Abstract
Kinetic analyses of bacterial growth, carbohydrate consumption, and metabolite production of 18 Bifidobacterium strains grown on fructose, oligofructose, or inulin were performed. A principal component analysis of the data sets, expanded with the results of a genetic screen concerning the presence of a β-fructofuranosidase gene previously encountered in Bifidobacterium animalis subsp. lactis DSM 10140T, revealed the existence of four clusters among the bifidobacteria tested. Strains belonging to a first cluster could not degrade oligofructose or inulin. Strains in a second cluster could degrade oligofructose, displaying a preferential breakdown mechanism, but did not grow on inulin. Fructose consumption was faster than oligofructose degradation. A third cluster was composed of strains that degraded all oligofructose fractions simultaneously and could partially break down inulin. Oligofructose degradation was substantially faster than fructose consumption. A fourth, smaller cluster consisted of strains that shared high fructose consumption and oligofructose degradation rates and were able to perform partial breakdown of inulin. For all strains, a metabolic shift toward more acetate, formate, and ethanol production, at the expense of lactate production, was observed during growth on less readily fermentable energy sources. No correlation between breakdown patterns and the presence of the β-fructofuranosidase gene could be detected. These variations indicate niche-specific adaptation of bifidobacteria and could have in vivo implications on the strain specificity of the stimulatory effect of inulin-type fructans on bifidobacteria.
More than a decade of intensive research has radically altered both scientists' and consumers' perceptions of the role and importance of the human colon microbiota and the gut ecosystem. The colonic microbiota is no longer regarded as only an intrinsic part of the digestive system but is considered a key element of an organ that influences body processes far beyond digestion (26, 38). The fundamentals of this new perception can be summarized as follows: (i) host health and well-being are influenced by the colon microbiota (26, 38), (ii) the nature of a healthy or balanced colon microbiota is definable (25, 27), and (iii) the composition and/or metabolic activity of the colon microbiota can be influenced (transiently) through changes in the diet (36). Notwithstanding the fact that the gut ecosystem remains largely unexplored (4, 6), different strategies to influence host health by managing the composition and/or activity of the colon microbiota through diet have emerged from these understandings (9, 12, 37). Although hard to define, the goal of such dietary interventions is to establish an optimally balanced colon microbiota, which is generally believed to be predominantly saccharolytic, comprising significant numbers of bifidobacteria and lactobacilli (27).
A well-established strategy to alter the gut ecosystem consists of the use of prebiotics, selectively fermented nondigestible food ingredients that allow specific changes in the composition and/or activity of the gastrointestinal microbiota, which confer benefits upon host well-being and health (11, 12). At the origin of the development of the prebiotic concept lies the observation of the stimulative effect of inulin-type fructans on the fecal Bifidobacterium population during in vitro experiments (12, 14, 48). Although the prebiotic properties of other food ingredients have been acknowledged (11), oligofructose and inulin still remain the best-studied ingredients and have gained a status of model prebiotics (2, 39). It has become clear that their stimulatory effect exceeds the colonic Bifidobacterium population and affects other genera, for instance, butyrate-producing colon bacteria belonging to clostridial cluster XIVa (5, 24), which have been reported to account for up to 3% of total fecal bacteria (17). However, due to their widely recognized beneficial effects on human health (12, 35), bifidobacteria and, to a lesser extent, lactobacilli are regarded as the main targets of prebiotic inulin-type fructans (27, 36).
Although stimulation of bifidobacteria by inulin-type fructans has been studied extensively for more than 15 years through both in vitro and in vivo trials, some rather elementary questions still await satisfactory answers (27). For example, it is not yet clear how bifidobacteria manage to gain the upper hand in the highly competitive human colon ecosystem when inulin or oligofructose is added to the diet, while it has been demonstrated that Lactobacillus spp. (15, 21, 22, 29), Roseburia spp. (3, 8), Bacteroides spp. (46), and some enterobacteria (16) can also use at least oligofructose as a substrate for fermentation (18). It has been suggested that (some) Bifidobacterium spp. are capable of intracellular or cell-associated degradation of oligofructose (8, 42, 46). Nonbifidobacterial species are thought to degrade oligofructose extracellularly, making them less competitive when growing on this substrate (8, 46). Moreover, since not all bifidobacteria are able to degrade inulin or even oligofructose to the same extent (18, 40), it seems likely that not all bifidobacterial species benefit in the same way from the presence of inulin-type fructans as energy sources in the colon. However, most studies concerning the bifidogenic effect of inulin-type fructans tend, unfortunately, to consider the bifidobacterial gut population as a whole, not taking into account the interspecies differences that exist between various bifidobacteria (27).
Recent in vitro studies focusing on detailed kinetic analyses of oligofructose degradation by bifidobacteria as well as Lactobacillus paracasei subsp. paracasei, Bacteroides thetaiotaomicron, Bacteroides fragilis, and Roseburia intestinalis have revealed remarkable differences between the bifidobacterial breakdown mechanism and that of species belonging to other genera (8, 29, 45, 46). While bifidobacteria seem to degrade oligofructose in a preferential way, only initiating the breakdown of a longer-chain-length fraction when shorter fractions are depleted, nonbifidobacterial species exhibit simultaneous degradation of all fractions, often combined with the release of large amounts of free fructose into the fermentation medium (8, 46). It has been suggested that the latter breakdown profile can be linked to extracellular degradation, which might prove to be less efficient than internal or cell wall-bound degradation in a highly competitive ecosystem such as the human colon (8, 46). The appearance in the growth environment of free fructose and short oligosaccharides, which become available to opportunistic competitors that are not able to break down inulin-type fructans themselves (10, 25), seems to be inherent to extracellular degradation. Bifidobacteria capable of intracellular degradation would not suffer from this drawback, which would explain their selective growth stimulation on inulin or oligofructose (8, 40, 46).
This study set out to kinetically investigate oligofructose and inulin degradation by a wide range of Bifidobacterium species, focusing in particular on the presence/absence of a preferential oligofructose breakdown mechanism, through intermediate-scale batch fermentation processes under strictly controlled conditions. Also, a genetic screen was performed to monitor the distribution of a selected β-fructofuranosidase gene among the bifidobacteria tested.
MATERIALS AND METHODS
Microorganisms and media.
