applied to the disks to obtain the following final 4-NQO concentrations: 1 μg, 2 μg, 4 μg, and 8 μg. A control without 4-NQO was prepared by adding only acetone. Once the acetone evaporated, the disks were aseptically placed on the centers of the pour plates. The plates were then incubated at 30°C for 3 days, and the zones of inhibition were measured. Replicate plates for each concentration of 4-NQO were assayed for each strain, and the assay was replicated four times. Assays with mitomycin C were done as described above but with the following concentrations: 0.25 μg, 0.5 μg, 1 μg, and 2 μg.
RESULTS
Identification of desiccation-sensitive mutants.
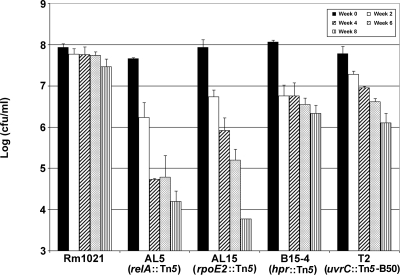
S. meliloti Rm1021 is very desiccation resistant. After 2 months of desiccation, 33% of Rm1021 cells were still viable (Fig. 1), and 4% and 2% were still viable after 3 and 4 months, respectively (data not shown). In order to identify genes that contribute to desiccation resistance, Rm1021 was mutagenized using Tn5 or Tn5-B50. An initial screen of ∼3,000 transposon mutants yielded 44 candidate desiccation-sensitive mutants. After a more-extensive and quantifiable drying assay, four desiccation-sensitive Tn5 mutants were confirmed. Mutants AL5, AL15, B15-4, and T2 all had decreased resistance to drying (Fig. 1). After 2 weeks, the viability of mutants AL5, AL15, and B15-4 was only 5 to 6%. Mutant T2 was less impaired, with about 31% survival at that time. The AL5 and AL15 mutants were the most sensitive, with cell viabilities of ≤1% after 4 weeks. After 4 weeks, about 2% of B15-4 and T2 cells were viable. To confirm that the transposon insertion caused the desiccation-sensitive phenotype, phage ΦM12 (7) was used to transduce the Tn5 and Tn5-B50 transposons from the mutants into Rm1021. The resulting transductants had desiccation sensitivities similar to those of the original mutants (data not shown). This showed that the desiccation-sensitive phenotypes correlated with the transposon insertions and were not due to other mutations that may have accumulated in the strains.
FIG. 1.
Desiccation sensitivities of S. meliloti mutants. Wild-type strain Rm1021 and desiccation-sensitive mutants AL5, AL15, B15-4, and T2 were tested as described in Materials and Methods. Viable cell counts (cfu/ml) were measured at time zero and every 2 weeks thereafter. Sample results without an error bar indicate that only one repetition had countable colonies at that time point. Error bars show standard deviations.
The locations of the transposons were determined by DNA sequencing to be in three regulatory genes (relA, rpoE2, and hpr) and one DNA repair gene (uvrC) (Table 1). The three S. meliloti regulatory genes have been studied previously, but the participation of these genes in desiccation resistance was not determined. RelA, a guanosine tetraphosphate (ppGpp) synthetase and hydrolase, is involved in the induction of the S. meliloti stringent response during amino acid starvation (41). S. meliloti RpoE2 is an extracytoplasmic function sigma factor that regulates genes during heat stress and entry into the stationary phase (26). The S. meliloti hpr gene is similar to the E. coli ptsH gene, which codes for a component of the glucose phosphotransferase system, but S. meliloti does not contain the genes needed for a complete phosphotransferase system (1). Recent research has demonstrated that Hpr has a role in succinate-mediated catabolite repression and most likely is involved in the regulation of other cellular processes (1). The annotated uvrC gene of S. meliloti has not been studied. In E. coli, UvrC is the endonuclease in the NER pathway that repairs bulky lesion damage caused by UV light and other agents (12). Since the three mutated regulatory genes are involved in regulating stress response pathways, the inactivation of these genes probably impairs the ability of the bacterium to effectively deal with the stresses associated with desiccation. In the uvrC mutant, we suggest that the repair of DNA damage caused by desiccation is impaired and this leads to desiccation sensitivity.
