Abstract
The detection and molecular characterization of pathogenic human viruses in urban sewage have been used extensively to derive information on circulating viruses in given populations throughout the world. In this study, a similar approach was applied to provide an overview of the epidemiology of waterborne gastroenteritis viruses circulating in urban areas of Caracas, the capital city of Venezuela in South America. Dry season sampling was conducted in sewers and in a major river severely polluted with urban sewage discharges. Nested PCR was used for detection of human adenoviruses (HAds), while reverse transcription plus nested or seminested PCR was used for detection of enteroviruses (HuEVs), rotaviruses (HRVs), noroviruses (HuNoVs), and astroviruses (HAstVs). HRVs were fully characterized with genotype-specific primers for VP4 (genotype P), VP7 (genotype G), and the rotavirus nonstructural protein 4 (NSP4). HuNoVs and HAstVs were characterized by sequencing and phylogenetic analysis. The detection rates of all viruses were ≥50%, and all sampling events were positive for at least one of the pathogenic viruses studied. The predominant HRV types found were G1, P[8], P[4], and NSP4A and -B. Genogroup II of HuNoVs and HAstV type 8 were frequently detected in sewage and sewage-polluted river waters. This study reveals relevant epidemiological data on the distribution and persistence of human pathogenic viruses in sewage-polluted waters and addresses the potential health risks associated with transmission of these viruses through water-related environmental routes.
Waterborne viral pathogens have a large socioeconomic impact in both developed and developing nations, but the magnitude of the impact and the burden of viral disease (i.e., severity and prevalence) are more severe in regions of the world with highly polluted environments (19, 47). Viral gastroenteritis resulting from exposure to contaminated drinking and recreational waters has been reported worldwide (28). The viruses of greatest significance in disease transmission by environmental water routes are shed with human fecal wastes, and their numbers and distribution in sewage-polluted waters depend both on the burden of viral disease in the population and on the availability of municipal sewage treatment processing (21, 51).
Gastrointestinal illnesses are the most common acute illnesses occurring among the different population strata living in the metropolitan area of Caracas, the capital city of Venezuela in South America. The privileged social strata of high-income families live in urbanized areas with adequate sanitation services, while the unprivileged social strata live in informal settlements with no access to basic sanitation services. In Venezuela, rotaviruses are the most frequent viral agents associated with gastrointestinal disease in children less than 5 years of age, independent of the socioeconomic status, with attack rates of 300 deaths and 39,000 hospitalizations per year (38). Past and current investigations of enteric viral infections have focused on the epidemiology and control of rotavirus (20, 35, 36, 37, 38, 55). Less attention has been given to other human pathogenic viruses that may be responsible for gastrointestinal disease in Venezuela, although a few studies exist for human noroviruses (HuNoVs), adenoviruses (HAds), hepatitis A virus, and astroviruses (HAstVs) (17, 30, 45, 46). Consequently, information on the burden of viral gastroenteritis in the Venezuelan population is incomplete, and not much research has been conducted on the potential transmission of human pathogenic viruses through water-related environmental routes.
The potential public health problems associated with viral pollution from discharge of treated and untreated sewage into receiving watersheds have not been extensively studied in Venezuela. The treatment of wastewater in Venezuela has been considered below average within the Latin American and Caribbean regions. Previous estimates indicated that more than 97% of the country's raw sewage was discharged as effluent into the environment (15). Numerous environmental problems linked to municipal sewage discharges persist because of the lack of investment in wastewater treatment systems or the failure of existing ones. This problem is compounded by additional pollution issues associated with informal settlements where wastes are dumped, without any control, into rivers and coastal environments.
The detection and molecular characterization of human pathogenic viruses in urban sewage have been used extensively to derive information on circulating viruses in given populations throughout the world (7, 40, 42, 58). In this study, a similar approach has been applied to provide an overview of the epidemiology of significant waterborne gastroenteritis viruses circulating in urban areas of Caracas. Additionally, this study addresses the public health risks associated with human pathogenic viruses transmitted through water-related environmental routes. For these purposes, the following two sets of samples were analyzed: (i) domestic sewage samples collected from urbanized areas with <1,000 middle- to high-income households connected to sewer systems and (ii) a river network in Caracas with severe sewage pollution resulting from the confluence of urban sewers of >40,000 households, including sewage from informal, high-population-density urban settlements.
The presence of human enteroviruses (HuEVs) and HAds was used to compare data on the occurrence of enteric viruses extensively studied in sewage-polluted aquatic environments worldwide. The detection and genotyping of human rotaviruses (HRVs) were used to seek a relationship between environmental occurrence and clinical isolates characterized in previous studies conducted in Venezuela. The genetic diversity of HuNoVs and HAstVs was determined by nucleotide sequencing and phylogenetic analysis of PCR products in order to identify genotype persistence and distribution in sewage-polluted waters. To the best of our knowledge, this is the first study of the molecular epidemiology of waterborne gastroenteritis viruses circulating in this geographical region.
MATERIALS AND METHODS
Sewage and sewage-polluted river water.
During a 6-month sampling period, domestic sewage and sewage-polluted river water samples (40 ml) were collected two to four times per month. Domestic sewage samples were derived from sewers of approximately 1,000 households from middle- to high-income families living in urbanized areas connected to both sewer and municipal water supply systems. Sewage-polluted river water samples were collected from the Guaire River, which is a highly polluted river (>106 fecal coliforms/100 ml) that receives uncontrolled sewage discharges from urbanized areas in addition to urban wastes from informal settlements and storm water runoff. The Guaire River is canalized and crosses the greater metropolitan area of the city of Caracas in a southeast direction. The Guaire River has a length of 72 km and is a major affluent of the River Tuy, which is a major river that empties into the Caribbean Sea. The Guaire River functions as the main collector of urban sewers from metropolitan Caracas, collecting along a 53-km length the domestic sewage generated by a population of approximately 5,000,000 inhabitants. The sampling period covered during the study corresponded to the dry season (October through March); thus, the pollution loads to Guaire River represented urban sewage discharges without any contribution from storm water runoff.
Recovery of viral particles and nucleic acid extraction.