The following 18 bifidobacterium strains belonging to 10 different species were included in this study: Bifidobacterium adolescentis LMG 10502T, Bifidobacterium adolescentis LMG 10733, Bifidobacterium adolescentis LMG 10734, Bifidobacterium angulatum LMG 11039T, Bifidobacterium bifidum DSM 20082, Bifidobacterium bifidum LMG 11583, Bifidobacterium breve LMG 11040, Bifidobacterium breve LMG 13194, Bifidobacterium breve Yakult, Bifidobacterium catenulatum LMG 11043T, Bifidobacterium dentium LMG 10507, Bifidobacterium gallicum LMG 11596T, Bifidobacterium longum LMG 11047, Bifidobacterium longum LMG 11570, Bifidobacterium longum LMG 11588, Bifidobacterium longum LMG 13196, Bifidobacterium pseudocatenulatum LMG 10505T, and Bifidobacterium thermophilum LMG 11574. Apart from B. thermophilum LMG 11574, which was originally isolated from the bovine rumen, all strains were human isolates, covering the range of bifidobacteria most commonly encountered in the gut (30, 31). All of them were obtained from the Belgian Co-Ordinated Collections of Micro-Organisms/Laboratory for Microbiology Ghent (Ghent, Belgium), with the exception of B. breve Yakult, which was kindly provided by Yakult Honsha Co. Ltd. (Tokyo, Japan), and B. bifidum DSM 20082, originating from the Deutsche Sammlung von Mikro-Organismen und Zellkulturen (Göttingen, Germany). For inoculum preparation and storage purposes, strains were grown anaerobically in reinforced clostridial medium (RCM; Oxoid Ltd., Basingstoke, United Kingdom), except for those belonging to the B. bifidum species, which were cultured in de Man-Rogosa-Sharpe medium (MRS medium; Oxoid). Strains were stored at −80°C in the appropriate medium supplemented with 25% (vol/vol) glycerol as a cryoprotectant.
Fermentation experiments were performed in a medium for colon bacteria (MCB) developed by Van der Meulen et al. (46) to support the growth of various human colon bacteria when supplemented with an adequate energy source. The medium was composed of the following (in g liter−1): bacteriological peptone (Oxoid), 6.5; soy peptone (Oxoid), 5.0; yeast extract (VWR International GmbH, Darmstadt, Germany), 3.0; tryptone (Oxoid), 2.5; NaCl, 4.5; KCl, 2.0; MgSO4·7H2O, 0.5; CaCl2·2H2O, 0.45; cysteine-HCl·H2O, 0.4; NaHCO3, 0.2; MnSO4·H2O, 0.2; FeSO4·7H2O, 0.005; ZnSO4·7H2O, 0.005; hemin, 0.005; and menadione, 0.005. It also contained H3PO4 at 0.5 ml liter−1 and Tween 80 at 2 ml liter−1. The pH of the medium was adjusted to 6.3 before being autoclaved at 210 kPa and 121°C for 20 min. After sterilization, fructose (VWR International), oligofructose (OraftiP95; BENEO-Orafti NV, Tienen, Belgium), or inulin (OraftiHP; BENEO-Orafti) was added aseptically as the sole energy source at a concentration of 50 mM fructose equivalents (FE). Fructose was autoclaved under the same conditions as the MCB; oligofructose and inulin were sterilized by membrane filtration, using Minisart filters (pore size, 0.2 μm; Sartorius AG, Göttingen, Germany). OraftiP95 and OraftiHP are commercial powders derived from chicory roots. OraftiP95 is obtained through enzymatic hydrolysis of chicory inulin. It consists mainly of oligofructose (≥93.2% [wt/wt]), with minor amounts of glucose, fructose, and sucrose (<6.8% [wt/wt]). The degree of polymerization (DP) of the oligofructose fractions varies between 2 and 8, with an average of 4. OraftiHP contains inulin (≥99.5% [wt/wt]), with a DP ranging from 12 to 65, and minor amounts of glucose, fructose, and sucrose (<0.5% [wt/wt]). The average DP of the inulin chains exceeds 23 due to removal of the smaller molecules during processing.
Solid RCM and MRS medium were prepared by adding 1.5% (wt/vol) agar (Oxoid) to the respective broths.
Fermentation experiments.
Fermentations were carried out in 2-liter Biostat B-DCU fermentors (Sartorius) containing 1.5 liters of MCB supplemented with the energy source under study. Inocula were prepared as follows. Strains were transferred from −80°C to the appropriate medium and incubated anaerobically at 37°C for 12 h in a modular atmosphere-controlled system (MG anaerobic work station; Don Whitley Scientific Ltd., West Yorkshire, United Kingdom) continuously sparged with a mixture of 80% nitrogen, 10% carbon dioxide, and 10% hydrogen (Air Liquide, Paris, France). Subsequently, the strains were propagated twice in MCB with fructose as the sole energy source and finally added to the fermentor. During inoculum build-up, the transferred volume was always 5% (vol/vol). Anaerobic conditions were ensured during fermentation experiments by continuously sparging the medium with a mixture of 90% nitrogen and 10% carbon dioxide (Air Liquide). The fermentation temperature was kept constant at 37°C. A constant pH of 6.3 was imposed and controlled automatically, using 1.5 M solutions of NaOH and H3PO4. To keep the medium homogeneous, a gentle stirring of 100 rpm was applied. Temperature, pH, and agitation speed were controlled online (MFCS/win 2.1; Sartorius). Fermentations were monitored for 48 h; samples for further analysis were taken at regular time intervals.
Analysis of bacterial growth, carbohydrate consumption, and metabolite production. (i) Bacterial growth.
Growth was followed throughout all fermentations by cell plating. Samples were plated on RCM or MRS agar as appropriate and incubated for 24 h under anaerobic conditions (modular atmosphere-controlled system).
(ii) Carbohydrate, organic acid, and ethanol determinations.
Residual concentrations of glucose, fructose, oligofructose, and inulin (with the last two expressed in mM FE), as well as concentrations of acetate, lactate, formate, and ethanol, were determined through high-performance liquid chromatography with a Waters chromatograph (Waters Corp., Milford, MA) equipped with a 2414 differential refractometer, a 600S controller, and a cooled 717 Plus autosampler. An ICSep ICE ORH-801 column (Interchim, Montluçon, France) was used with 10 mM of H2SO4 as the mobile phase at a flow rate of 0.4 ml min−1. The column temperature was kept constant at 35°C. Samples were centrifuged (21,036 × g for 20 min), and an equal volume of 20% (wt/vol) trichloroacetic acid was added for protein removal and complete acid hydrolysis of the β-(2→1) glycosidic bonds of oligofructose and inulin. After centrifugation (21,036 × g, 20 min), the supernatant was filtered (pore size, 0.2 μm) (Minisart RC4 filters; Sartorius) before injection (30 μl). All samples were analyzed in triplicate.