Phenotype and symbiosis tests.
The phenotypes of all the mutants were determined by using several common phenotype tests for S. meliloti (Table 1). The AL5 (relA) and B15-4 (hpr) mutants had increased cytochrome respiration in the Nadi cytochrome oxidase assay. These two mutants also shared similar phenotypes in the other tests. When these mutants were cultured on YMB medium containing Congo red dye, which stains exopolysaccharides, the AL5 and B15-4 mutants stained darker than wild-type cells, indicating a change in cell surface polysaccharides. Calcofluor binds EPSI, the exopolysaccharide also known as succinoglycan, and colonies producing EPSI are fluorescent under UV light (17). Rm1021 normally has low fluorescence on LB-calcofluor agar medium, but the AL5 and B15-4 mutants and their transductants had brighter fluorescence, indicating increased EPSI production (Table 1). Increased EPSI production was previously described for relA mutants of Rm1021 (41, 42). The AL5 and B15-4 strains also had increased glycogen accumulation, as evidenced by increased iodine staining of the colonies versus that of the wild type (Table 1). The AL15 (rpoE2) and T2 (uvrC) mutants all exhibited phenotypes similar to that of Rm1021 in the Congo red, calcofluor, iodine, and Nadi tests (Table 1). The transductants had the same phenotypes as the parent mutant strains (data not shown).
All of the mutants formed symbioses with alfalfa but with different degrees of success. All of the mutants induced nodules that were similar to those formed by wild-type Rm1021. However, in addition to wild-type nodules, the plants nodulated with AL5 also had large, white nodules (Table 1). Plants inoculated with AL5 were not quite as green as plants inoculated with Rm1021, and there was a significant difference in the dry weight of the shoots and leaves between plants nodulated with Rm1021 and AL5 (Table 1). The root nodules formed by AL5 and Rm1021 had similar nitrogenase (acetylene reduction) activities (data not shown). The transductants had the same symbiotic phenotypes as the original mutant strains (data not shown).
The desiccation-sensitive strains were tested for sensitivity to salt and osmotic stress. AL5 and the AL5 transductant were inhibited in TY medium supplemented with 0.4 M NaCl, as indicated by a reduced growth curve slope between 6 and 24 h, while the other mutants grew similarly to Rm1021 (data not shown). In TY medium with 0.2 M NaCl, all the strains had growth rates similar to that of Rm1021. Conditions of high osmolarity were created by using TY medium supplemented with 0.2 M PEG 200 and 0.4 M PEG 200. All of the strains had similar growth characteristics under conditions of high osmolarity (data not shown).
relA mutant characterization.
Other tests were performed to further characterize the relA mutants from this study and others. The previously characterized S. meliloti relA mutant DW186 does not grow on defined medium in the presence of AT (41). AT is a histidine analog that induces histidine starvation, which induces RelA and, as a result, the stringent response (2). relA mutants cannot grow on medium with AT because the stringent response is not initiated. As with the previous relA mutant, our mutant (AL5) and the corresponding transductant had impaired growth on medium with AT (data not shown).
In addition to DW186, four suppressor mutants of DW186 have been identified (41, 42). These suppressor mutants have accumulated mutations in subunits of RNA polymerase that partially restore the wild-type phenotype (42). Since AL5 exhibited sensitivity to salt, we also tested DW186 and the four suppressor mutants for this phenotype. All of these strains exhibit salt sensitivity (data not shown). We also tested these mutants, along with Rm1021 and AL5, for desiccation sensitivity and found that all the mutants were sensitive to desiccation (data not shown).
Further characterization of uvr genes.