The concentration of viral particles was accomplished by ultracentrifugation and elution with 0.25 N glycine buffer, following procedures adapted from Pina et al. (41). Briefly, 35 ml of each water sample was ultracentrifuged at 140,000 × g using an SW28 rotor for 2 h 30 min at 4°C in a Beckman ultracentrifuge. Elution of viral particles was achieved by adding 5 ml of 0.25 N glycine buffer (pH 9.5) to the sediment and incubating on ice for 30 min. The solution was neutralized by adding 5 ml of 2× phosphate-buffered saline. The suspended solids were removed by centrifugation (12,000 × g for 15 min), and the viruses were finally recovered by ultracentrifugation at 229,600 × g for 1 h at 4°C in an SW41 Ti rotor. Viral particles were resuspended in 100 μl of 1× phosphate-buffered saline and immediately processed for nucleic acid extraction or were stored at −80°C until use.
Viral RNA was extracted from sample concentrates with Trizol (Invitrogen, Inc., Carlsbad, CA), following the manufacturer's instructions. RNA was finally resuspended in 50 μl of RNase-free H2O. Following RNA extraction, guanidinthiocyanate was used to extract the DNA. Briefly, after removing the aqueous phase containing RNA, the tubes were spun at 12,000 × g for 5 min at 4°C and the aqueous phase was removed to avoid RNA contamination. Back extraction buffer (250 μl), consisting of 4 M guanidinthiocyanate, 50 mM sodium citrate, and 1 M Tris, was added and mixed intensively for at least 3 min. Tubes were then centrifuged at 12,000 × g for 30 min at room temperature. The upper aqueous phase containing the DNA was transferred to a new tube. DNA was precipitated with isopropanol and washed with 70% ethanol. DNA was finally resuspended in 50 μl of DNase-free H2O and stored at −80°C until use.
Molecular detection and characterization of enteric viruses.
The detection and characterization of waterborne gastroenteritis viruses were performed with a combination of several molecular techniques. Nested PCR was used for detection of HAds, while reverse transcription-PCR (RT-PCR) and nested or seminested PCR was used for detection of HuEVs, HRVs, HuNoVs, and HAstVs. For RT, 39 μl of extracted RNA was incubated with 1 μl of RNasin at 95°C for 5 min and placed immediately on ice for RNA denaturalization. All 40 μl was added to an RT-PCR Master Mix for a final volume of 70 μl containing 1× RT buffer, 5 mM MgCl2, 300 nM of random primers, 0.3 mM deoxynucleoside triphosphate, and 800 U reverse transcriptase SuperScript II. All reagents were purchased from Invitrogen (Carlsbad, CA). Multiplex PCR with genotype-specific primers for VP4 (genotype P), VP7 (genotype G), and the rotavirus nonstructural protein 4 (NSP4) was used for detection and characterization of HRVs. The molecular characterization of HuNoV and HAstV was performed by sequencing and phylogenetic analysis of the second round of PCR amplicons. For the first PCR round, 5 μl of DNA, equivalent to 3.5 ml of the original sample, or 5 μl of cDNA, equivalent to 2 ml of water, was added to a reaction mixture (25 μl) consisting of 1× PCR buffer, 0.2 mM deoxynucleoside triphosphate, 0.2 μM of each primer, 2.5 U Taq polymerase, and 1 to 2 mM MgCl2. For the second PCR round, the same concentration of reagents was used with 1 μl of the first PCR product added to the PCR tube. Primer sequences and positions, MgCl2 final concentrations, and cycling conditions for detection and characterization of each viral group are shown in Table 1. Positive and negative controls (clinical samples for each virus type and RNA/DNA-free water) were included in all PCR runs. PCR products were analyzed by gel electrophoresis on a 2% strength agarose gel.
TABLE 1.
Primers and amplification conditions for detection and molecular characterization of waterborne gastroenteritis viruses
| Viruses | Protein(s) (genotype) | Round of PCR | Primer | Sequence (5′-3′) | Cycling protocola | MgCl2 concn (mM) | Location of amplicon (bp positions) | Amplicon size (bp) | Reference(s) |
|---|---|---|---|---|---|---|---|---|---|
| HRVs | VP7 (G) | 1st | VP7 F | ATGTATGGTATTGAATATACCAC | A | 1 | 49-71 | ||
| VP7 (G) | 1st | VP7 R | AACTTGCCACCATTTTTTCC | A | 1 | 914-933 | 884 | 21 | |
| VP7 (G) | 2nd (multiplex) | VP7 R | AACTTGCCACCATTTTTTCC | B | 1 | 914-933 | |||
| G1 | 2nd (multiplex) | aBT1 | CAAGTACTCAAATCAATGATGG | B | 1 | 314-335 | 619 | ||
| G2 | 2nd (multiplex) | ACT2 | CAATGATATTAACACATTTTCTGTG | B | 1 | 411-435 | 522 | ||
| G3 | 2nd (multiplex) | G3 | ACGAACTCAACACGAGAGG | B | 1 | 250-269 | 683 | ||
| G4 | 2nd (multiplex) | aDT4 | CGTTTCTGGTGAGGAGTTG | B | 1 | 480-499 | 453 | ||
| G8 | 2nd (multiplex) | aAT8 | GTCACACCATTTGTAAATTCG | B | 1 | 178-198 | 755 | ||
| G9 | 2nd (multiplex) | aFT9 | CTAGATGTAACTACAACTAC | B | 1 | 757-776 | 176 | ||
| G10 | 2nd (multiplex) | G10 | ATGTCAGACTACARATACTGG | B | 1 | 666-687 | 267 | 16, 21, 23 | |
| VP4 (P) | 1st | A | TGGCTTCGTTCATTTATAGACA | C | 1.5 | 11-32 | |||
| VP4 (P) | 1st | B | CTAAATGCTTTTGAATCATCCCA | C | 1.5 | 1071-1094 | 1,084 | ||
| P(8) | 2nd (multiplex) | C | ATATTCCTACGAGTTTAGTATC | C | 1.5 | 487-508 | 498 | ||
| P(4) | 2nd (multiplex) | D | ACTAACATGTGGTTCAACTGCGAT | C | 1.5 | 325-348 | 338 | ||