(iii) Breakdown of oligofructose and inulin.
Detailed analysis of the breakdown of the different fractions of oligofructose was performed by gas chromatography using a 5300-HT HRGC instrument (Carlo Erba, Rodina, Italy). The gas chromatograph was equipped with an SGE Clad-5 aluminum capillary column (Achrom NV, Zulte, Belgium), a cooled on-column AS-550 autoinjector, and a flame ionization detector (detector temperature of 447°C). The oven temperature varied linearly from 105 to 440°C at 10°C min−1. Samples were derivatized following a procedure involving oximation and silylation (20). The oxime-trimethylsilyl sugar derivatives were extracted using iso-octane; the resulting iso-octane phase was injected (1 μl) into the gas chromatography column. The same procedure was carried out for reference samples containing reference oligofructose (RaftiloseP95X; BENEO-Orafti), glucose, fructose, and sucrose as external standards.
Qualitative analysis of inulin breakdown in fermentations with OraftiHP as the sole added energy source was performed using high-performance anion-exchange chromatography (HPAEC) with pulsed amperometric detection (PAD), using a DX500 chromatograph (Dionex Corp., Sunnyvale, CA). A CarboPac PA100 column (Dionex) was used with a gradient of the following three eluent solutions as the mobile phase: 0.1 M NaOH (eluent A), 0.1 M NaOH and 0.4 M NaCH3COOH (eluent B), and 1 M NaOH (eluent C). The following gradient was applied: 0 min, 96% A and 4% B; 5 min, 96% A and 4% B; 15 min, 60% A and 40% B; 35 min, 30% A and 70% B; 50 min, 10% A and 90% B; 60 min, 100% B; 85 min, 100% B; 90 min, 100% C; 95 min, 100% C; 96 min, 96% A and 4% B; and 101 min, 96% A and 4% B. The injection volume was 50 μl. Sample preparation involved centrifugation (21,036 × g, 20 min), followed by protein removal using an isovolume of acetonitrile, centrifugation (21,036 × g, 20 min), appropriate dilution, and filtration (Minisart RC4 filters). Samples were analyzed in duplicate.
Genetic screen for a β-fructofuranosidase gene.
For DNA isolation, strains were transferred from −80°C to 10 ml of the appropriate medium and incubated anaerobically at 37°C for 12 h. DNA was extracted following a method previously described by Vanhoutte et al. (47). Primers were designed based on the β-fructofuranosidase gene sequence of Bifidobacterium animalis subsp. lactis DSM 10140T (7), updated with sequence information obtained from the European Molecular Biology Laboratory Nucleotide Sequence Database (entries AAN23970, CAD26970, BAF39931, AAT28190, and ABN04092). Degenerate primers BFRA1-F (5′-TGGATCAACGAYCCGAACGG-3′) and BFRA1-R (5′-ACCTTCGGGTCGCGGWAGTG-3′) were used for the amplification of a 400-bp region of the bifidobacterial β-fructofuranosidase gene mentioned above. PCR amplifications were performed using a DNA T3 thermocycler (Biometra GmbH, Göttingen, Germany) and a total volume of 50 μl, containing 500 ng of genomic DNA, 0.25 μg of bovine serum albumin, a 100 μM concentration of each deoxyribonucleotide triphosphate, 10 pmol of each primer, and 1.25 IU of Taq polymerase (Roche Diagnostics GmbH, Mannheim, Germany). PCR conditions were set as follows: initial denaturation at 95°C for 300 s; 30 cycles of denaturation at 95°C for 45 s, annealing at 60°C for 45 s, and extension at 72°C for 60 s; and a final extension at 72°C for 600 s. Amplicons were sequenced using the primers mentioned above in a commercial facility making use of capillary sequencing technology (VIB Genetic Service Facility, Antwerp, Belgium), and the forward and reverse sequences were assembled for each strain. The resulting contig sequences were aligned with the protein sequences of the six genes whose accession numbers are mentioned above, using BLASTX (1), to determine conformity.
Statistical analysis.
A principal component analysis (PCA) was performed on the outcomes of the fermentation experiments. For all strains, the quantitative data (growth, carbohydrate consumption, and metabolite production) obtained after 48 h of growth on each of the energy sources applied were listed and completed with qualitative observations regarding the nature of oligofructose degradation (preferential/nonpreferential, faster/slower than fructose consumption), the ability to break down inulin, and the presence/absence of a β-fructofuranosidase gene. After PCA followed by varimax rotation with Kaiser normalization, cluster analysis by the unweighted-pair group method using average linkages, based on Euclidian distance, was performed. The software package SPSS 13.0 (SPSS Inc., Chicago, IL) was used for statistical analysis; results were visualized with BioNumerics 4.5 (Applied Maths NV, Sint-Martens-Latem, Belgium).
RESULTS
Fermentation experiments.
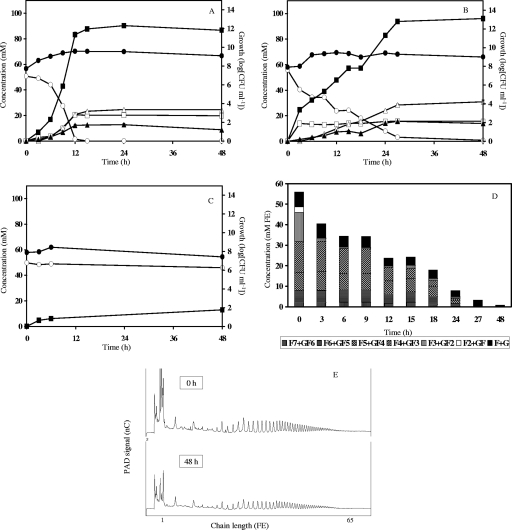
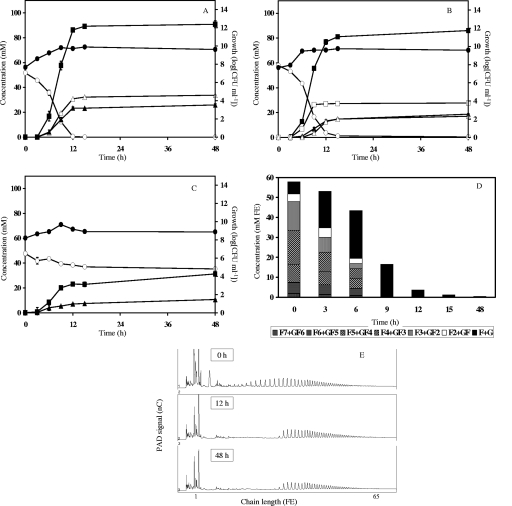
Fifty-four fermentations with 18 Bifidobacterium strains were followed for 48 h. The kinetics of bacterial growth, carbohydrate consumption, and metabolite production were analyzed in detail. Only four of the strains included in this study were not able to grow on either oligofructose or inulin. Among the strains that could degrade oligofructose, two distinct groups could be identified. The first group, including B. adolescentis LMG 10734, exhibited a preferential oligofructose degradation pattern (Fig. 1). The second group initiated degradation of all oligofructose fractions simultaneously, as was the case for B. longum LMG 11570 (Fig. 2).
