Abstract
Listeria monocytogenes is a gram-positive, food-borne pathogen that causes disease in both humans and animals. There are three major genetic lineages of L. monocytogenes and 13 serovars. To further our understanding of the differences that exist between different genetic lineages/serovars of L. monocytogenes, we analyzed the global protein expression of the serotype 1/2a strain EGD and the serotype 4b strain F2365 during early-stationary-phase growth at 37°C. Using multidimensional protein identification technology with electrospray ionization tandem mass spectrometry, we identified 1,754 proteins from EGD and 1,427 proteins from F2365, of which 1,077 were common to both. Analysis of proteins that had significantly altered expression between strains revealed potential biological differences between these two L. monocytogenes strains. In particular, the strains differed in expression of proteins involved in cell wall physiology and flagellar biosynthesis, as well as DNA repair proteins and stress response proteins.
Listeria monocytogenes is a gram-positive, facultative intracellular pathogen and is the causative agent of listeriosis. It is an opportunistic food-borne pathogen that can cause life-threatening infections, including meningitis, septicemia, miscarriage, and fetal death (52). L. monocytogenes is responsible for nearly 28% of all food-related deaths in the United States (31, 33, 52). Those that are most susceptible include immunocompromised individuals, the elderly, pregnant women, and neonates (44).
L. monocytogenes has been divided into three genetic lineages using ribotyping, sequence variations of known virulence genes, and multilocus sequence typing (34, 47, 55). Lineage I contains serovars 1/2b, 3b, and 4b, and lineage II contains serovars 1/2a, 1/2c, and 3c. Almost all of the major food-borne epidemics of listeriosis have been caused by strains in serovar 4b (41, 55), and lineage I contains a significantly higher proportion of human isolates than do the other divisions (21, 35, 41). Many human clinical isolates are also present in lineage II, and in particular, serotype 1/2a is prevalent among food isolates (19, 29). However, listeriosis cases caused by lineage II isolates tend to be more sporadic and not associated with epidemics.
DNA microarrays using L. monocytogenes strains from different lineage groups demonstrated that many genes have diverged between the lineages and revealed that lineage I is a more clonal population than lineage II (3, 6, 12). The genomes of two strains representing epidemic clonal groups within serovar 4b (F2365 and H7858) have been sequenced, along with two serovar 1/2a strains (EGD and F6854) (20, 39). Comparative genomics between lineage I (serovar 4b) and lineage II (serovar 1/2a) showed that all of the previously identified virulence factors were common to all L. monocytogenes strains; thus, many of these 4b-specific genes encode either unknown proteins or poorly characterized surface proteins or transcriptional regulators (39). Most of the genomic differences between the strains were phage insertions, transposable elements, and single nucleotide polymorphisms. Comparison of how listerial strains respond to stationary phase at the transcriptome level revealed differences between the two lineages in cell wall synthesis, the stress-related sigma B regulon, and virulence-related genes (50).
Conducting comparisons between lineage I and lineage II listerial strains at the genome sequence level and at the transcriptome level is not sufficient to allow identification of the virulence factors that enable strains to cause epidemic listeriosis. To better understand the differences between epidemic clonal groups and sporadic disease isolates, there is a need to compare how they respond to the environment at the protein level. In the current study, we examined the expressed proteomes of serovar 4b isolate F2365 and serovar 1/2a isolate EGD during early stationary phase at 37°C using multidimensional protein identification technology (MuDPIT) (56). Our results demonstrate that excellent coverage of the L. monocytogenes proteome can be achieved using MuDPIT, and they provide a map of the expressed proteomes of these strains during early stationary phase. By identifying proteins that are orthologous between the strains, our results further show that MuDPIT may be an effective tool for conducting quantitative comparisons of protein expression between F2365 and EGD. The results of our quantitative comparisons reveal similarities and differences in how the two strains respond to the same environment, and they lay the groundwork for future work to compare how lineage I and lineage II listerial isolates respond to changes in their environment.
MATERIALS AND METHODS
Bacterial growth conditions and protein isolations.
Colonies from freshly streaked plates of L. monocytogenes strains F2365 (serotype 4b) and EGD (serotype 1/2a) were used to inoculate 2-ml starter cultures of each strain in brain heart infusion (BHI) medium, which were then grown for 8 h aerobically at 37°C with rotary aeration to ensure that the two strains were equally acclimated to medium conditions. Following this initial period, cultures were diluted 1:100 in 20 ml of BHI medium and growth was allowed to continue aerobically at 37°C overnight (17 h) with rotary aeration to an optical density at 600 nm of 1.3 to 1.4 (cell concentrations were ∼5 × 109 CFU/ml for both strains). At this point, the entire culture was pelleted by centrifugation (5,656 × g for 10 min at 4°C) and resuspended in 4 ml of lysis solution (2% Triton X-100, 2.6 mg/ml sodium azide, 0.1 M Tris [pH 8.0], 8 mM phenylmethanesulfonyl fluoride). Lysozyme (2 mg/ml) was added, followed by incubation for 2 h at 37°C. Bacteria were then sonicated with four 30-s pulses (Fisher Scientific Model 100 Sonic Dismembrator, setting 3) on ice with 1 min of cooling between pulses. Samples were treated with 85 μg/ml DNase I and 20 μg/ml RNase A for 30 min at 37°C, and cell debris was pelleted by centrifugation (18,000 × g for 5 min at 4°C). Supernatant containing the protein was precipitated with an equal volume of 50% trichloroacetic acid overnight at −20°C. Precipitated protein was pelleted by centrifugation at 18,000 × g for 5 min at 10°C, washed with ice-cold acetone (Chromosolv for high-pressure liquid chromatography; Sigma-Aldrich), and then dried at room temperature.