| P(6) | 2nd (multiplex) | E | CTGAGCACGTTGATAAGTCAC | C | 1.5 | 733-755 | 745 | ||
| P(9) | 2nd (multiplex) | 4T-1 | TGAGACATGCAATTGGAC | C | 1.5 | 385-402 | 391 | ||
| P(14) | 2nd (multiplex) | SE-1 | CTCTGCTACTCTACCTATTTG | C | 1.5 | 271-291 | 281 | 2, 14, 60 | |
| NSP4 | 1st | SE | GGCTTTTAAAAGTTCTGTTCCGAG | D | 2 | 1-24 | |||
| NSP4 | 1st | AS | GGTCACATCAAGACCATTCC | D | 2 | 730-750 | 750 | 9, 48 | |
| NSP4A, -B, and -C | 2nd (multiplex) | NSP4FW | GGAATGGCGTATTTTCC | E | 2 | 126-142 | |||
| NSP4A | 2nd (multiplex) | NSP4A | TGTTCTTTGTAACCTGTC | E | 2 | 286-305 | 179 | ||
| NSP4B | 2nd (multiplex) | NSP4B | CTTGCGGTGAAGAGTTCGG | E | 2 | 605-623 | 497 | ||
| NSP4C | 2nd (multiplex) | NSP4C | TTAAATTATCAGCATATCATGAATTCG | E | 2 | 426-452 | 326 | 48 | |
| HuEVs | 1st | EV1 | CGGCCCCTGAATGCGGC | F | 2 | 450-466 | |||
| 1st | EV2 | CACCGGATGGCCAATCCA | F | 2 | 643-626 | 196 | 24 | ||
| 2nd | EVD2 | CCCCTGAATGCGGCTAAT | G | 1 | 454-471 | ||||
| 2nd | EVU2 | ATTGTCACCATAAGCAGCCA | G | 1 | 600-581 | 146 | 24 | ||
| HuNoVs | 1st | MJV12 | TAYCAYTATGATGCHGAYTA | H | 2 | 4553-4572 | |||
| 1st | RegA | CTCRTCATCICCATARAAIGA | H | 2 | 4859-4879 | 327 | 59 | ||
| 2nd (multiplex) | p290 | GATTACTCCAAGTGGGACTCCAC | I | 2 | 4568-4590 | ||||
| 2nd (multiplex) | Mp290 | GATTATACTSSMTGGGAYTCMAC | I | 2 | 4568-4590 | ||||
| 2nd (multiplex) | rev SR46 | CCAGTGGGCGATGGAATTCCA | I | 2 | 4754-4773 | 204 | |||
| 2nd (multiplex) | rev SR48-52 | CCARTGRTTTATRCTGTTCAC | I | 2 | 4754-4773 | 204 | 1, 25, 32 | ||
| HAstVs | 1st | Mon 340 | CGTCATTATTTGTTGTCATACT | J | 2 | 1182-1203 | |||
| 1st | Mon 348 | ACATGTGCTGCTGTTACTATG | J | 2 | 1450-1470 | 289 | 5 | ||
| 2nd | Mon 394 | GARATCCGTGATGCTAATGG | K | 2 | 1250-1269 | ||||
| 2nd | Mon 348 | ACATGTGCTGCTGTTACTATG | K | 2 | 1450-1470 | 220 | 6 | ||
| HAds | 1st | hexAA1885 | GCCGCAGTGGTCTTACATGCACATC | L | 1.5 | 18858-18883 | |||
| 1st | hexAA1913 | CAGCACGCCGCGGATGTCAAAGT | L | 1.5 | 19136-19158 | 301 | 41 | ||
| 2nd | nehexAA1893 | GCCACCGAGACGTACTTCAGCCTG | L | 1.5 | 18937-18960 | ||||
| 2nd | nehexAA1905 | TTGTACGAGTACGCGGTATCCTCGCGGTC | L | 1.5 | 19051-19079 | 143 | 41 |
A, 95°C for 2 min; 40 cycles of 94°C for 1 min, 56°C for 30 s, and 72°C for 1 min; and 72°C for 10 min; B, 94°C for 2 min; 40 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 1 min; and 72°C for 10 min; C, 95°C for 2 min; 40 cycles of 94°C for 1 min, 54°C for 30 s, and 72°C 1 min; and 72°C for 10 min; D, 95°C for 5 min; 25 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 1 min; and 72°C for 10 min; E, 95°C for 5 min; 25 cycles of 94°C for 30 s, 44°C for 30 s, and 72°C for 30 s; and 72°C for 10 min; F, 94°C for 2 min; 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s; and 72°C for 10 min; G, 94°C 2 min; 40 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s; and 72°C for 10 min; H, 94°C for 2 min; 40 cycles of 94°C 1 min, 37°C for 1 min, and 72°C for 1 min; and 72°C for 10 min; I, 94°C for 2 min; 40 cycles of 94°C for 30 s, 49°C for 1 min, and 72°C for 30 s; and 72°C for 10 min; J, 94°C for 2 min; 30 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 30 s; and 72°C for 10 min; K, 94°C for 2 min; 40 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 30 s; and 72°C for 10 min; L, 94°C for 4 min; 30 cycles of 94°C for 90 s, 55°C for 90 s, and 72°C for 2 min; and 72°C for 10 min.
Sequencing and phylogenetic analysis.
PCR products obtained from the second round of amplification for HuNoVs and HAstVs were excised from the gel and sequenced in both directions using the BigDye Terminator cycle chemistry and 3130XL DNA analyzer (Applied Biosystems, Foster City, CA). The sequences obtained were assembled using the DNAMan software 5.2.2 (Lynnon Biosoft, Québec, Canada) and manually corrected after electropherogram observation. Sequences were aligned with the ClustalW method, and phylogenetic analyses were conducted using MEGA4 software (54). The evolutionary distances were inferred by the neighbor-joining method (49) using a bootstrap test of 2,000 replicates to assess tree reliability.
Nucleotide sequence accession numbers.
The nucleotide sequences corresponding to fragments of the RNA polymerase gene of HuNoVs and the ORF1 genome of HAstVs have been deposited in the GenBank database under accession no. FJ430532 to FJ430549 and FJ430550 to FJ430562, respectively.
RESULTS
Virus distribution in sewage and sewage-polluted river waters.
In the present study, epidemiological features of waterborne gastroenteritis viruses circulating in urban areas of Caracas were investigated with a combination of molecular methods. Sewage and sewage-polluted river waters were selected for molecular surveillance of these viruses. The distribution of viruses recovered from urban sewage is summarized in Table 2. At the sewage sites, HuEVs and HuNoVs were found in 75% (9 of 12) of the samples, HRVs and HAstVs in 67% (8 of 12) of the samples, and HAds in 50% (6 of 12) of the samples. Table 3 summarizes the distribution of waterborne gastroenteritis viruses in the sewage-polluted Guaire River. For the 18 samples collected, the detection rates were 89% (16 of 18) for HAstVs, 83% (15 of 18) for HAds and HRVs, and 72% (13 of 18) for HuEVs and HuNoVs.
TABLE 2.