FIG. 1.
Inulin-type fructan degradation fingerprint of Bifidobacterium adolescentis LMG 10734. Growth, carbohydrate consumption, and metabolite production in MCB supplemented with 50 mM FE of fructose (A), oligofructose (OraftiP95) (B), or inulin (OraftiHP) (C) are shown. ○, substrate (FE); ▪, acetate; □, lactate; ▵, formate; ▴, ethanol; •, growth. (D) Oligofructose degradation. F, fructose; G, glucose. (E) Qualitative inulin degradation. An HPAEC-PAD chromatogram is shown.
FIG. 2.
Inulin-type fructan degradation fingerprint of Bifidobacterium longum LMG 11570. Growth, carbohydrate consumption, and metabolite production in MCB supplemented with 50 mM FE of fructose (A), oligofructose (OraftiP95) (B), or inulin (OraftiHP) (C) are shown. ○, substrate (FE); ▪, acetate; □, lactate; ▵, formate; ▴, ethanol; •, growth. (D) Oligofructose degradation. F, fructose; G, glucose. (E) Qualitative inulin degradation. An HPAEC-PAD chromatogram is shown.
Inulin-type fructan degradation fingerprints, consisting of growth, carbohydrate consumption, and metabolite production profiles for each strain-substrate combination, detailed quantitative analysis of oligofructose degradation, and HPAEC-PAD patterns providing qualitative data concerning inulin degradation, were constructed for all strains. Graphical representations of all data are provided as supplemental material.
Genetic screen for a β-fructofuranosidase gene.
A genetic screen for the distribution of a selected β-fructofuranosidase gene among the bifidobacteria included in the present study was performed. Thirteen of the 18 strains tested positive for the presence of the gene, with BLASTX identities ranging between 87% and 98%. Strains lacking the selected gene included B. bifidum DSM 20082, B. bifidum LMG 11583, B. gallicum LMG 11596T, B. longum LMG 11047, and B. thermophilum LMG 11574.
Statistical analysis.
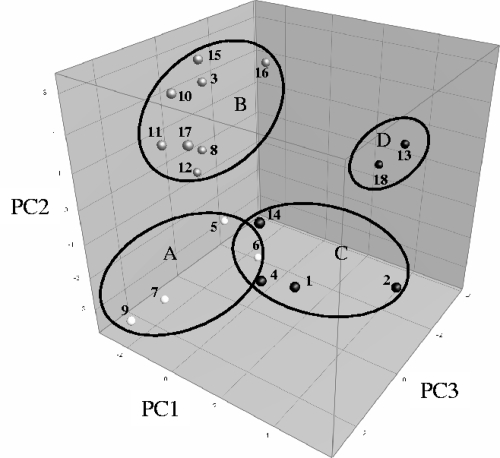
A PCA was performed on the extended data set generated by the results of the fermentation experiments after 48 h, supplemented with additional qualitative information concerning the preferential/nonpreferential character of oligofructose consumption, the rate of oligofructose degradation (faster/slower than fructose consumption), the breakdown of longer inulin chains, and the presence of the β-fructofuranosidase gene under study (see the supplemental material). The four main principal components (PC) accounted for 81.3% of the total variance. High scores for PC1, representing 31.6% of the variance, corresponded with fast degradation of oligofructose (compared with fructose consumption) and breakdown of the shorter fractions of long-chain inulin. High values on the PC2 axis, accounting for 20.5% of the variance, reflected preferential oligofructose degradation, combined with high acetate, formate, and ethanol production levels. Low lactate production levels during growth on fructose and the presence of the β-fructofuranosidase gene selected were translated into high scores for PC3, reflecting 18.6% of the variance. Although representing 10.6% of the total variation, PC4 was not taken into account for further analysis, as it expressed less significant variability, such as fluctuations in the minor residual concentrations of fructose after 48 h of fermentation. However, because other variables had a less pronounced but not negligible impact on the positions of the strains in the three-dimensional score plot (Fig. 3), care needs to be taken in interpreting the graph; the combination with the data provided as supplemental material remains indispensable.
FIG. 3.
Three-dimensional score plot of a PCA of the data obtained from the inulin-type fructan degradation fingerprints of 18 Bifidobacterium strains. Dots: 1, B. adolescentis LMG 10502T; 2, B. adolescentis LMG 10733; 3, B. adolescentis LMG 10734; 4, B. angulatum LMG 11039T; 5, B. bifidum DSM 20082; 6, B. bifidum LMG 11583; 7, B. breve LMG 11040; 8, B. breve LMG 13194; 9, B. breve Yakult; 10, B. catenulatum LMG 11043T; 11, B. dentium LMG 10507; 12, B. gallicum LMG 11596T; 13, B. longum LMG 11047; 14, B. longum LMG 11570; 15, B. longum LMG 11588; 16, B. longum LMG 13196; 17, B. pseudocatenulatum LMG 10505T; 18, B. thermophilum LMG 11574.