Protein was resuspended in 0.5 ml of solubilization solution {7 M urea, 20 mM Tris-Cl, pH 8.0, 5 mM EDTA, 5 mM MgCl2, 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 1 mM phenylmethanesulfonyl fluoride}. Following quantitation of solubilized protein using the 2-D Quant kit (Amersham Biosciences), 0.1 mg of protein was precipitated with an equal volume of 50% trichloroacetic acid for 1 h at −20°C. Precipitated protein was pelleted, washed twice with ice-cold acetone, and resuspended in 0.1 ml of 100 mM ammonium bicarbonate and 5% acetonitrile. Protein was treated with 5 mM dithiothreitol for 10 min at 65°C, followed by treatment with 10 mM iodoacetamide for 30 min at 30°C. Protein was digested for 15 h with 2 μg sequencing-grade trypsin at 37°C. Tryptic digestions were stopped by decreasing the pH to 4.0 with acetic acid. Peptides were desalted using a peptide macrotrap (Michrom Bioresources, Inc.), and eluted samples were dried at room temperature and stored at −80°C until further needed. Before samples were processed through two-dimensional (2D) liquid chromatography (LC)-tandem mass spectrometry (MS-MS), purified peptides were resuspended in 0.02 ml of 0.1% formic acid and 5% acetonitrile. Proteins were isolated from EGD and F2365 from three independent experiments.
Protein analysis.
MuDPIT analysis was performed on three replicates of EGD and F2365 by using strong cation exchange followed by reverse-phase chromatography coupled directly in line with an electrospray ionization (ESI) ion trap tandem mass spectrometer (LCQ; ThermoElectron Corp.) exactly as previously described (37). The salt gradient applied in this study was applied in steps of 0, 10, 15, 20, 25, 30, 35, 40, 45, 50, 57, 64, 90, and 700 mM ammonium acetate in 5% acetonitrile-0.1% formic acid. The reverse-phase gradient used 0.1% formic acid in acetonitrile. The acetonitrile concentration was increased in a linear gradient from 5% to 30% in 20 min and then 30% to 95% in 7 min, followed by 5% for 10 min for the 0, 10, 15, 25, 30, 45, 64, 90, and 700 mM salt gradient steps. For the 20, 35, 40, 50, and 57 mM salt gradient steps, acetonitrile concentration was increased in a linear gradient from 5% to 40% in 65 min, 95% for 15 min, and 5% for 20 min.
All database searches of tandem mass spectra were performed using TurboSEQUEST (Bioworks Browser 3.2; ThermoElectron) (14). Mass spectra and tandem mass spectra were searched against the appropriate in silico trypsin-digested protein database of L. monocytogenes strain F2365 or EGD downloaded from the National Center for Biotechnology Information (NCBI). Cysteine carbamidomethylation and methionine oxidation (single and double) were included in the search criteria. We used the reverse database functionality in Bioworks 3.2 and searched MS-MS data for each strain against the appropriate reversed database using the same search criteria as described above. We assigned probabilities to peptide identifications by calculating the composite score derived from the SEQUEST Xcorr and ΔCn for all peptides from the real and reverse database searches (32, 40). We combined real and reverse database peptides and ranked them based on the composite score. We used the distribution of the scores to calculate the probability for each peptide identification to be derived from the real database (D. Kunec, B. Nanduri, L. A. Hanson, and S. C. Burgess, presented at the 54th ASMS Conference on Mass Spectrometry, Seattle, WA, 2006). Protein probabilities were then calculated as previously described using only peptides with P ≤ 0.01 (32, 40).
Protein comparisons.
Because F2365 and EGD were sequenced by two different research groups (20, 39), the annotation (protein names) varied greatly between the two strains. Therefore, to facilitate differential expression analysis, we first identified orthologous proteins between EGD and F2365 by identifying reciprocal-best-BLAST hits. To estimate the quantity of each protein, we used our published label-free quantification method based on the sum of Xcorrs of all the peptides from each protein (38). ProtQuant was used to identify significant changes in orthologous protein expression between EGD and F2365 (4). ProtQuant is a custom program that calculates the mean sum of Xcorrs of all the identified peptides from all three replicates for each protein and then conducts one-way analysis of variance (α ≤ 0.05) to identify statistically significant differences in protein expression between treatments.
RESULTS
Proteome profiles of EGD and F2365.