Distribution of waterborne gastroenteritis viruses in domestic sewage from the city of Caracas, Venezuelaa
| Sample collection date (sample designation) | HRVs
|
HuEVs | HAds | HuNoVs | HAstVs | ||
|---|---|---|---|---|---|---|---|
| VP4 | VP7 | NSP4 | |||||
| October 2007 (SewCI) | P[8], P[4] | G1, G9 | A, B | + | + | + | − |
| October 2007 (SewVillI) | P[4] | G1 | A, B | + | + | + | + |
| October 2007 (SewIvicI) | NT | G1 | A | + | + | + | − |
| October 2007 (SewCCI) | P[8] | G1 | B | + | + | + | − |
| November 2007 (SewCII) | P[4] | NT | NT | + | − | + | + |
| December 2007 (SewCIII) | P[4] | NT | NT | + | − | + | + |
| December 2007 (SewCIV) | P[4] | G1 | NT | + | − | + | + |
| January 2008 (SewCV) | − | − | − | + | − | − | + |
| January 2008 (SewCVI) | − | − | − | − | + | + | − |
| February 2008 (SewCVII) | − | − | − | − | − | − | + |
| February 2008 (SewCVIII) | − | − | − | − | − | − | + |
| February 2008 (SewCIX) | P[4] | G1 | B | + | + | + | + |
+, found; −, not found; NT, nontypeable.
TABLE 3.
Distribution of waterborne gastroenteritis viruses in the highly sewage-polluted Guaire River in Caracas, Venezuelaa
| Sample collection date (sample designation) | HRVs
|
HuEVs | HAds | HuNoVs | HAstV | ||
|---|---|---|---|---|---|---|---|
| VP4 | VP7 | NSP4 | |||||
| October 2007 (GuRI) | P[4] | NT | B, C | + | + | − | − |
| October 2007 (GuRII) | P[4] | G1, G10 | B | + | + | + | − |
| November 2007 (GuRIII) | NT | NT | B | − | + | − | + |
| December 2007 (GuRIV) | P[4] | G1 | B | + | + | + | + |
| December 2007 (GuRV) | P[4] | NT | B | + | + | + | + |
| January 2008 (GuRVI) | NT | G1 | NT | − | − | − | + |
| January 2008 (GuRVII) | − | − | − | − | − | + | + |
| February 2008 (GuRVIII) | − | − | − | − | + | − | + |
| February 2008 (GuRIX) | − | − | − | − | − | − | + |
| February 2008 (GuRX) | P[4] | G1 | B, C | + | + | + | + |
| February 2008 (GuRXI) | P[4], P[8] | G1 | B, A | + | + | + | + |
| February 2008 (GuRXII) | P[4], P[8] | G1 | B, A | + | + | + | + |
| March 2008 (GuRXIII) | P[4], P[8] | G1 | B | + | + | + | + |
| March 2008 (GuRXIV) | P[4], P[8] | G1 | B, A | + | + | + | + |
| April 2008 (GuRXV) | P[4] | G1 | B, A | + | + | + | + |
| April 2008 (GuRXVI) | P[4] | G1 | B, A | + | + | + | + |
| April 2008 (GuRXVII) | P[4], P[8] | G1 | B, A | + | + | + | + |
| April 2008 (GuRXVIII) | P[4], P[8] | G1 | B, A | + | + | + | + |
+, found; −, not found; NT, nontypeable.
Molecular characterization of HRVs.
HRVs and HAstVs were the waterborne gastroenteritis viruses most frequently detected both in sewage and in the highly polluted Guaire River. Twenty-three samples that were positive for HRVs represented 77% (23 of 30) of the total samples processed and included both sewage and sewage-polluted river waters. In 23% (7 of 30) of the samples, none of the three rotaviral genes was detected, and these samples were therefore negative.
The distribution and predominance of HRV G, P, and NSP4 genotypes varied among the two sets of samples analyzed (Table 2). The frequencies of detection of G and P types in urban sewage were relatively similar, with genotypes G1 and P[4] occurring in 50% (6 of 12) of the samples. The frequency of detection of the P[8] type was 17% (2 of 12). A G9 genotype strain was detected only once, along with the G1 genotype. NSP4 genotypes A and B were more frequently found in sewage.
The distribution of predominant HRV G, P, and NSP4 genotypes in the Guaire River is shown in Table 3. The most frequent G type detected was again genotype G1, found in 66% (12 of 18) of the samples. The G10 type was detected only once, along with the predominant G1 type. The frequencies of detection of P genotypes were 72% (13 of 18) for P[4], followed by 33% (6 of 13) for the P[8] type. NSP4B was present in 78% (14 of 18) of the samples and was the predominant genotype, followed by genotype NSP4A, which was found in 39% (7 of 18) of the samples, always in combination with genotype NSP4B. The genotype NSP4C, mostly associated with infections in animals, was detected twice, along with genotype NSP4B. Four samples from the Guaire River could not be typed for the NSP4 gene.
Molecular characterization of HuNoVs and HAstVs.
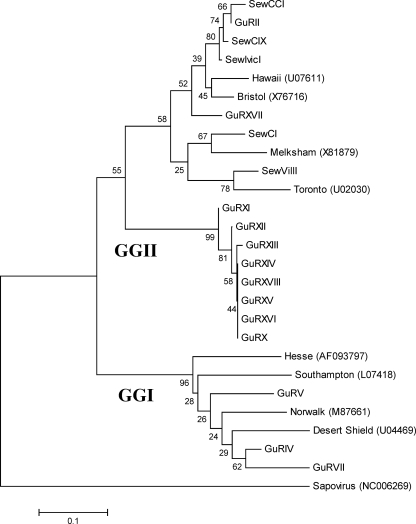
The most predominant genogroup of HuNoVs found in both sewage and Guaire River belonged to genogroup II (GGII), found in 83% (15 of 18) of sequenced samples (Fig. 1). Although not all of the samples that were positive for HuNoVs could be successfully sequenced, due mainly to insufficient PCR products, a slightly different distribution of the HuNoV genogroups was observed. All HuNoV sequences found in sewage belonged exclusively to GGII, while the sequences detected in the Guaire River were distributed between the two genogroups; 23% (3 of 13) of the sequences were similar to GGI, while 77% (10 of 13) belonged to GGII.
FIG. 1.