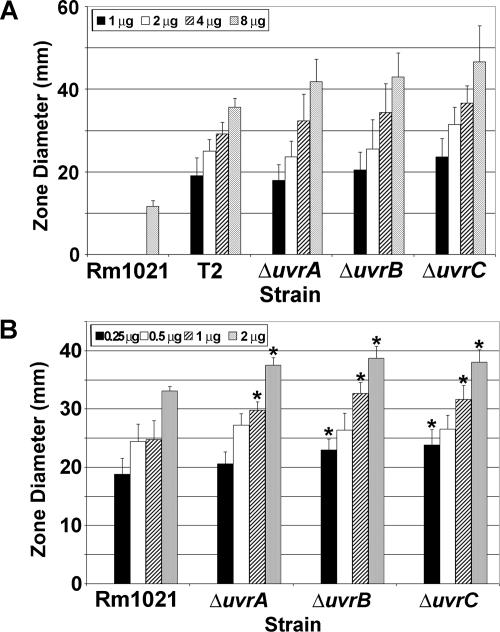
uvrC mutants of E. coli are very sensitive to UV light (37). Since the activity of the uvrC gene in S. meliloti had not been previously studied, the T2 mutant was exposed to UV light to determine if the mutation affected UV resistance (Table 3). Complete killing of Rm1021 occurred after exposure to 6.0 J/cm2 of UV light. Complete killing of T2 occurred after 0.2 J/cm2 of UV exposure, which means T2 is 30 times more sensitive than Rm1021. The increased UV sensitivity of mutant T2 confirms that uvrC plays a role in the repair of UV-induced DNA damage in Rm1021. Zone-of-inhibition assays were also performed by using 4-NQO, a compound that leads to adducts in DNA and has a sensitivity profile with DNA repair mutants similar to that of UV exposure (Fig. 2A). With 8 μg of 4-NQO, the zone of inhibition radius for T2 was 3.1 times larger than the zone for Rm1021. The transductant of T2 exhibited similar sensitivities to UV and 4-NQO (data not shown).
TABLE 3.
UV sensitivity of T2 and uvr gene deletion mutants
| Strain | Growtha after exposure to UV light at indicated no. of J/cm2
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.05 | 0.1 | 0.15 | 0.2 | 0.25 | 0.3 | 3.0 | 4.5 | 6.0 | |
| Rm1021 | + | + | + | + | + | + | + | + | +/− | − |
| T2 | + | + | + | +/− | − | − | − | − | − | − |
| Rm1021ΔuvrA | + | +/− | − | − | − | − | − | − | − | − |
| Rm1021ΔuvrB | + | +/− | − | − | − | − | − | − | − | − |
| Rm1021ΔuvrC | + | +/− | − | − | − | − | − | − | − | − |
+, solid growth; +/−, patchy growth; −, no growth.
FIG. 2.
Sensitivities of mutants to 4-NQO and mitomycin C. (A) 4-NQO was placed in the center of a plate as described in Materials and Methods. The average diameter (mm) of the zone of inhibition (n = 6) was plotted. All uvr gene mutants exhibited increased sensitivity to 4-NQO compared to that of wild-type Rm1021. There was no inhibition for any strain at 0 μg 4-NQO. (B) Sensitivity to mitomycin C was determined as indicated above. Zones of inhibition for 0 μg mitomycin C were zero for all strains. An asterisk indicates a significant difference (P < 0.005) between the results for the mutant and for Rm1021 in a Student's t test. Error bars show standard deviations.
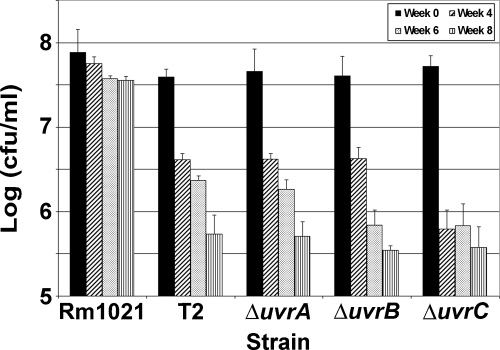
To study the link between the NER pathway and desiccation resistance in Rm1021 further, deletion mutants lacking uvrA (SMc01235), uvrB (SMc04231), or uvrC were constructed. Each mutant was slightly more sensitive to UV light than T2 was (Table 3). These data show that all three genes were critical for repairing UV-induced damage in Rm1021. Assays with 4-NQO were also done on the three uvr deletion mutants. The uvrA, -B, and -C deletion mutants had about the same zone size for 8 μg 4-NQO as observed for T2 (Fig. 2A). All three deletion mutants had similar zones of inhibition when tested with 2 μg of mitomycin C, and these were significantly different from that of Rm1021 (Fig. 2B). The uvr deletion mutants were also screened for desiccation sensitivity. Since T2 was sensitive to desiccation, we expected the uvr deletion mutants to exhibit similar sensitivities. As expected, all three of the uvr deletion mutants were desiccation sensitive (Fig. 3). Since the samples were not exposed to DNA-damaging agents during the drying assay, the DNA damage that is recognized by the NER pathway must be desiccation induced.