Based on their respective genome annotations, EGD (NC_003210) has 2,846 putative protein-encoding genes and F2365 (NC_002973) has 2,821 putative protein-encoding genes. However, the names for the protein-encoding genes from one strain did not correspond to the names provided for the other strain. We therefore generated a list of orthologous proteins between EGD and F2365 by conducting reciprocal-BLAST searches. We identified 2,608 orthologous proteins (reciprocal-best-BLAST hits) between EGD and F2365. However, we could not identify F2365 orthologs for 238 EGD proteins, and 213 F2365 proteins had no EGD orthologs.
To determine the proteins expressed during stationary phase for F2365 and EGD, we conducted 2D LC ESI MS-MS with proteins isolated from these strains. Using this method, we identified 1,427 proteins in F2365 (50.5% of the predicted proteome) and 1,754 proteins in EGD (61.6% of the predicted proteome). Together, a total of 2,104 L. monocytogenes proteins were identified. Based on the list of EGD-F2365 orthologous proteins, we determined that the two strains expressed a common set of 1,077 proteins during growth in BHI medium.
A thorough analysis of the biological functions represented by the identified proteins requires identification of the functions of these proteins. We evaluated three different sources for obtaining protein function: Clusters of Orthologous Groups (COGs), Gene Ontology (GO), and the functional classification codes from ListiList (http://genolist.pasteur.fr/ListiList/index.html). We did not find COG assignments for 24% of identified proteins from EGD and F2365. Furthermore, 19% of EGD and F2365 proteins that did have a COG designation were assigned to either the category “general function prediction only” (COG-R) or the category “function unknown” (COG-S) (data not shown). Overall, COGs did not contain functional information for nearly 41% of our data (854 out of the total protein set of 2,104).
Using GO annotation from Uniprot (http://beta.uniprot.org/), we found that fewer proteins from our data set had assigned functions with GO than when they were analyzed by COGs. GO was available for approximately 63% of identified EGD and F2365 proteins. Additionally, we found that many of these classifications were based on eukaryotic functions and cellular locations and were therefore not relevant for our analysis (data not shown).
By contrast, using the functional classification codes provided by ListiList, we were able to assign a classification code to 92% of the identified proteins from F2365 and 96% of the identified proteins from EGD. Approximately 28% of ListiList classification codes were similar to unknown proteins (category 5) or not similar to other proteins (category 6). Overall, the ListiList database provided information for 68% of our total proteins (1,435 out of the total 2,104 proteins). This functional coverage was better than that of COGs or GO. Furthermore, because ListiList classification codes are specific for listerial species, they are more relevant than those generated by GO or COGs for analysis of L. monocytogenes proteins. Therefore, we used ListiList functional categories for data analysis in this study.
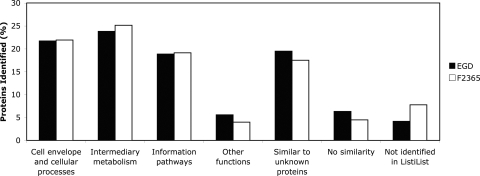
Using ListiList's functional classifications, the proteins identified in EGD and F2365 were divided into six general categories (Fig. 1). These six categories were as follows: 1, cell envelope and cellular processes; 2, intermediary metabolism; 3, information pathways; 4, other functions; 5, proteins that are similar to unknown proteins; and 6, proteins with no similarity to other proteins. We added an additional category (7) to indicate proteins whose functions were not identified through ListiList. The two strains showed similar patterns in coverage of these seven functional categories relative to the total number of proteins present in that category for that strain. The best coverage for both EGD and F2365 was in intermediary metabolism, and the lowest coverage was in the category labeled “other functions” (Fig. 1).
FIG. 1.
Percentages of proteins identified relative to the set of proteins with ListiList classification. Percentages (y axis) were calculated for both strains and classified according to the six categories provided by ListiList and a seventh category including those proteins that were not identified by ListiList.
The genome annotation of EGD had a larger number of proteins classified as “hypothetical” than did that of F2365. This could be because the EGD genome was sequenced earlier than F2365 (2001 versus 2004, respectively); therefore, less protein function information was available when the EGD sequence was annotated. However, for many of the proteins that were classified as hypothetical in EGD, we were able to transfer annotation from orthologous genes in F2365. There was one protein classified as hypothetical in F2365 for which we were able to transfer annotation from EGD; this protein was classified as a putative cell surface protein in EGD.
Proteins common to EGD and F2365.
A common set of 1,077 proteins was expressed by both EGD and F2365 when cultivated at 37°C in BHI broth. Of these proteins, 5% were not classified by ListiList (59 out of 1,077 proteins). In addition, 223 of the common set of proteins that were classified by ListiList were assigned to either category 5 or 6, and both categories represent unknown function classifications. The intermediary metabolism category accounted for 286 (26%) of the proteins from the set common to both EGD and F2365. The cell envelope and cellular processes category accounted for 240 (22%) of the common proteins (Table 1, common set).
TABLE 1.