Phylogenetic analysis of the HuNoV sequences recovered from sewage and the sewage-polluted river. The following reference strains were used in the analysis: Norwalk (M87661), Southampton (L07418), and Hesse (AF093797) for HuNoV GGI; and Hawaii (U07611), Melksham (X81879), Bristol (X7616), and Toronto (U02030) for HuNoV GGII. Sapovirus sequence NC006269 was used as an out group. The phylogenetic analysis was based on a 112-nucleotide fragment of the RNA polymerase gene.
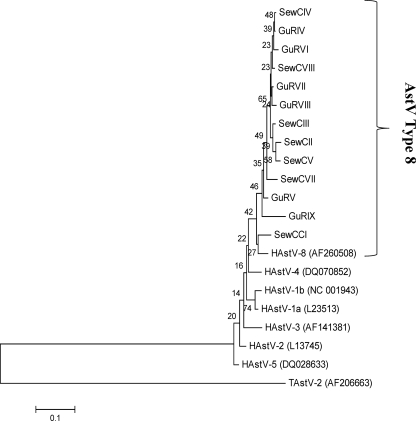
HAstVs were among the most predominant viral types found both in sewage and in the sewage-polluted Guaire River during the sampling period covered in this research. However, more HAstV sequences were recovered from the Guaire River than from sewers. The detection rate of HAstVs in the Guaire River was 89% (16 of 18), while 67% (8 of 12) of HAstVs corresponded to sewage samples. Thirteen of 23 samples positive for HAstVs were successfully analyzed, and all the sequences obtained from sewage and from the Guaire River clustered together with HAstV type 8 (Fig. 2).
FIG. 2.
Phylogenetic analysis of the HAstV sequences recovered from sewage and the sewage-polluted river. The following type 1 to type 8 HAstV reference strains were used in the analysis: HAstV-1a (L23513), HAstV-1b (NC001943), HAstV-2 (L13745), HAstV-3 (AF141381), HAstV-5 (DQ028633), and HAstV-8 (AF260508). The Turkey HAstV sequence, TAstV-2 (AF206663), was used as an out group. The phylogenetic analysis was based on a 219-nucleotide fragment of the open reading frame 1 genome of HAstVs.
DISCUSSION
The present study provides an overview of the epidemiology of waterborne gastroenteritis viruses circulating in the metropolitan city of Caracas, the capital city of Venezuela in South America. The presence of all viruses was investigated by molecular methods, which allowed both sensitive and precise identification of predominant human pathogenic viruses occurring in urban sewers and in the Guaire River, which crosses the city of Caracas and receives raw sewage discharges along much of its length. Molecular surveillance of waterborne gastroenteritis viruses was possible by examining sample volumes of 35 ml, which allowed us to obtain location-specific data on predominant types of viruses of public health concern. There were no previous investigations addressing the presence and genetic diversity of multiple pathogenic HuEVs in this geographical region.
The detection of HuEVs and HAds allowed us to compare occurrence data of enteric viruses in Caracas and in sewage and sewage-polluted aquatic environments worldwide. Our results indicate a relatively high prevalence of HuEVs and HAds in the sewage-polluted waters of Caracas, similar to the results derived from other geographical regions (3, 27, 32, 41, 42, 43, 44, 52). Overall, the most frequently detected viruses in the two set of samples analyzed were HuEVs, HRVs, and HAstVs. The only major difference between sampling sites was the more frequent detection of the enteric viruses in the highly polluted Guaire River, which receives uncontrolled household waste from underserved central and marginal urban areas of Caracas. The organic and pathogen loads this river receives are from a population of 5,000,000 inhabitants, mostly low-income households.
HRVs are among the most predominant viral agents responsible for infantile acute gastroenteritis in Venezuela; therefore, these viruses are considered of epidemiological importance in this country. The PCR-based methods applied in this study allowed full characterization of predominant group A RVs in urban sewage and in the highly sewage-polluted river using genotype-specific primers for the VP7, VP4, and NSP4 genes. The predominance of G1 strains in these samples is in accordance with the results from a previous survey conducted in stool specimens of children from an urban area of Caracas (20). G1 plays a major role in infantile diarrhea worldwide and is the HRV genotype most frequently isolated from sewers in other geographical regions (4, 12, 33, 34, 50, 57, 58). Similarly, the frequent isolation of HRV strains P[4] and P[8] in our study corresponds well with the reported occurrence and distribution of these pathogenic strains worldwide (33). The finding of a G9 strain in one sewage site sampled indicates that the circulation of such a strain may not be uncommon in Venezuela, which is not surprising considering the global emergence of the G9 strain in clinical cases as reported by researchers from other countries (33, 50, 57). There were also prevalent combinations of G and P types occurring simultaneously in sewage and Guaire River samples that corresponded to G1P[4] and G1P[8]. The G1P[8] strain is employed in Venezuela as a live attenuated vaccine (a RIX4414 strain of G1P[8] specificity) in infants and young children under 1 year; the G1P[4] strain has been reported to be an unusual genotype combination occurring among children with acute gastroenteritis in clinical studies conducted in Japan (39).
There are no previous epidemiological data on NSP4 genotypes in Venezuela; therefore, our data represent the first documentation of NSP4 genotypes circulating in this geographical region. The genotyping of the NSP4 gene was accomplished by a recently developed typing strategy that allows accurate discrimination among NSP4 genotypes A to C (48). Studies of worldwide circulation of HRV NSP4 genotypes indicate that types A and B are frequently linked to disease in humans (8, 11, 23, 26), and these two genotypes were predominantly detected in the sewage-related water samples analyzed in this investigation. More interesting was the presence of two samples from the Guaire River carrying mixed NSP4 genotypes, including genotypes B and C; the NSP4 genotype C is more often associated with canine and feline rotaviruses than with HRVs (9). Due to the nature of the fecal contamination of Guaire River, we cannot rule out the possibility that these two samples belonged to animal rotaviruses.
HuNoV and HAstV are responsible for a large number of gastroenteritis cases worldwide (29, 51). In the present study, we found these viruses in more than 50% of the samples analyzed from sewage and from the sewage-impacted river. The molecular characterization of HuNoVs showed that all HuNoV sequences recovered from the samples analyzed in the present study belonged to GGII, the genogroup that includes the globally circulating genotype (GGII.4) (29). Considering the wide distribution and frequent occurrence of waterborne and food-borne outbreaks worldwide, it is of epidemiological relevance to document predominant HuNoV genotypes and strains circulating in different geographical areas. The first approach we tried for the detection and characterization of HuNoVs and HAstVs was based in primers and PCR conditions that amplified the capsid gene that is commonly used for proper genotype identification (61). Nevertheless, that strategy was less sensitive in our study than the primer combination and nested-PCR method based on the amplification of the polymerase region, which allowed us to detect HuNoVs and HAstVs more efficiently.