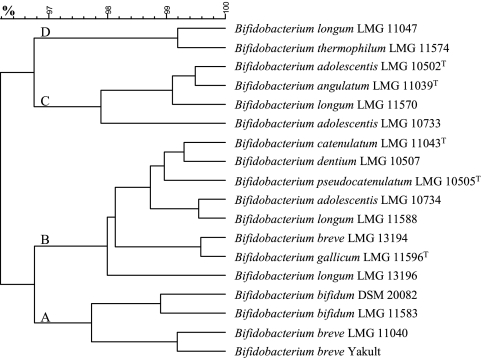
Clustering led to the identification of four distinct groups among the bifidobacterial strains tested (Fig. 3 and 4). Cluster A (cluster center, [−2.1, −3.2, 0.1]) consisted only of B. bifidum and B. breve. All strains were characterized by the lack of the ability to degrade oligofructose or inulin; variance within the cluster was due mainly to the presence/absence of the β-fructofuranosidase gene. Both B. bifidum strains tested shared another feature; besides acetate and lactate, both strains produced more ethanol than formate, whereas all other strains maintained an approximate ethanol/formate ratio of 1:2.
FIG. 4.
Dendrogram representing cluster analysis by the unweighted-pair group method using average linkages, based on Euclidian distance, after PCA of the data obtained from the inulin-type fructan degradation fingerprints of 18 Bifidobacterium strains.
The largest cluster, cluster B (cluster center, [−1.6, 1.8, 0.1]), was composed of eight strains belonging to seven different species (Fig. 3 and 4). None of these strains was able to degrade inulin. Oligofructose degradation succeeded in a preferential way for most of the members of this cluster, except for B. dentium LMG 10507 (nonpreferential). Fructose consumption was generally faster than oligofructose breakdown; exceptions here were B. pseudocatenulatum LMG 10505T and, again, B. dentium LMG 10507. The latter two strains were also the only ones belonging to cluster B that exhibited a high lactate production rate on oligofructose. All strains tested positive for the presence of the β-fructofuranosidase gene, except for B. gallicum LMG 11596T. Figure 1 shows the inulin-type fructan degradation fingerprint of B. adolescentis LMG 10734, a typical representative of cluster B.
Cluster C (cluster center, [3.8, −0.4, 1.6]) included strains belonging to the species B. adolescentis, B. angulatum, and B. longum (Fig. 3 and 4). All of the strains belonging to this cluster shared the ability to partially degrade inulin; after approximately 12 h of fermentation, the substrate was consumed up to a particular chain length, whereas fractions with a higher DP were subjected to only slow and minor degradation in the remaining 36 h of the fermentation experiments. Degradation of the different oligofructose fractions was clearly nonpreferential and faster than fructose consumption. Another feature common to all members of cluster C was a metabolic shift toward more ethanol and formate production at the expense of lactate production when cells were growing on fructose compared to growth on oligofructose. All strains also tested positive for the presence of the β-fructofuranosidase gene. The inulin-type fructan degradation fingerprint of B. longum LMG 11570 (Fig. 2) is representative of this cluster.
Cluster D (cluster center, [2.9, 0.1, −3.7]) contained only two strains, namely, B. longum LMG 11047 and B. thermophilum LMG 11574 (Fig. 3 and 4). Concerning the degradation of inulin, both strains showed a profile comparable to that of the bifidobacteria belonging to cluster C. However, since fructose and oligofructose both were depleted after 6 h of fermentation, no distinction could be made between oligofructose degradation and fructose consumption rates. The fast oligofructose degradation displayed by both strains also prevented differentiation of the preferential or nonpreferential character of their respective breakdown patterns. A metabolic shift toward more production of formate and ethanol on oligofructose was observed for both strains, although only trace concentrations could be detected for B. thermophilum LMG 11574. Neither B. longum LMG 11047 nor B. thermophilum LMG 11574 possessed the β-fructofuranosidase gene under investigation.
DISCUSSION
The large variety that exists among bifidobacteria concerning the ability to degrade oligofructose and inulin has been intriguing scientists for more than 15 years (13, 40). Variations in bifidobacterial fructan degradation mechanisms are translated on a physiological level to differences in substrate suitability and fermentation rate (7, 19, 41, 49). A better understanding of this variability would allow a more accurate prediction of the prebiotic impact of different inulin-type fructans on the colon microbiota and would facilitate the design of prebiotics with a higher degree of selectivity (27, 36).
In the present study, the kinetics of growth, carbohydrate consumption, and metabolite production of 18 Bifidobacterium strains, growing on fructose, oligofructose, or inulin, were monitored and analyzed in detail. These results, combined with the outcome of a genetic screen for the presence of a β-fructofuranosidase gene that was first encountered in B. animalis subsp. lactis DSM 10140T, led—after statistical analysis—to the identification of four main clusters of bifidobacteria. The boundaries of these clusters did not correspond with species limits, confirming the previously reported observation that interspecies variations in bifidobacterial fructan breakdown abilities are common (40).
A first cluster (cluster A), comprised of B. bifidum and B. breve, was not able to metabolize inulin-type fructans. Bacteria belonging to this cluster might benefit indirectly from the fermentation of oligofructose and inulin by other members of the gut microbiota, for example, through the lowering of the intestinal pH (39) or the increased availability of monosaccharides as short-living substrates for cross-feeding in the colon as a result of fructan degradation by others (10, 25). However, it seems unlikely that their numbers would increase substantially after the intake of these prebiotics (10). Nevertheless, caution remains needed in extrapolating results of in vitro studies under optimal conditions to the complexity of the highly competitive gut ecosystem (43).
The presence of B. bifidum DSM 20082 in this cluster is remarkable, as this strain was recently identified as being able to degrade inulin (40). Previous studies confirmed the poor growth of B. bifidum on inulin-type fructans (13, 33), but strain-to-strain differences in β-fructofuranosidase production levels have been reported (40). The species B. bifidum appeared to occupy a singular position in the Bifidobacterium genus, reflected by the fact that its members were the only bifidobacteria included in the present study that grew poorly on RCM. Also, they were the only strains that did not produce ethanol and formate in the expected 1:2 molar ratio (44), suggesting the use of a sugar breakdown pathway uncommon among bifidobacteria. As mucin degraders, B. bifidum strains have been reported to hold a specific ecological niche in the human colon (32).
Another noteworthy constituent of cluster A is the commercial probiotic strain B. breve Yakult. Although the findings of this study by no means affect the beneficial probiotic properties attributed to this strain, they suggest that B. breve Yakult might not be suited for the development of a synbiotic in combination with inulin-type fructans. Indeed, the ability of bifidobacteria to metabolize oligofructose intracellularly or at the cell surface is speculated to be a conditio sine qua non for prebiotic stimulation (8, 18, 46).