FIG. 3.
Sensitivity of uvr mutants to desiccation. The number of viable cells was measured at 0, 4, 6, and 8 weeks of drying for Rm1021, T2, and the uvrA, uvrB, and uvrC deletion mutants of Rm1021. All of the uvr mutant strains were sensitive to desiccation. Error bars show standard deviations.
DISCUSSION
For a soil bacterium, resistance to desiccation is likely to be an important survival trait. Some gram-positive soil bacteria, such as the Bacillus species, survive dry conditions by making spores (29). The gram-negative bacterium Azotobacter has an encystment process that promotes survival (32). Rhizobia are gram-negative bacteria that do not make spores or form cysts but can survive within the soil environment. Desiccation stress has been studied in Rhizobium species for many years, but only recently at the molecular level (6, 38). Some strains, such as Rm1021, are very desiccation resistant. After 2 months of drying, 33% of the Rm1021 cells were still viable (Fig. 1). For other strains, such as R. leguminosarum 3841, only 37% of the cells are still viable after 16 h of drying (9).
Vriezen et al. (39) described three stages of the desiccation process. Stage I is the drying of the cells, which results in the loss of water and the accumulation of salts and solutes and leads to osmotic stress. Metabolic processes are also slowed and eventually stopped due to the lack of water. The lack of water in the cells also leads to increased DNA damage since enzymes are not active to repair damage. Stage II is the storage phase, where the cells have lost all water and are just waiting to be rehydrated. Stage II is usually where the decline of cell viability occurs. The final step, stage III, is the rehydration of the cells. During this phase, bacterial metabolism is revived and enzymes repair the cell damage (39). The work presented in this report identified four genes that play a role in desiccation resistance in Rm1021. Three of these are regulatory genes (relA, rpoE2, and hpr), and one gene is involved in DNA repair (uvrC). The roles of the gene products and stages of drying in which they are most critical will be discussed below.
rpoE2, a stress-induced sigma factor, is critical during the drying phase (stage I) of desiccation. In a previous study, the S. meliloti rpoE2 gene was found to be upregulated during salt stress, entry into stationary phase after carbon or nitrogen starvation, and heat shock (26). Forty-four genes were shown to be induced by rpoE2 under heat stress conditions (26). In addition, a study of genes induced during desiccation in B. japonicum USDA 110 showed the upregulation of an rpoE gene (blr7797) (6) which has 61% identity with SMc01506, the S. meliloti Rm1021 rpoE2 gene. From the results of the above studies, it is plausible that RpoE2 in Rm1021 may also regulate genes involved in desiccation-induced stress. The results for mutant AL15 (Table 1) from this study demonstrate that rpoE2 is needed for desiccation resistance (Fig. 1).
The role of the hpr gene in a stage of desiccation was more difficult to identify. Since HPr is a regulator of succinate-mediated catabolite repression (1), it may be critical at any stage of the drying process. An hpr mutant of S. meliloti Rm1021 had altered carbon catabolism, lower survival in stationary phase, and early production of low-molecular-weight succinoglycan (1); these physiological changes may lead to increased desiccation susceptibility during drying. Alternatively, the change in succinoglycan production may provide less protection of the cell during the storage phase. The homolog of S. meliloti hpr (SMc02754) in B. japonicum, ptsH (blr8148), is upregulated during desiccation (6), indicating that hpr may be involved in the drying stage of desiccation. The results for the desiccation-sensitive hpr mutant (B15-4) from this work support the data for B. japonicum.