Protein classifications of EGD-specific proteins, F2365-specific proteins, and the common set of proteins based on ListiList categories
| Function or description | ListiList category no. | No. of proteins
|
||
|---|---|---|---|---|
| EGD | F2365 | Common | ||
| Cell envelope and cellular processes | 1 | 141 | 73 | 240 |
| Cell wall | 1.1 | 24 | 10 | 43 |
| Transport/binding proteins and lipoproteins | 1.2 | 77 | 40 | 105 |
| Sensors (signal transduction) | 1.3 | 5 | 0 | 10 |
| Membrane bioenergetics | 1.4 | 7 | 4 | 21 |
| Mobility and chemotaxis | 1.5 | 3 | 3 | 12 |
| Protein secretion | 1.6 | 0 | 1 | 7 |
| Cell division | 1.7 | 6 | 3 | 9 |
| Cell surface proteins | 1.8 | 19 | 12 | 31 |
| Soluble internalin | 1.9 | 0 | 0 | 2 |
| Transformation/competence | 1.10 | 0 | 0 | 0 |
| Intermediary metabolism | 2 | 132 | 73 | 286 |
| Metabolism of carbohydrates and related molecules | 2.1 | 53 | 24 | 125 |
| Specific pathways | 2.1.1 | 50 | 24 | 104 |
| Main glycolytic pathways | 2.1.2 | 2 | 0 | 16 |
| Tricarboxylic acid cycle | 2.1.3 | 1 | 0 | 3 |
| Metabolism of amino acids and related molecules | 2.2 | 32 | 27 | 63 |
| Metabolism of nucleotides and nucleic acids | 2.3 | 6 | 6 | 35 |
| Metabolism of lipids | 2.4 | 16 | 6 | 23 |
| Metabolism of coenzymes and prosthetic groups | 2.5 | 23 | 10 | 36 |
| Metabolism of phosphate | 2.6 | 2 | 0 | 4 |
| Information pathways | 3 | 107 | 49 | 224 |
| DNA replication | 3.1 | 7 | 4 | 11 |
| DNA restriction/modification and repair | 3.2 | 12 | 2 | 17 |
| DNA recombination | 3.3 | 5 | 1 | 10 |
| DNA packaging and segregation | 3.4 | 1 | 0 | 10 |
| RNA synthesis | 3.5 | 54 | 26 | 91 |
| Initiation | 3.5.1 | 3 | 1 | 0 |
| Regulation | 3.5.2 | 48 | 25 | 81 |
| Elongation | 3.5.3 | 2 | 0 | 8 |
| Termination | 3.5.4 | 1 | 0 | 2 |
| RNA modification | 3.6 | 8 | 4 | 14 |
| Protein synthesis | 3.7 | 15 | 9 | 57 |
| Ribosomal proteins | 3.7.1 | 11 | 6 | 27 |
| Aminoacyl-tRNA synthetases | 3.7.2 | 3 | 2 | 19 |
| Initiation | 3.7.3 | 0 | 0 | 5 |
| Elongation | 3.7.4 | 1 | 1 | 4 |
| Termination | 3.7.5 | 0 | 0 | 2 |
| Protein modification | 3.8 | 4 | 3 | 9 |
| Protein folding | 3.9 | 1 | 0 | 5 |
| Other functions | 4 | 53 | 12 | 45 |
| Adaptation to atypical conditions | 4.1 | 2 | 6 | 22 |
| Detoxification | 4.2 | 9 | 2 | 9 |
| Phage-related functions | 4.3 | 28 | 2 | 3 |
| Transposon and insertion sequence | 4.4 | 10 | 1 | 1 |
| Miscellaneous | 4.5 | 4 | 1 | 10 |
| Similar to unknown proteins | 5 | 163 | 71 | 179 |
| From Listeria | 5.1 | 20 | 5 | 11 |
| From other organisms | 5.2 | 143 | 66 | 168 |
| No similarity | 6 | 67 | 20 | 44 |
| Not identified in ListiList | 7 | 14 | 52 | 59 |
Of the proteins common to the two strains that have intermediary metabolic functions, 86 were involved in fatty acid, amino acid, and lipid metabolism. In addition, 125 of the proteins were involved in carbohydrate metabolism. Proteins involved in glycolysis, the pentose phosphate pathway, and electron transport were expressed by both strains. Nucleotide/nucleic acid biosynthesis was also found to be conserved in both strains (Table 1).
Several information pathways were found to be similar for both EGD and F2365. For instance, 148 proteins in both strains were found to be involved in RNA synthesis and protein synthesis (Table 1). Additionally, DNA replication was conserved; DNA polymerase I, DNA polymerase III, primase, and DNA gyrase were expressed in both strains.
A set of proteins that corresponded to the phosphoenolpyruvate:carbohydrate phosphotransferase (PTS) system was expressed in both EGD and F2365. In fact, 3% of the commonly expressed proteins were classified as being part of the PTS system (35 out of 1,077). The PTS system functions to transport sugars into the cell and phosphorylate sugars during this transport process (43). Cytoplasmic IIA components, as well as membrane-bound components IIB and IIC, were identified in both strains. The heat-stable protein HPr, which is required for the initial transport and phosphorylation of the carbohydrates, was also expressed by both strains (43).