Strains belonging to clusters B, C, and D shared the ability to degrade oligofructose, but intercluster variation was obvious. Cluster B was composed mainly of strains that displayed preferential degradation of the short fractions of oligofructose, a breakdown pattern that has previously been reported as characteristic for bifidobacteria and linked with cell-associated fructan degradation (8, 45, 46). The somewhat particular fructan breakdown characteristics of B. dentium LMG 10507—a cluster B strain—are possibly related to the oral cavity, the particular ecological habitat where this strain is commonly encountered. Detailed analysis of oligofructose degradation by cluster C bifidobacteria revealed a nondiscriminative breakdown mechanism, acting on all different chain length fractions simultaneously. In some cases, an increase in the free fructose concentration in the fermentation medium was observed. Similar breakdown patterns have up to now been reported only for nonbifidobacterial species and are generally associated with extracellular oligofructose degradation (8, 29, 46). In the present study, results of metabolite production analyses indicate a different mechanism. As reported previously, a metabolic shift toward more acetate, formate, and ethanol production—at the expense of lactate production—can be noted when bifidobacteria are grown on less readily fermentable substrates (8, 44-46). The latter is associated with more ATP production, resulting in a more efficient use of the available energy source (44, 45). Although the present study largely confirmed these findings, it was demonstrated that the nature of these less readily fermentable substrates differed between various clusters of bifidobacteria. Whereas strains belonging to cluster B produced substantially more lactate and less acetate, formate, and ethanol on fructose, oligofructose proved to be the preferential energy source for cluster C bifidobacteria. The latter observations contradict the idea of initial extracellular oligofructose degradation followed by the uptake of free fructose. They seem to indicate the occurrence of simultaneous extracellular degradation of the longest oligofructose fractions—as revealed by increasing free fructose concentrations—and uptake and subsequent intracellular degradation of the shorter ones. The capacity to partially degrade inulin, shared by all cluster C bifidobacteria, could offer an additional advantage to these strains for survival and proliferation in a highly competitive ecosystem such as the human colon (10, 25).
Cluster D bifidobacteria were characterized by short substrate depletion times on fructose or oligofructose, in the latter case not allowing discrimination between preferential and nonpreferential degradation mechanisms. Analysis of metabolite production revealed a minor preference for fructose as an energy source. Strains belonging to this group appeared to combine the efficient fructose consumption system displayed by cluster B bifidobacteria with the ability to partially degrade inulin, a typical cluster C characteristic, making them promising study objects for further research regarding the bifidogenic effect of inulin-type fructans.
None of the strains included in the present study was able to degrade all chain length fractions of inulin. This confirms earlier findings of low bifidobacterial β-fructofuranosidase activity toward long-chain inulin molecules (19, 25, 41). A recent study including 55 strains belonging to 11 Bifidobacterium species reported on the partial breakdown of inulin, with the noteworthy exception of B. adolescentis ALB 1, a strain capable of degrading all fractions (40).
In a previous study, the kinetics of growth, carbohydrate consumption, and metabolite production by B. animalis subsp. lactis DN-173 010, a commercial probiotic strain, were investigated on a range of different substrates under circumstances comparable to the ones applied here (45). The strain has been shown to possess a preferential oligofructose degradation mechanism and proven unable to metabolize inulin, two characteristics typical of cluster B bifidobacteria. However, its inability to grow on monosaccharides such as fructose would ensure a high rating on the PC1 axis, possibly granting B. animalis subsp. lactis DN-173 010 an outlier position.
The distribution of the strains possessing the β-fructofuranosidase gene under investigation (7) was random over all clusters. Even some of the strains that were not able to degrade oligofructose tested positive for the gene, probably indicating low levels of gene expression or the lack of an adequate fructan uptake system. A large variety exists among bifidobacteria, both in fructofuranosidases (7, 19, 41, 49) and in transport systems for oligofructose (23, 34, 42). Moreover, various fructan degradation mechanisms are thought to be active in the same strain. Study of the genome of B. longum NCC2705 revealed that this strain is equipped with more than 40 glycosylhydrolases that are predicted to be involved in the degradation of higher-order oligosaccharides (23, 34, 42). Also, this strain possesses at least nine transport systems involved in the uptake of oligofructose. In spite of this large genotypic variety, less variation seems to exist on the phenotypic level, as revealed by the present study. Since this phenotypic differentiation probably reflects niche-specific adaptation, it seems rather unlikely that each of the previously described clusters is equally sensitive toward stimulation by prebiotic inulin-type fructans. However, the question of which strains will benefit most from oligofructose or inulin consumption remains unanswered. The use of species-specific primers for the quantitative monitoring of the bifidobacterial gut population during in vivo trials with prebiotics is therefore recommended (27, 30).
A thorough study of bifidobacterial fructan metabolism will provide a solid framework for the determination/prediction of which bifidobacterial species and even strains will be stimulated by the addition of inulin or oligofructose to the diet (27). This might prove of great importance if prebiotics were to be applied for immunomodulative purposes (36). Furthermore, as many commercialized probiotics are bifidobacteria (28), it has always seemed tempting to combine both a prebiotic and a probiotic in a so-called synbiotic, a term derived from the word “synergy,” expressing the hope that the combined effect of both functional food ingredients will be larger than the sum of their individual contributions (37). However, it remains unclear which Bifidobacterium species are fit to be used in a synbiotic preparation (18). The framework mentioned here could turn out to be a valuable tool for the rational selection of a suitable probiotic-prebiotic combination.
The present study is the first kinetic approach to investigate growth, carbohydrate consumption, and metabolite production of bifidobacteria growing on fructose, oligofructose, and inulin. Combined with detailed analyses of both oligofructose and inulin degradation, it revealed the existence of a limited number of phenotypically distinct clusters among the tested strains. None of the species included in this study was able to degrade inulin completely. Two clusters could degrade inulin partially, varying among each other in the efficiency of fructose consumption. One group displayed preferential degradation of the short fractions of oligofructose, a feature previously reported for bifidobacteria, but was not able to metabolize inulin. Another group was not capable of degrading inulin-type fructans and displayed growth only on fructose. Further in vivo studies using strain- or species-specific primers to investigate stimulation of bifidobacteria by inulin-type fructans are needed to clarify the significance of the variation observed.
Supplementary Material
Acknowledgments
This research was funded by the Research Council of the Vrije Universiteit Brussel, the Fund for Scientific Research-Flanders (FWO-AL418), the Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT-Vlaanderen; GBOU project IWT-010054 [Development of a fast, noninvasive technological tool to investigate the functionality and effectiveness of pro- and prebiotics in normal healthy humans: the use of a labeled biomarker]), Institute Danone, and Yakult Belgium. Gwen Falony was the recipient of a Ph.D. grant from the IWT-Vlaanderen.