UvrC is essential to DNA repair by the NER pathway. The NER pathway targets lesions that distort the DNA helix, such as UV light-induced pyrimidine dimers and 6,4-photoproducts (36) or bulky adducts caused by the reaction of DNA with various chemicals, like 4-NQO. The NER pathway appears to be important for normal desiccation resistance in S. meliloti. A transposon insertion into uvrC (T2) and deletion mutations of uvrA, uvrB, or uvrC all resulted in strains that were desiccation sensitive (Fig. 3). The Rm1021 uvr mutants were also sensitive to UV light (Table 3) and 4-NQO (Fig. 2A), suggesting that all of these genes have functions in wild-type S. meliloti that correspond to their roles in repair in other bacteria. Supporting this, the uvrA, uvrB, and uvrC mutants also have increased sensitivity to mitomycin C (Fig. 2B), which has also been observed in E. coli uvrA, uvrB, and uvrC mutants (37). The mitomycin C sensitivity of wild-type Rm1021 was not as dramatic as the sensitivity observed with 4-NOQ. This difference may be due to the fact that mitomycin C predominately forms cross-links between the strands of a DNA helix and forms cross-links between bases on a single strand less often (35).
The data collected in this study show that the deletion of uvrA, -B, or -C results in increased sensitivity to desiccation (Fig. 3). Since most DNA damage probably occurs during stage II of drying, the uvr genes would be the most critical during stage III when the cells are being revived. uvrA, -B, and -C mutants cannot repair large DNA adducts, which lead to fatal mutations. During the desiccation assays in this work, the samples were not exposed to UV light or specific chemicals that induce DNA damage. Thus, while the desiccation-induced DNA damage is recognized by the NER pathway, the exact nature of the DNA lesions is unknown. Other DNA repair pathways may also be critical for desiccation resistance. Nine DNA repair and modification genes were induced under desiccating conditions in B. japonicum (6). We are currently investigating the role of other DNA repair genes in the desiccation resistance of S. meliloti Rm1021.
relA, which regulates the stringent response, is most likely involved in the drying phase (stage I) of desiccation. In a relA mutant, the stringent response is not activated and, as a result, the cell cannot adjust to starvation as quickly and goes through a period of no or very slow protein production (13). In the case of desiccation, the RelA-defective cell would most likely be unable to adjust to dry conditions because the lack of RelA activity does not allow the cell to switch to a “survival mode.” AL5, the RelA mutant isolated in this study, was desiccation sensitive, which indicates that RelA was needed for survival during desiccation. Four other RelA mutants have been characterized in S. meliloti and Rhizobium etli (4, 23, 40, 41). A defect in RelA activity was detected by sensitivity to AT or by the observation of low ppGpp accumulation for three of the mutants (4, 23, 41). AL5 was also unable to grow in the presence of AT, indicating that RelA was inactive (data not shown).
Wells and Long (41) identified a S. meliloti relA mutant, DW186, that was deficient in symbiosis. DW186 produces very few nodules, and those that do form are small and white. The symbiosis is ineffective and produces plants that look like uninoculated plants (41). Suppressor mutants of DW186, which have mutations in rpoC (DW362 and DW366) and rpoB (DW364 and DW371), were able to restore nodulation on alfalfa but did not produce an effective symbiosis (41, 42). The RelA mutant from our study, AL5, shared some phenotypes with DW186 and its suppressor mutants but differed in symbiotic effectiveness. AL5 produced a mix of nodule phenotypes, and the plants were reduced in size (Table 1), but the effect on symbiosis was not as severe as observed for DW186. We also tested DW186 and the four suppressor mutants for desiccation sensitivity, and all strains showed desiccation sensitivities similar to that of AL5 (data not shown). This supports the idea that the relA suppressors are not able to overcome all of the defects caused by the original relA mutation. The differences between DW186 and AL5 in symbiosis could be due to the type of mutation—DW186 is an internal deletion and AL5 is a transposon insertion.