The molecular characterization of HAstVs revealed that all the sequences recovered from the sewage-polluted waters analyzed in our study belonged to HAstV type 8. This was surprising, since type 1 has been described as the most globally prevalent and was also the most predominant type detected in previous clinical studies conducted in Colombia and in Venezuela (30). In the present study, HAstV type 8 circulation is documented for the first time in Venezuela. Further studies of infectivity and molecular surveillance of sewage-borne pathogenic viruses in Venezuela are warranted in order to obtain not only occurrence data of potential waterborne viral pathogens but also knowledge about environmental persistence and quantitative estimations of viral loads in surface waters used for recreation and water supply.
In the absence of reliable disease surveillance programs in Venezuela, the molecular detection and characterization of predominant sewage-borne viral pathogens circulating among the population not only provide relevant location-specific epidemiological data on potential waterborne pathogenic viruses but also represent an alternative strategy that may used in subsequent studies to establish health-based targets (i.e., water quality targets for pathogens), as recommended by the World Health Organization Guidelines for Drinking Water Quality to control microbial hazards in drinking water as discussed in detail by Roda-Husman and Bartram (47).
The increasing problem of raw sewage disposal into surface waters, with the subsequent adverse effects on the human health and ecology of aquatic environments, is not uncommon for countries in South America. Therefore, the public health risks associated with waterborne transmission of viruses and the environmental impacts resulting from sewage discharges need to be addressed through adequate monitoring programs. There have been no regulations for the monitoring of pathogenic viruses in waters in Venezuela and in South America such as those that have existed among countries from the European Union and in the United States. In these countries, HuEVs were the focus of extensive monitoring regulations and water quality guidelines intended to protect human health as they directly addressed the public health risks associated with viruses in water (10, 53, 56). Among the enteric viruses, HuEVs and HAds have been proposed as viral markers to identify and to monitor human fecal contamination sources (13, 41). Few studies addressing viral pathogens in environmental waters have been conducted in Venezuela and other countries in South America. Miagostovich et al. (31) evaluated the presence of viruses in river water of the hydrographic basin of the Amazon and reported frequent detection of human gastroenteritis viruses associated with disordered urbanization processes. Similar data obtained from our study indicate that prevailing anthropogenic impacts on aquatic environments may play an important role in the epidemiology of waterborne disease occurrence in this region. The identification of common pollution problems associated with urban sewage discharges in South America could be addressed through regional and international scientific cooperation, including strong governmental commitment and investment in adequate wastewater treatment processes in order to reduce the risks of adverse effects to human health and the environment in the region, thus aiding in decreasing the global burden of waterborne diseases and environmental degradation.
Acknowledgments
This research was supported by the Venezuelan Institute for Scientific Research (IVIC-883) and by TOTAL Venezuela S.A. (LOCTI program).
Footnotes
Published ahead of print on 21 November 2008.
REFERENCES
- 1.Ando, T., S. S. Monroe, J. R. Gentsch, Q. Jin, D. C. Lewis, and R. I. Glass. 1995. Detection and differentiation of antigenically distinct small round structured viruses (Norwalk-like viruses) by reverse transcription-PCR and Southern hybridization. J. Clin. Microbiol. 33:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arista, S., E. Vizzi, C. Alaimo, D. Palermo, and A. Cascio. 1999. Identification of human rotavirus strains with the P[14] genotype by PCR. J. Clin. Microbiol. 37:2706-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arraj, A., J. Bohatier, C. Aumeran, J. L. Bailly, H. Laveran, and O. Traoré. 2008. An epidemiological study of enteric viruses in sewage with molecular characterization by RT-PCR and sequence analysis. J. Water Health 6:351-358. [DOI] [PubMed] [Google Scholar]
- 4.Baggi, F., and R. Peduzzi. 2000. Genotyping of rotaviruses in environmental water and stool samples in southern Switzerland by nucleotide sequence analysis of 189 base pairs at the 5′ end of the VP7 gene. J. Clin. Microbiol. 38:3681-3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belliot, G., H. Laveran, and S. S. Monroe. 1997. Detection and genetic differentiation of human astroviruses: phylogenetic grouping varies by coding region. Arch. Virol. 142:1323-1334. [DOI] [PubMed] [Google Scholar]
- 6.Belliot, G. M., R. L. Fankhauser, and S. S. Monroe. 2001. Characterization of ‘Norwalk-like viruses’ and astroviruses by liquid hybridization assay. J. Virol. Methods 91:119-123. [DOI] [PubMed] [Google Scholar]
- 7.Bofill-Mas, S., S. Pina, and R. Girones. 2000. Documenting the epidemiologic patterns of polyomaviruses in human populations by studying their presence in urban sewage. Appl. Environ. Microbiol. 66:238-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao, X. R., S. Akihara, Z. Y. Fang, O. Nakagomi, and H. Ushijima. 1999. Genetic variation in the VP4 and NSP4 genes of human rotavirus serotype 3 (G3 type) isolated in China and Japan. Microbiol. Immunol. 43:171-175. [DOI] [PubMed] [Google Scholar]
- 9.Ciarlet, M., F. Liprandi, M. E. Conner, and M. K. Estes. 2000. Species specificity and interspecies relatedness of NSP4 genetic groups by comparative NSP4 sequence analyses of animal rotaviruses. Arch. Virol. 145:371-383. [DOI] [PubMed] [Google Scholar]
- 10.Council of European Directives. 1975. Council Directive 76/160/EEC of 8 December 1975 concerning the quality of bathing water. European Commission, Brussels, Belgium.
- 11.Cunliffe, N. A., P. A. Woods, J. P. Leite, B. K. Das, M. Ramachandran, M. K. Bhan, C. A. Hart, R. I. Glass, and J. R. Gentsch. 1997. Sequence analysis of NSP4 gene of human rotavirus allows classification into two main genetic groups. J. Med. Virol. 53:41-50. [PubMed] [Google Scholar]
- 12.Espínola, E. E., G. I. Parra, G. Russomando, and J. Arbiza. 2008. Genetic diversity of the VP4 and VP7 genes affects the genotyping of rotaviruses: analysis of Paraguayan strains. Infect. Genet. Evol. 8:94-99. [DOI] [PubMed] [Google Scholar]
- 13.Fong, T. T., D. W. Griffin, and E. K. Lipp. 2005. Molecular assays for targeting human and bovine enteric viruses in coastal waters and application for library-independent source tracking. Appl. Environ. Microbiol. 71:2070-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gentsch, J. R., R. I. Glass, P. Woods, V. Gouvea, M. Gorziglia, J. Flores, B. K. Das, and M. K. Bhan. 1992. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 30:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Global Environment Outlook 2000. 2000. United Nations Environment Programme, p.432. Earthscan Publications, Ltd., London, United Kingdom.