We thank Thomas Calmeyn and the analytical laboratory team of BENEO-Orafti NV for their support.
Footnotes
Published ahead of print on 14 November 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bosscher, D., J. Van Loo, and A. Franck. 2006. Inulin and oligofructose as prebiotics in the prevention of intestinal infections and diseases. Nutr. Res. Rev. 19:216-226. [DOI] [PubMed] [Google Scholar]
- 3.Duncan, S. H., R. I. Aminov, K. P. Scott, P. Louis, T. B. Stanton, and H. J. Flint. 2006. Proposal of Roseburia faecis sp. nov., Roseburia hominis sp. nov., and Roseburia inulinovorans sp. nov., based on isolates from human feces. Int. J. Syst. Evol. Microbiol. 56:2437-2441. [DOI] [PubMed] [Google Scholar]
- 4.Duncan, S. H., P. Louis, and H. J. Flint. 2007. Cultivable bacterial diversity from the human colon. Lett. Appl. Microbiol. 44:343-350. [DOI] [PubMed] [Google Scholar]
- 5.Duncan, S. H., K. P. Scott, A. G. Ramsay, H. J. M. Harmsen, G. W. Welling, C. S. Stewart, and H. J. Flint. 2003. Effects of alternative dietary substrates on competition between human colonic bacteria in an anaerobic fermentor system. Appl. Environ. Microbiol. 69:1136-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ehrmann, M. A., M. Korakli, and R. F. Vogel. 2003. Identification of the gene for beta-fructofuranosidase of Bifidobacterium lactis DSM10140T and characterization of the enzyme expressed in Escherichia coli. Curr. Microbiol. 46:391-397. [DOI] [PubMed] [Google Scholar]
- 8.Falony, G., A. Vlachou, K. Verbrugghe, and L. De Vuyst. 2006. Cross-feeding between Bifidobacterium longum BB536 and acetate-converting, butyrate-producing colon bacteria during growth on oligofructose. Appl. Environ. Microbiol. 72:7835-7841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.FAO/WHO. 2001. Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. Report of a joint FAO/WHO expert consultation. World Health Organization, Geneva, Switzerland. http://www.who.int/foodsafety/publications/fs_management/en/probiotics.pdf.
- 10.Flint, H. J., S. H. Duncan, K. P. Scott, and P. Louis. 2007. Interactions and competition within the microbial community of the human colon: links between diet and health. Environ. Microbiol. 9:1101-1111. [DOI] [PubMed] [Google Scholar]
- 11.Gibson, G. R., H. M. Probert, J. A. E. Van Loo, R. A. Rastall, and M. B. Roberfroid. 2004. Dietary modulation of the human colonic microbiota: updating the concept of prebiotics. Nutr. Res. Rev. 17:259-275. [DOI] [PubMed] [Google Scholar]
- 12.Gibson, G. R., and M. B. Roberfroid. 1995. Dietary modulation of the human colonic microbiota—introducing the concept of prebiotics. J. Nutr. 125:1401-1412. [DOI] [PubMed] [Google Scholar]
- 13.Gibson, G. R., and X. Wang. 1994. Bifidogenic properties of different types of fructo-oligosaccharides. Food Microbiol. 11:491-498. [Google Scholar]
- 14.Gibson, G. R., and X. Wang. 1994. Enrichment of bifidobacteria from human gut contents by oligofructose using continuous culture. FEMS Microbiol. Lett. 118:121-127. [DOI] [PubMed] [Google Scholar]
- 15.Goh, Y. J., C. Zhang, A. K. Benson, V. Schlegel, J.-H. Lee, and R. W. Hutkins. 2006. Identification of a putative operon involved in fructooligosaccharide utilization by Lactobacillus paracasei. Appl. Environ. Microbiol. 72:7518-7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartemink, R., K. M. J. VanLaere, and F. M. Rombouts. 1997. Growth of enterobacteria on fructo-oligosaccharides. J. Appl. Microbiol. 83:367-374. [DOI] [PubMed] [Google Scholar]
- 17.Hold, G. L., A. Schwiertz, R. I. Aminov, M. Blaut, and H. J. Flint. 2003. Oligonucleotide probes that detect quantitatively significant groups of butyrate-producing bacteria in human feces. Appl. Environ. Microbiol. 69:4320-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huebner, J., R. L. Wehling, and R. W. Hutkins. 2007. Functional activity of commercial prebiotics. Int. Dairy J. 17:770-775. [Google Scholar]
- 19.Janer, C., L. M. Rohr, C. Pelaez, M. Laloi, V. Cleusix, T. Requena, and L. Meile. 2004. Hydrolysis of oligofructoses by the recombinant β-fructofuranosidase from Bifidobacterium lactis. Syst. Appl. Microbiol. 27:279-285. [DOI] [PubMed] [Google Scholar]
- 20.Joye, D., and H. Hoebregs. 2000. Determination of oligofructose, a soluble dietary fiber, by high-temperature capillary gas chromatography. J. AOAC Int. 83:1020-1025. [PubMed] [Google Scholar]
- 21.Kaplan, H., and R. W. Hutkins. 2000. Fermentation of fructooligosaccharides by lactic acid bacteria and bifidobacteria. Appl. Environ. Microbiol. 66:2682-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaplan, H., and R. W. Hutkins. 2003. Metabolism of fructooligosaccharides by Lactobacillus paracasei 1195. Appl. Environ. Microbiol. 69:2217-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klijn, A., A. Mercenier, and F. Arigoni. 2005. Lessons from the genomes of bifidobacteria. FEMS Microbiol. Rev. 29:491-509. [DOI] [PubMed] [Google Scholar]
- 24.Langlands, S. J., M. J. Hopkins, N. Coleman, and J. H. Cummings. 2004. Prebiotic carbohydrates modify the mucosa-associated microflora of the human large bowel. Gut 53:1610-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Louis, P., K. P. Scott, S. H. Duncan, and H. J. Flint. 2007. Understanding the effects of diet on bacterial metabolism in the large intestine. J. Appl. Microbiol. 102:1197-1208. [DOI] [PubMed] [Google Scholar]
- 26.Macfarlane, G. T., and J. H. Cummings. 1991. The colonic flora, fermentation and large bowel digestive function, p. 51-92. In S. F. Phillips, J. H. Pemberton, and R. G. Shorter (ed.), The large intestine: physiology, pathophysiology and disease. Raven Press Ltd., New York, NY.