A R. etli relA mutant, CMPG8705, exhibited a symbiotic phenotype that was less severe than that of S. meliloti DW186 and more similar to that of AL5 on bean plants (4, 23). CMPG8705 has 75% less nitrogen fixation activity than the wild type (CNPAF512) on bean plants. The leaves of the plants have a yellowish color but are not completely chlorotic, and the pigment difference is noticeable only at the later stages of symbiosis. CMPG8705 also nodulates beans at the same rate as the wild type, but the mutant-induced nodules had a slightly different color than wild-type nodules (23). The R. etli CNPAF512 (wild type of CMPG8705) RelA protein is 83% identical to RelA of Rm1021. CMPG8705 has an insertion in RelA at amino acid 137, which is close to the insertion in AL5 (Table 1), and the symbiotic phenotypes of these two mutants were similar. This indicates that a transposon mutation early in the protein (before amino acid 137) may be responsible for the difference in symbiotic effectiveness between AL5 and DW186, which has amino acids 182 to 495 deleted.
Another RelA mutant of S. meliloti, W11, was isolated because of a salt-sensitive phenotype (40). The RelA of W11 and parental strain S. meliloti 042BM is identical to RelA of Rm1021. AL5 was salt sensitive (data not shown). We also tested DW186 and the four suppressor mutants of DW186, and all of these strains were also salt sensitive (data not shown). Since mutant AL5 was the only desiccation-sensitive mutant from this study that was also sensitive to salt, and none of the strains were sensitive to osmotic stress, these data indicate that desiccation sensitivity does not always result in increased susceptibility to these other stresses.
The data from this study and previous studies (6, 9, 21) demonstrate that desiccation resistance of rhizobia is a complex process that involves many cellular activities. While it is easy to assign a role for the uvr genes in desiccation resistance, the three regulatory genes identified in this work (hpr, relA, and rpoE2) directly and indirectly control numerous cellular processes. As a result, pinpointing the exact role of these genes in desiccation resistance is difficult, and only further study of these genes and the genes they regulate will solve the mystery of desiccation resistance.
Acknowledgments
Washington State University undergraduate students Helen Yi, Jennifer Gruhn, Michelle (Mace) Collins, and Mary Wen provided technical assistance for the project.
This work was supported by grant DE-FG03-96ER20225 from the Energy Biosciences Program at the United States Department of Energy.
Footnotes
Published ahead of print on 21 November 2008.
REFERENCES
- 1.Arango Pinedo, C., R. M. Bringhurst, and D. J. Gage. 2008. Sinorhizobium meliloti mutants lacking phosphotransferase system enzyme HPr or EIIA are altered in diverse processes, including carbon metabolism, cobalt requirements, and succinoglycan production. J. Bacteriol. 190:2947-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker, A., H. Bergès, E. Krol, C. Bruand, S. Rüberg, D. Capela, E. Lauber, E. Meilhoc, F. Ampe, F. J. de Bruijn, J. Fourment, A. Francez-Charlot, D. Kahn, H. Küster, C. Liebe, A. Pühler, S. Weidner, and J. Batut. 2004. Global changes in gene expression in Sinorhizobium meliloti 1021 under microoxic and symbiotic conditions. Mol. Plant-Microbe Interact. 17:292-303. [DOI] [PubMed] [Google Scholar]
- 3.Beringer, J. E. 1974. R factor transfer in Rhizobium leguminosarum. Microbiology 84:188-198. [DOI] [PubMed] [Google Scholar]