The two strains shared many similarities in membrane protein expression. For instance, 13 cell wall surface anchor proteins were found in both EGD and F2365, nine of which were classified as having the LPXTG motif (5). Two cell wall-bound internalins with LPXTG motifs were expressed by both EGD and F2365: internalin A and another internalin family protein (gi: 46908017). Internalins are virulence proteins involved in cell invasion. There were also three cell division proteins that were shared between the two strains (DivIC, FtsY, and FtsZ). Additionally, the cell shape-determining protein MreB was also expressed in both F2365 and EGD. Another cell division-related protein that both strains expressed was the septation ring formation regulator EzrA, even though expression levels differed between these two strains.
There were several proteins with activities related to oxidative stress that were expressed in both EGD and F2365. Some of these proteins included thiol peroxidase, catalase, superoxide dismutase, coproporphyrinogen III oxidase, peroxide resistance protein, and several different oxidoreductases. The expression of these proteins during stationary phase indicates that these strains are prepared for surviving environments where oxidative damage is likely to occur.
Another interesting feature shared between EGD and F2365 was that both strains expressed several types of helicases. Both EGD and F2365 expressed the DNA helicases PcrA, RecG, RecQ, and DinG and the RNA helicases DbpA and DeaD. A majority of these identified helicases have important roles in bacterial DNA repair (RecG, RecQ, DinG, and DbpA) (1, 23, 28, 53). PcrA has been suggested to work with the recombination proteins RecFOR, yet its precise function has not yet been identified (42). Other repair proteins that were expressed in both EGD and F2365 were RecO, RecD, RecR, RecJ, RecN, and photolyase.
Finally, expression of several PrfA-regulated proteins was detected in both strains, including listeriolysin, actin-assembly inducing protein (ActA), phosphatidylinositol phospholipase C (PI-PLC), and phosphatidylcholine phospholipase C.
Analysis of proteins specific to either EGD or F2365.
The results of our reciprocal BLAST searches between EGD and F2365 showed that 141 of the expressed EGD proteins did not have orthologous protein-encoding genes in F2365. Of these, 77% (109 out of 141) were classified as hypothetical proteins in EGD. F2365 had a total of 79 expressed proteins that did not have an orthologous protein-encoding gene in EGD, and 49 (62%) of these were classified as hypothetical (see the table in the supplemental material).
Several of the expressed EGD proteins that lacked orthologous protein-encoding genes in the F2365 genome were bacteriophage proteins. In total, EGD expressed 28 bacteriophage proteins (Table 1). By contrast, F2365 expressed five phage-related proteins according to the ListiList classification codes (three of these were also expressed by EGD). However, the F2365 “phage-related” proteins were named as hypothetical proteins in the F2365 annotation, so it is not certain that they are truly phage proteins. Eleven of the EGD bacteriophage proteins were A118-specific proteins (30), which are known to be encoded in the EGD genome (39).
Another protein detected in EGD that does not have an ortholog in F2365 is internalin B, which is truncated in F2365 because inlB has a premature stop codon due to a point mutation.
Analysis of orthologous proteins differentially expressed by EGD or F2365.
Of the proteins that had orthologous protein-encoding genes in EGD and F2365, expression of 413 was significantly different between the two strains under the culture conditions that we tested (see the table in the supplemental material). Expression of 322 proteins was significantly increased in EGD relative to F2365, and expression of 91 was significantly decreased in EGD. The predominant differences in protein expression were in the cell envelope and cellular processes category; approximately 22% of the proteins differentially expressed by EGD and F2365 were in this category. Differences in this category were mostly seen in transport proteins, lipoproteins, and cell surface proteins (Table 1).
F2365 and EGD had several differences in the expression of cell wall physiology proteins (cell division, cell shape, and cell wall biosynthesis proteins). EGD had a significantly higher expression of several cell division proteins than did F2365, including DNA segregation ATPase FtsK/SpoIIIE protein, septation ring formation regulator EzrA, a cell wall hydrolase (autolysin), and N-acetylmuramoyl-l-alanine amidase protein. EGD also expressed significantly more of the cell shape-determining protein MreB. F2365 expressed significantly more of the cell division protein FtsQ. Several cell wall biosynthesis enzymes had increased expression in EGD compared to F2365, including GlmS, which catalyzes the first step in hexosamine biosynthesis; d-alanyl-d-alanine carboxypeptidase; UDP-N-acetylglucosamine 1-carboxyvinyltransferase (MurA); and a penicillin binding protein.
Another difference in expression of cell wall physiology proteins was observed with cell wall anchor proteins. EGD expressed significantly more of five of these cell wall anchor proteins, while F2365 expressed significantly more of one cell wall anchor protein. All of these cell wall-bound proteins have a predicted LPXTG motif. While the exact function of these cell wall-associated proteins is not currently known, it is reasonable to expect that the differences in cell wall-associated protein expression patterns between strains could be related to differences in pathogenicity or antigenicity between strains due to their location at the cell surface.
There was also good evidence that EGD expressed more flagellar structural proteins than did F2365. EGD expressed significantly more flagellar hook-associated protein, flagellar motor switch protein, flagellar M-ring protein, and flagellum-specific ATP synthase than did F2365. By contrast, F2365 expressed significantly more flagellar biosynthesis protein (gi: 46906930). EGD also expressed more of the chemotaxis protein CheY than did F2365. Both EGD and F2365 had nine flagellar proteins that were expressed by the two strains.