- 27.Macfarlane, S., G. T. Macfarlane, and J. H. Cummings. 2006. Prebiotics in the gastrointestinal tract. Aliment. Pharmacol. Ther. 24:701-714. [DOI] [PubMed] [Google Scholar]
- 28.Makras, L., L. Avonts, and L. De Vuyst. 2004. Probiotics, prebiotics, and gut health, p. 416-482. In C. Remacle and B. Reusens (ed.), Functional foods: ageing and degenerative disease. Woodhead Publishing Ltd., Cambridge, United Kingdom.
- 29.Makras, L., G. Van Acker, and L. De Vuyst. 2005. Lactobacillus paracasei subsp. paracasei 8700:2 degrades inulin-type fructans exhibiting different degrees of polymerization. Appl. Environ. Microbiol. 71:6531-6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuki, T., K. Watanabe, J. Fujimoto, Y. Kado, T. Takada, K. Matsumoto, and R. Tanaka. 2004. Quantitative PCR with 16S rRNA-gene-targeted species-specific primers for analysis of human intestinal bifidobacteria. Appl. Environ. Microbiol. 70:167-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCartney, A. L., W. Z. Wang, and G. W. Tannock. 1996. Molecular analysis of the composition of the bifidobacterial and Lactobacillus microflora of humans. Appl. Environ. Microbiol. 62:4608-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McWilliam Leitch, E. C., A. W. Walker, S. H. Duncan, G. Holtrop, and H. J. Flint. 2007. Selective colonization of insoluble substrates by human faecal bacteria. Environ. Microbiol. 9:667-679. [DOI] [PubMed] [Google Scholar]
- 33.Mitsuoka, T. 1984. Taxonomy and ecology of bifidobacteria. Bifidobacteria Microflora 3:11-28. [Google Scholar]
- 34.Parche, S., J. Amon, I. Jankovic, E. Rezzonico, M. Beleut, H. Barutcu, I. Schendel, M. P. Eddy, A. Burkovski, F. Arigoni, and F. Titgemeyer. 2007. Sugar transport systems of Bifidobacterium longum NCC2705. J. Mol. Microbiol. Biotechnol. 12:9-19. [DOI] [PubMed] [Google Scholar]
- 35.Picard, C., J. Fioramonti, A. Francois, T. Robinson, F. Neant, and C. Matuchansky. 2005. Bifidobacteria as probiotic agents—physiological effects and clinical benefits. Aliment. Pharmacol. Ther. 22:495-512. [DOI] [PubMed] [Google Scholar]
- 36.Rastall, R. A., G. R. Gibson, H. S. Gill, F. Guarner, T. R. Klaenhammer, B. Pot, G. Reid, I. R. Rowland, and M. E. Sanders. 2005. Modulation of the microbial ecology of the human colon by probiotics, prebiotics and synbiotics to enhance human health: an overview of enabling science and potential applications. FEMS Microbiol. Ecol. 52:145-152. [DOI] [PubMed] [Google Scholar]
- 37.Rastall, R. A., and V. Maitin. 2002. Prebiotics and synbiotics: towards the next generation. Curr. Opin. Biotechnol. 13:490-496. [DOI] [PubMed] [Google Scholar]
- 38.Roberfroid, M. B. 2005. The gastrointestinal system: a major target for functional foods, p. 17-36. In M. B. Roberfroid and I. Wolinsky (ed.), Inulin-type fructans: functional food ingredients. CRC Press LCC, Boca Raton, FL.
- 39.Roberfroid, M. B. 2005. Inulin-type fructans and the modulation of the intestinal microflora, p. 151-181. In M. B. Roberfroid and I. Wolinsky (ed.), Inulin-type fructans: functional food ingredients. CRC Press LCC, Boca Raton, FL.
- 40.Rossi, M., C. Corradini, A. Amaretti, M. Nicolini, A. Pompei, S. Zanoni, and D. Matteuzzi. 2005. Fermentation of fructooligosaccharides and inulin by bifidobacteria: a comparative study of pure and fecal cultures. Appl. Environ. Microbiol. 71:6150-6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ryan, S. M., G. F. Fitzgerald, and D. van Sinderen. 2005. Transcriptional regulation and characterization of a novel β-fructofuranosidase-encoding gene from Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 71:3475-3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M. C. Zwahlen, F. Desiere, P. Bork, M. Delley, R. D. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steer, T., H. Carpenter, K. Tuohy, and G. R. Gibson. 2000. Perspectives on the role of the human gut microbiota and its modulation by pro- and prebiotics. Nutr. Res. Rev. 13:229-254. [DOI] [PubMed] [Google Scholar]
- 44.Van der Meulen, R., T. Adriany, K. Verbrugghe, and L. De Vuyst. 2006. Kinetic analysis of bifidobacterial metabolism reveals a minor role for succinic acid in the regeneration of NAD+ through its growth-associated production. Appl. Environ. Microbiol. 72:5204-5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van der Meulen, R., L. Avonts, and L. De Vuyst. 2004. Short fractions of oligofructose are preferentially metabolized by Bifidobacterium animalis DN-173 010. Appl. Environ. Microbiol. 70:1923-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van der Meulen, R., L. Makras, K. Verbrugghe, T. Adriany, and L. De Vuyst. 2006. In vitro kinetic analysis of oligofructose consumption by Bacteroides and Bifidobacterium spp. indicates different degradation mechanisms. Appl. Environ. Microbiol. 72:1006-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vanhoutte, T., G. Huys, E. De Brandt, and J. Swings. 2004. Temporal stability analysis of the microbiota in human feces by denaturing gradient gel electrophoresis using universal and group-specific 16S rRNA gene primers. FEMS Microbiol. Ecol. 48:437-446. [DOI] [PubMed] [Google Scholar]
- 48.Wang, X., and G. R. Gibson. 1993. Effects of the in vitro fermentation of oligofructose and inulin by bacteria growing in the human large intestine. J. Appl. Bacteriol. 75:373-380. [DOI] [PubMed] [Google Scholar]
- 49.Warchol, M., S. Perrin, J. P. Grill, and F. Schneider. 2002. Characterization of a purified β-fructofuranosidase from Bifidobacterium infantis ATCC 15697. Lett. Appl. Microbiol. 35:462-467. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.