- 4.Calderón-Flores, A., G. Du Pont, A. Huerta-Saquero, H. Merchant-Larios, L. Servín-González, and S. Durán. 2005. The stringent response is required for amino acid and nitrate utilization, Nod factor regulation, nodulation, and nitrogen fixation in Rhizobium etli. J. Bacteriol. 187:5075-5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chao, W.-L., and M. Alexander. 1984. Mineral soils as carriers for Rhizobium inoculants. Appl. Environ. Microbiol. 47:94-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cytryn, E. J., D. P. Sangurdekar, J. G. Streeter, W. L. Franck, W. Chang, G. Stacey, D. W. Emerich, T. Joshi, D. Xu, and M. J. Sadowsky. 2007. Transcriptional and physiological responses of Bradyrhizobium japonicum to desiccation-induced stress. J. Bacteriol. 189:6751-6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finan, T. M., E. Hartwieg, K. LeMieux, K. Bergman, G. C. Walker, and E. R. Signer. 1984. General transduction in Rhizobium meliloti. J. Bacteriol. 159:120-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibbons, R. J. 1964. Metabolism of intracellular polysaccharide by Streptococcus mitis and its relation to inducible enzyme formation. J. Bacteriol. 87:1512-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilbert, K. B., E. M. Vanderlinde, and C. K. Yost. 2007. Mutagenesis of the carboxy terminal protease CtpA decreases desiccation tolerance in Rhizobium leguminosarum. FEMS Microbiol. Lett. 272:65-74. [DOI] [PubMed] [Google Scholar]
- 10.Griffitts, J. S., and S. R. Long. 2008. A symbiotic mutant of Sinorhizobium meliloti reveals a novel genetic pathway involving succinoglycan biosynthetic functions. Mol. Microbiol. 67:1292-1306. [DOI] [PubMed] [Google Scholar]
- 11.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 12.Howard-Flanders, P., R. P. Boyce, and L. Theriot. 1966. Three loci in Escherichia coli K-12 that control the excision of pyrimidine dimers and certain other mutagen products from DNA. Genetics 53:1119-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jensen, K. F., and S. Pedersen. 1990. Metabolic growth rate control in Escherichia coli may be a consequence of subsaturation of the macromolecular biosynthetic apparatus with substrates and catalytic components. Microbiol. Mol. Biol. Rev. 54:89-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones, K. M., H. Kobayashi, B. W. Davies, M. E. Taga, and G. C. Walker. 2007. How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat. Rev. Microbiol. 5:619-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kneen, B. E., and T. A. LaRue. 1983. Congo red absorption by Rhizobium leguminosarum. Appl. Environ. Microbiol. 45:340-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knobloch, J. K. M., M. Nedelmann, K. Kiel, K. Bartscht, M. A. Horstkotte, S. Dobinsky, H. Rohde, and D. Mack. 2003. Establishment of an arbitrary PCR for rapid identification of Tn917 insertion sites in Staphylococcus epidermidis: characterization of biofilm-negative and nonmucoid mutants. Appl. Environ. Microbiol. 69:5812-5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leigh, J. A., E. R. Signer, and G. C. Walker. 1985. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc. Natl. Acad. Sci. USA 82:6231-6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marrs, B., and H. Gest. 1973. Genetic mutations affecting the respiratory electron transport system of the photosynthetic bacterium Rhodopseudomonas capsulata. J. Bacteriol. 114:1045-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mary, P., N. Dupuy, C. Dolhem-Biremon, C. Defives, and R. Talliez. 1994. Differences among Rhizobium meliloti and Bradyrhizobium japonicum strains in tolerance to desiccation and storage at different relative humidities. Soil Biol. Biochem. 26:1125-1132. [Google Scholar]
- 20.Mary, P., D. Ochin, and R. Talliez. 1985. Rates of drying and survival of Rhizobium meliloti strains during storage at different relative humidities. Appl. Environ. Microbiol. 50:207-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McIntyre, H. J., H. Davies, T. A. Hore, S. H. Miller, J.-P. Dufour, and C. W. Ronson. 2007. Trehalose biosynthesis in Rhizobium leguminosarum bv. trifolii and its role in desiccation tolerance. Appl. Environ. Microbiol. 73:3984-3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meade, H. M., S. R. Long, G. B. Ruvkun, S. E. Brown, and M. Ausubel. 1982. Physical and genetic characteristics of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moris, M., K. Braeken, E. Schoeters, C. Verreth, S. Beullens, J. Vanderleyden, and J. Michiels. 2005. Effective symbiosis between Rhizobium etli and Phaseolus vulgaris requires the alarmone ppGpp. J. Bacteriol. 