EGD had a higher expression of DNA repair proteins than did F2365. For instance, EGD had a significantly higher amount of UvrA and UvrC expressed than did F2365. UvrA and UvrC are components of an endonuclease enzyme complex involved in nucleotide excision repair (49). EGD expressed significantly more 3-methyladenine DNA glycosylase, which catalyzes the first step in base excision repair by cleaving damaged DNA bases within double-stranded DNA to produce an abasic site. EGD had higher expression of the base excision repair protein uracil DNA glycosylase, and it expressed more PriA, which helps DNA polymerase II restart DNA synthesis immediately following UV exposure (46). EGD also expressed significantly more MutS, which recognizes mismatches to initiate the DNA repair process, and RecN, which repairs double-strand breaks. EGD had a higher amount of MutY expressed, which is part of the GO repair system that is largely involved in repair of oxidative damage (36). F2365 had a higher level of expression of the MutT repair protein, which is also involved in the GO repair system.
In addition to DNA repair enzymes, EGD expressed significantly more of several DNA metabolism enzymes than did F2365, including DNA ligase, DNA gyrase, DNA polymerase III, and DNA topoisomerase ParC. Three DNA helicases had significantly greater expression in EGD than in F2365: PcrA, which is a homolog of UvrD; a helicase that serves as the epsilon subunit of DNA polymerase III; and a helicase in the Snf2 family. EGD also expressed significantly more SbcC, which is part of a two-protein complex that cleaves hairpin structures that cause DNA replication to stall (10) and inserts double-strand breaks to remove DNA-bound protein (9).
EGD also had stronger expression of stress response proteins than did F2365 during stationary phase. There were significantly more of the chaperones GroEL, GroES, DnaK, ClpB, and GrpE in EGD, and there was significantly higher expression of a protein in the PfpI superfamily, which combines molecular chaperone and aminopeptidase activities and performs a protective function under a wide range of stress conditions. EGD also expressed a significantly higher amount of RsbR, which helps induce sigma B-mediated bacterial response to physical stress (48). EGD had significantly increased expression of osmotic stress proteins OsmC and Ctc (17) compared to F2365, and it had increased expression of oxidative stress proteins Dpr (involved in peroxide resistance), thioredoxin, and methionine sulfoxide reductase A (which reduces methionine sulfoxide). Two cold shock domain proteins also had increased expression in EGD compared to F2365. In addition, two acid tolerance proteins, glutamate decarboxylase (gamma subunit) and a putative lactoylglutathione lyase, also had significantly increased expression in EGD. F2365, on the other hand, expressed significantly higher amounts of sigma B activator RsbT as well as the oxidative stress proteins catalase and thiol peroxidase.
Interestingly, EGD expressed significantly higher amounts of HPr and phosphoenolpyruvate protein phosphotransferase. The genes encoding these proteins are cotranscribed, and expression is increased in the presence of glucose (8). Increased expression of these proteins induces expression of glucose catabolic enzymes and inhibits expression of PrfA-regulated virulence genes (11). Another interesting protein that was expressed only by EGD was the fibronectin binding protein. Fibronectin binding proteins allow L. monocytogenes to bind to fibronectin found associated with eukaryotic cells, which is an important step in establishing infection (18).
DISCUSSION
Listeria monocytogenes is a major threat to both the medical and food industries. Here, we characterized the proteins expressed during early stationary phase in BHI at 37°C in L. monocytogenes strains EGD and F2365. Strain F2365 is representative of serovar 4b epidemic isolates from lineage I, and strain EGD is representative of serovar 1/2a sporadic disease isolates from lineage II. Together, these two serovars contribute to a majority of reported cases of listeriosis (25).
Previous investigations of the L. monocytogenes proteome have largely used 2D gel electrophoresis followed by identification of individual spots by mass spectrometry (13, 16, 22, 24, 45, 54). In the current study, we used MuDPIT (2D LC directly inline with ESI and MS-MS) to evaluate the expressed proteome of EGD and F2365. Using this method, we achieved approximately 50 to 60% coverage of the predicted proteomes, which to our knowledge is the best-reported coverage of the L. monocytogenes proteome. Our protein isolation technique also allowed good coverage of membrane proteins. Importantly, this coverage of the L. monocytogenes proteome was achieved by analyzing 2 μg of trypsin-digested protein per replicate by 2D LC ESI MS-MS. By comparison, ≥60 to 100 μg is required for analysis by 2D gel electrophoresis (13, 22, 54). Our results demonstrate that MuDPIT is a practical and effective method for evaluating the L. monocytogenes proteome.
Because the two strains were cultivated under the same bacterial growth condition, we found that many proteins that conduct core metabolic functions were expressed in both EGD and F2365. Additionally, the presence of similar transport proteins suggests that the two strains utilize similar nutrients. Common DNA replication proteins and helicases indicate that DNA replication and chromosome segregation also proceed similarly between these two strains under these growth conditions.