- 16.Gómara, M. I., D. Cubitt, U. Desselberger, and J. Gray. 2001. Amino acid substitution within the VP7 protein of G2 rotavirus strains associated with failure to serotype. J. Clin. Microbiol. 39:3796-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.González, G. G., F. H. Pujol, F. Liprandi, L. Deibis, and J. E. Ludert. 1998. Prevalence of enteric viruses in human immunodeficiency virus seropositive patients in Venezuela. J. Med. Virol. 55:288-292. [DOI] [PubMed] [Google Scholar]
- 18.Gouvea, V., R. I. Glass, P. Woods, K. Taniguchi, H. F. Clark, B. Forrester, and Z. Y. Fang. 1990. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J. Clin. Microbiol. 28:276-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grabow, W. O. K. 2007. Overview of health-related water virology, p. 1-25. In A. Bosh (ed.), Human viruses in water, 1st ed. Elsevier, Amsterdam, The Netherlands.
- 20.Hoshino, Y., M. Wagner, X. Yan, I. Perez-Schael, and A. Z. Kapikian. 2003. Horizontal transmission of rhesus monkey rotavirus-based quadrivalent vaccine during a phase 3 clinical trial in Caracas, Venezuela. J. Infect. Dis. 187:791-800. [DOI] [PubMed] [Google Scholar]
- 21.Hurst, C. J., and P. A. Murphy. 1997. The transmission and prevention of infectious disease, p. 3-54. In C. J. Hurst (ed.), Modeling disease transmission and its prevention by disinfection, 1st ed. Cambridge University Press, New York, NY.
- 22.Iturriza-Gómara, M., G. Kang, and J. Gray. 2004. Rotavirus genotyping: keeping up with an evolving population of human rotaviruses. J. Clin. Virol. 31:259-265. [DOI] [PubMed] [Google Scholar]
- 23.Iturriza-Gòmara, M., E. Anderton, G. Kang, C. Gallimore, W. Phillips, U. Desselberger, and J. Gray. 2003. Evidence for genetic linkage between the gene segments encoding NSP4 and VP6 proteins in common and reassortant human rotavirus strains. J. Clin. Microbiol. 41:3566-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iturriza-Gómara, M., B. Megson, and J. Gray. 2006. Molecular detection and characterization of human enteroviruses directly from clinical samples using RT-PCR and DNA sequencing. J. Med. Virol. 78:243-253. [DOI] [PubMed] [Google Scholar]
- 25.Jiang, X., C. Espul, W. M. Zhong, H. Cuello, and D. O. Matson. 1999. Characterization of a novel human calicivirus that may be a naturally occurring recombinant. Arch. Virol. 144:2377-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirkwood, C. D., and E. A. Palombo. 1997. Genetic characterization of the rotavirus nonstructural protein, NSP4. Virology 236:258-265. [DOI] [PubMed] [Google Scholar]
- 27.Krikelis, V., N. Spyrou, P. Markoulatos, and C. Serie. 1985. Seasonal distribution of enteroviruses and adenoviruses in domestic sewage. Can. J. Microbiol. 31:24-25. [DOI] [PubMed] [Google Scholar]
- 28.Leclerc, H., L. Schwartzbrod, and E. Dei-Cas. 2002. Microbial agents associated with waterborne diseases. Crit. Rev. Microbiol. 28:371-409. [DOI] [PubMed] [Google Scholar]
- 29.Lopman, B., M. Zambon, and D. W. Brown. 2008. The evolution of norovirus, the “gastric flu.” PLoS Med. 5:e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Medina, S. M., M. F. Gutierrez, F. Liprandi, and J. E. Ludert. 2000. Identification and type distribution of astroviruses among children with gastroenteritis in Colombia and Venezuela. J. Clin. Microbiol. 38:3481-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miagostovich, M. P., F. F. Ferreira, F. R. Guimaraes, T. M. Fumina, L. Diniz-Mendes, S. L. Luz, L. A. Silva, and P. G. Leite. 2008. Molecular detection and characterization of gastroenteritis viruses occurring naturally in the stream waters of Manaus, Central Amazonia, Brazil. Appl. Environ. Microbiol. 74:375-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Myrmel, M., E. M. Berg, B. Grinde, and E. Rimstad. 2006. Enteric viruses in inlet and outlet samples from sewage treatment plants. J. Water Health 4:197-209. [PubMed] [Google Scholar]
- 33.Parez, N. 2008. Rotavirus gastroenteritis: why to back up the development of new vaccines? Comparative immunology. Microbiol. Infect. Dis. 31:253-269. [DOI] [PubMed] [Google Scholar]
- 34.Pérez-Schael, I. 1996. The impact of rotavirus disease in Venezuela. J. Infect. Dis. 174(Suppl. 1):S19-S21. [DOI] [PubMed] [Google Scholar]
- 35.Pérez-Schael, I., R. Gonzalez, R. Fernandez, E. Alfonzo, D. Inaty, Y. Boher, and L. Sarmiento. 1999. Epidemiological features of rotavirus infection in Caracas, Venezuela: implications for rotavirus immunization programs. J. Med. Virol. 59:520-526. [DOI] [PubMed] [Google Scholar]
- 36.Pérez-Schael, I., M. J. Guntiñas, M. Pérez, V. Pagone, A. M. Rojas, R. González, W. Cunto, Y. Hoshino, and A. Z. Kapikian. 1997. Efficacy of the rhesus rotavirus-based quadrivalent vaccine in infants and young children in Venezuela. N. Engl. J. Med. 337:1181-1187. [DOI] [PubMed] [Google Scholar]
- 37.Pérez-Schael, I., B. Salinas, R. González, H. Salas, J. E. Ludert, M. Escalona, A. Alcalá, and M. A. Rosas. 2007. Rotavirus mortality confirmed by etiologic identification in Venezuelan children with diarrhea. Pediatr. Infect. Dis. J. 26:393-397. [DOI] [PubMed] [Google Scholar]
- 38.Perez-Schael. I., B. Salinas, M. Tomat, A. C. Linhares, M. L. Guerrero, G. M. Ruiz-Palacios, A. Bouckenooghe, and J. P. Yarzabal. 2007. Efficacy of the human rotavirus vaccine RIX4414 in malnourished children. J. Infect. Dis. 196:537-540. [DOI] [PubMed] [Google Scholar]
- 39.Phan, T. G., P. Khamrin, T. D. Quang, S. K. Dey, F. Yagyu, S. Okitsu, O. Nishio, and H. Ushijima. 2007. Genetic characterization of group A rotavirus strains circulating among children with acute gastroenteritis in Japan in 2004-2005. Infect. Genet. Evol. 7:247-253. [DOI] [PubMed] [Google Scholar]
- 40.Pina, S., M. Buti, R. Jard, P. Clemente-Casares, J. Jofre, and R. Girones. 2001. Genetic analysis of hepatitis A virus strains recovered from the environment and from patients with acute hepatitis. J. Gen. Virol. 82:2955-2963. [DOI] [PubMed] [Google Scholar]
- 41.Pina, S., P. Monserrat, F. Lucena, J. Jofre, and R. Girones. 1998. Viral pollution in the environment and in shellfish: human adenovirus detection by PCR as an index of human viruses. Appl. Environ. Microbiol. 64:3376-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pintó, R. M., D. Alegre, A. Domínguez, W. M. El-Senousy, G. Sánchez, C. Villena, M. I. Costafedra, L. Aragonés, and A. Bosch. 2007. Hepatitis A virus in urban sewage from two Mediterranean countries. Epidemiol. Infect. 135:270-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pintó, R. M., and J. C. Saiz. 2007. Enteric hepatitis viruses, p. 39-67. In A. Bosh (ed.), Human viruses in water, 1st ed. Elsevier, Amsterdam, The Netherlands.