187:5460-5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Revellin, C., G. Meunier, J. J. Giraud, G. Sommer, P. Wadoux, and G. Catroux. 2000. Changes in the physiological and agricultural characteristics of peat-based Bradyrhizobium japonicum inoculants after long-term storage. Appl. Microbiol. Biotechnol. 54:206-211. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 26.Sauviac, L., H. Philippe, K. Phok, and C. Bruand. 2007. An extracytoplasmic function sigma factor acts as a general stress response regulator in Sinorhizobium meliloti. J. Bacteriol. 189:4204-4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 28.Schroeder, B. K., B. L. House, M. W. Mortimer, S. N. Yurgel, S. C. Maloney, K. L. Ward, and M. L. Kahn. 2005. Development of a functional genomics platform for Sinorhizobium meliloti: construction of an ORFeome. Appl. Environ. Microbiol. 71:5858-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Setlow, P. 1995. Mechanisms for the prevention of damage to DNA in spores of Bacillus species. Annu. Rev. Microbiol. 49:29-54. [DOI] [PubMed] [Google Scholar]
- 30.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat. Biotechnol. 1:784-791. [Google Scholar]
- 31.Simon, R., J. Quandt, and W. Klipp. 1989. New derivatives of transposon Tn5 suitable for mobilization of replicons, generation of operon fusions and induction of genes in Gram-negative bacteria. Gene 80:161-169. [DOI] [PubMed] [Google Scholar]
- 32.Socolofsky, M. D., and O. Wyss. 1962. Resistance of the Azotobacter cyst. J. Bacteriol. 84:119-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Somerville, J. E., and M. L. Kahn. 1983. Cloning of the glutamine synthetase I gene from Rhizobium meliloti. J. Bacteriol. 156:168-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas, D. C., I. Husain, S. G. Chaney, G. B. Panigrahi, and I. G. Walker. 1991. Sequence effect on incision by (A)BC excinuclease of 4NQO adducts and UV photoproducts. Nucleic Acids Res. 19:365-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomasz, M. 1995. Mitomycin C: small, fast and deadly (but very selective). Chem. Biol. 2:575-579. [DOI] [PubMed] [Google Scholar]
- 36.Van Houten, B. 1990. Nucleotide excision repair in Escherichia coli. Microbiol. Rev. 54:18-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vidal, L. S., L. B. Santos, C. Lage, and A. C. Leitão. 2006. Enhanced sensitivity of Escherichia coli uvrB mutants to mitomycin C points to a UV-C distinct repair for DNA adducts. Chem. Res. Toxicol. 19:1351-1356. [DOI] [PubMed] [Google Scholar]
- 38.Vriezen, J. A. C., F. J. de Bruijn, and K. Nüsslein. 2006. Desiccation responses and survival of Sinorhizobium meliloti USDA 1021 in relation to growth phase, temperature, chloride and sulfate availability. Lett. Appl. Microbiol. 42:172-178. [DOI] [PubMed] [Google Scholar]
- 39.Vriezen, J. A. C., F. J. de Bruijn, and K. Nüsslein. 2007. Responses of rhizobia to desiccation in relation to osmotic stress, oxygen, and temperature. Appl. Environ. Microbiol. 73:3451-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei, W., J. Jiang, X. Li, L. Wang, and S. S. Yang. 2004. Isolation of salt-sensitive mutants from Sinorhizobium meliloti and characterization of genes involved in salt tolerance. Lett. Appl. Microbiol. 39:278-283. [DOI] [PubMed] [Google Scholar]
- 41.Wells, D. H., and S. R. Long. 2002. The Sinorhizobium meliloti stringent response affects multiple aspects of symbiosis. Mol. Microbiol. 43:1115-1127. [DOI] [PubMed] [Google Scholar]
- 42.Wells, D. H., and S. R. Long. 2003. Mutations in rpoBC suppress the defects of a Sinorhizobium meliloti relA mutant. J. Bacteriol. 185:5602-5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yurgel, S. N., J. Berrocal, C. Wilson, and M. L. Kahn. 2007. Pleiotropic effects of mutations that alter the Sinorhizobium meliloti cytochrome c respiratory system. Microbiology 153:399-410. [DOI] [PubMed] [Google Scholar]
- 44.Yurgel, S. N., and M. L. Kahn. 2005. Sinorhizobium meliloti dctA mutants with partial ability to transport dicarboxylic acids. J. Bacteriol. 187:1161-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]