Expression of several PrfA-regulated virulence proteins was detected, including listeriolysin, ActA, PI-PLC, and phosphatidylcholine phospholipase C. Typically, cultivation under either nutrient-restricted or iron-restricted medium is required to induce higher levels of expression of the genes encoding these proteins, particularly for hlyA (encoding listeriolysin) and plcA (encoding PI-PLC) (2, 15, 51). One possible explanation for the expression of these virulence proteins is that we harvested proteins during stationary phase, which is known to stimulate sigma B-mediated expression of PrfA-regulated virulence genes (26). In fact, there was evidence that a sigma B-mediated stress response was present in the current study; sigma B was detected from EGD, and several sigma B-activating proteins, including RbsR, RsbS, RsbT, and RsbU (7), were also detected.
The protein expression patterns detected in the current study are consistent with previously reported studies of Listeria monocytogenes during stationary phase. One study found a large number of differentially expressed protein spots in strain Scott A between exponential and stationary phase; however, only 10 spots were identified by peptide mass fingerprinting (54). In this previous study, LacI, GrpE, and superoxide dismutase were reported to have higher expression during stationary phase, and we detected all three of these proteins in the current study. Interestingly, we also detected expression of several proteins that Weeks et al. reported were either downregulated or not expressed during stationary phase, including elongation factor Ts, DNA polymerase III, HPr, Smc, and ATP synthase. Another study reported increased expression of some cellular metabolism proteins during stationary phase as well as proteins involved in stress adaptation (16). Many of the stress response proteins that they found with increased expression in stationary phase were detected in the current study, including ferritin, GroES, and listeriolysin.
Comparisons of the EGD and F2365 genomes have provided information on the genes that may play vital roles in strain-specific functions (20, 39). A previously reported comparison of strains from serovars 1/2a and 4b at the genome level showed that 83 genes were specific to serotype 1/2a and 51 genes were specific to the 4b serotype (39), and our reciprocal-BLAST searches between the EGD and F2365 genomes identified 238 proteins unique to EGD and 213 proteins unique to F2365. It could be predicted that many of these strain-specific genes allow the strains to adapt to their respective ecologic niches. However, to our surprise, we found that many strain-specific genes are expressed at the protein level during stationary phase: specifically, 141 EGD-specific proteins and 79 F2365-specific proteins were identified in the current study. Not surprisingly, approximately two-thirds of the EGD- and F2365-specific expressed proteins were hypothetical. More work remains to be done to determine the functions of these strain-specific proteins.
There were also a surprising number of proteins differentially expressed between the two strains, considering that they were grown under the same culture condition. Even though the two strains may perform basic cellular processes similarly, they are different in other characteristics. In particular, our data show that these two strains differ in expression of cell envelope proteins and flagellar proteins. Differences between outer structures of these two serotypes may help to determine the type of environment that is best suited for the microorganism or how it is presented to the host's immune response. These differences may help explain why serotype 4b strains are associated with outbreaks of human listeriosis while serotype 1/2a strains tend to cause isolated cases of human listeriosis (25).
Additionally, the two strains had differences in DNA repair and stress response proteins that were expressed during stationary phase. Although both strains expressed these proteins, many of them had significantly higher expression in EGD, suggesting that stationary phase may induce a stronger stress response in EGD than in F2365. Such proteins may play an important role in survival under stressful conditions associated with food processing. Further experimental analysis needs to be performed to verify whether the increased expression of these stress response proteins provides a greater tolerance to stress.
The data presented here provide the most comprehensive expressed proteome map to date for L. monocytogenes during early stationary phase. The current study also demonstrates that protein expression comparisons between lineage I and lineage II listerial strains can reveal similarities and differences in how they respond to the environment. Information from this type of study complements comparisons made at the genome sequence level and at the transcriptome level to identify virulence factors responsible for the epidemic potential of some listerial strains. The current report demonstrates the feasibility of using MuDPIT to analyze listerial protein expression in response to the host. In the future, we will use this method to study lineage I and lineage II adaptation to growth in the host environment to identify host-induced virulence factor expression.
Supplementary Material
Acknowledgments
This project was supported by the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service, grant 2007-35201-17732.
We thank Michelle Banes, Susan Bridges, Ranjit Kumar, and Chamali Thanthiriwatte for their assistance with this project. We also thank Tibor Pechan for his technical assistance with mass spectrometry. Also, we thank Fiona McCarthy and the AgBase group at MSU for their support and assistance with GO classifications.