- 44.Puig, M., J. Jofre, F. Lucena, A. Allard, G. Wadell, and R. Girones. 1994. Detection of adenoviruses and enteroviruses in polluted waters by nested PCR amplification. Appl. Environ. Microbiol. 60:2963-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pujol, F. H., I. Rodríguez, N. Martínez, C. Borberg, M. O. Favorov, H. A. Fields, and F. Liprandi. 1994. Viral hepatitis serological markers among pregnant women in Caracas, Venezuela: implication for perinatal transmission of hepatitis B and C. GEN (Gastroenterol. Endocrinol. Nutr.) (Caracas) 48:25-28. [PubMed] [Google Scholar]
- 46.Pujol, F. H., G. Vásquez, A. M. Rojas, M. E. Fuenmayor, C. L. Loureiro, I. Pérez-Schael, M. K. Estes, and F. Liprandi. 1998. Norwalk virus infection in Venezuela. Ann. Trop. Med. Parasitol. 92:205-211. [PubMed] [Google Scholar]
- 47.Roda-Husman, A. M., and J. Bartram. 2007. Global supply of virus-safe drinking water, p. 127-162. In A. Bosh (ed.), Human viruses in water, 1st ed. Elsevier, Amsterdam, The Netherlands. [DOI] [PMC free article] [PubMed]
- 48.Rodríguez-Díaz, J., E. Rubilar-Abreu, M. Spitzner, K. Hedlund, F. Liprandi, and L. Svensson. 2008. Design of a multiplex nested PCR for genotyping of the NSP4 from group A rotavirus. J. Virol. Methods 149:240-245. [DOI] [PubMed] [Google Scholar]
- 49.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 50.Santos, N., and Y. Hoshino. 2005. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev. Med. Virol. 15:29-56. [DOI] [PubMed] [Google Scholar]
- 51.Schwab, K. 2007. Waterborne gastroenteritis viruses, p. 27-38. In A. Bosh (ed.), Human viruses in water, 1st ed. Elsevier, Amsterdam, The Netherlands.
- 52.Sedmak, G., D. Bina, and J. MacDonald. 2003. Assessment of an enterovirus sewage surveillance system by comparison of clinical isolates with sewage isolates from Milwaukee, Wisconsin, collected August 1994 to December 2002. Appl. Environ. Microbiol. 69:7181-7187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Standing Committee of Analysts. 1995. Methods for the isolation and identification of human enteric viruses from waters and associated materials. Methods for the examination of waters and associated materials. HMSO Publications, London, United Kingdom.
- 54.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 55.Urrestarazu, M. I., F. Liprandi, E. Pérez de Suárez, R. González, and I. Pérez-Schael. 1999. Características etiológicas, clínicas y sociodemográficas de la diarrea aguda en Venezuela. Panam. Salud Publica Pan Am. J. Public Health 6:149-156. [DOI] [PubMed] [Google Scholar]
- 56.U.S. Environmental Protection Agency. 1995. Virus monitoring protocol for the Information Collection Requirements Rule. Publication EPA/814-B-95-002. Government Printing Office, U.S. Environmental Protection Agency, Cincinnati, OH.
- 57.van Zyl, W. B., N. A. Page, W. O. K. Grabow, A. D. Steele, and M. B. Taylor. 2006. Molecular epidemiology of group A rotaviruses in water sources and selected raw vegetables in southern Africa. Appl. Environ. Microbiol. 72:4554-4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Villena, C., W. M. El-Senousy, F. X. Abad, R. M. Pinto, and A. Bosch. 2003. Group A rotavirus in sewage samples from Barcelona and Cairo: emergence of unusual genotypes. Appl. Environ. Microbiol. 69:3919-3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vinjé, J., R. A. Hamidjaja, and M. D. Sobsey. 2004. Development and application of a capsid VP1 (region D) based reverse transcription PCR assay for genotyping of genogroup I and II noroviruses. J. Virol. Methods 116:109-117. [DOI] [PubMed] [Google Scholar]
- 60.Wu, H., K. Taniguchi, F. Wakasugi, S. Ukae, S. Chiba, M. Ohseto, A. Hasegawa, T. Urasawa, and S. Urasawa. 1994. Survey on the distribution of the gene 4 alleles of human rotaviruses by polymerase chain reaction. Epidemiol. Infect. 112:615-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yan, H., F. Yagyu, S. Okitsu, O. Nishio, and H. Ushijima. 2003. Detection of norovirus (GI, GII), sapovirus and astrovirus in fecal samples using reverse transcription single-round multiplex PCR. J. Virol. Methods 114:37-44. [DOI] [PubMed] [Google Scholar]