Footnotes
Published ahead of print on 21 November 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bockmann, R., C. Dickneite, B. Middendorf, W. Goebel, and Z. Sokolovic. 1996. Specific binding of the Listeria monocytogenes transcriptional regulator PrfA to target sequences requires additional factor(s) and is influenced by iron. Mol. Microbiol. 22:643-653. [DOI] [PubMed] [Google Scholar]
- 2.Bohne, J., Z. Sokolovic, and W. Goebel. 1994. Transcriptional regulation of prfA and PrfA-regulated virulence genes in Listeria monocytogenes. Mol. Microbiol. 11:1141-1150. [DOI] [PubMed] [Google Scholar]
- 3.Borucki, M. K., M. J. Krug, W. T. Muraoka, and D. R. Call. 2003. Discrimination among Listeria monocytogenes isolates using a mixed genome DNA microarray. Vet. Microbiol. 92:351-362. [DOI] [PubMed] [Google Scholar]
- 4.Bridges, S. M., G. B. Magee, N. Wang, W. P. Williams, S. C. Burgess, and B. Nanduri. 2007. ProtQuant: a tool for the label-free quantification of MudPIT proteomics data. BMC Bioinformatics 8(Suppl. 7):S24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cabanes, D., P. Dehoux, O. Dussurget, L. Frangeul, and P. Cossart. 2002. Surface proteins and the pathogenic potential of Listeria monocytogenes. Trends Microbiol. 10:238-245. [DOI] [PubMed] [Google Scholar]
- 6.Call, D. R., M. K. Borucki, and T. E. Besser. 2003. Mixed-genome microarrays reveal multiple serotype and lineage-specific differences among strains of Listeria monocytogenes. J. Clin. Microbiol. 41:632-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaturongakul, S., and K. J. Boor. 2004. RsbT and RsbV contribute to σB-dependent survival under environmental, energy, and intracellular stress conditions in Listeria monocytogenes. Appl. Environ. Microbiol. 70:5349-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen, D. P., A. K. Benson, and R. W. Hutkins. 1998. Cloning and expression of the Listeria monocytogenes Scott A ptsH and ptsI genes, coding for HPr and enzyme I, respectively, of the phosphotransferase system. Appl. Environ. Microbiol. 64:3147-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connelly, J. C., E. S. de Leau, and D. R. Leach. 2003. Nucleolytic processing of a protein-bound DNA end by the E. coli SbcCD (MR) complex. DNA Repair (Amsterdam) 2:795-807. [DOI] [PubMed] [Google Scholar]
- 10.Connelly, J. C., L. A. Kirkham, and D. R. Leach. 1998. The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc. Natl. Acad. Sci. USA 95:7969-7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deutscher, J., R. Herro, A. Bourand, I. Mijakovic, and S. Poncet. 2005. P-Ser-HPr—a link between carbon metabolism and the virulence of some pathogenic bacteria. Biochim. Biophys. Acta 1754:118-125. [DOI] [PubMed] [Google Scholar]
- 12.Doumith, M., C. Cazalet, N. Simoes, L. Frangeul, C. Jacquet, F. Kunst, P. Martin, P. Cossart, P. Glaser, and C. Buchrieser. 2004. New aspects regarding evolution and virulence of Listeria monocytogenes revealed by comparative genomics and DNA arrays. Infect. Immun. 72:1072-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duche, O., F. Tremoulet, A. Namane, and J. Labadie. 2002. A proteomic analysis of the salt stress response of Listeria monocytogenes. FEMS Microbiol. Lett. 215:183-188. [DOI] [PubMed] [Google Scholar]
- 14.Eng, J. K., A. L. McCormack, and J. R. Yates III. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976-989. [DOI] [PubMed] [Google Scholar]
- 15.Erdenlig, S., A. J. Ainsworth, and F. W. Austin. 2000. Pathogenicity and production of virulence factors by Listeria monocytogenes isolates from channel catfish. J. Food Prot. 63:613-619. [DOI] [PubMed] [Google Scholar]
- 16.Folio, P., P. Chavant, I. Chafsey, A. Belkorchia, C. Chambon, and M. Hebraud. 2004. Two-dimensional electrophoresis database of Listeria monocytogenes EGDe proteome and proteomic analysis of mid-log and stationary growth phase cells. Proteomics 4:3187-3201. [DOI] [PubMed] [Google Scholar]
- 17.Gardan, R., O. Duche, S. Leroy-Setrin, and J. Labadie. 2003. Role of ctc from Listeria monocytogenes in osmotolerance. Appl. Environ. Microbiol. 69:154-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilot, P., P. Andre, and J. Content. 1999. Listeria monocytogenes possesses adhesins for fibronectin. Infect. Immun. 67:6698-6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilot, P., A. Genicot, and P. Andre. 1996. Serotyping and esterase typing for analysis of Listeria monocytogenes populations recovered from foodstuffs and from human patients with listeriosis in Belgium. J. Clin. Microbiol. 34:1007-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 21.Gray, M. J., R. N. Zadoks, E. D. Fortes, B. Dogan, S. Cai, Y. Chen, V. N. Scott, D. E. Gombas, K. J. Boor, and M. Wiedmann. 2004. Listeria monocytogenes isolates from foods and humans form distinct but overlapping populations. Appl. Environ. Microbiol. 70:5833-5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guilbaud, M., I. Chafsey, M. F. Pilet, F. Leroi, H. Prevost, M. Hebraud, and X. Dousset. 2008. Response of Listeria monocytogenes to liquid smoke. J. Appl. Microbiol. 104:1744-1753. [DOI] [PubMed] [Google Scholar]
- 23.Hasegawa, S. L., P. W. Doetsch, K. K. Hamilton, A. M. Martin, S. A. Okenquist, J. Lenz, and J. M. Boss. 1991. DNA binding properties of YB-1 and dbpA: binding to double-stranded, single-stranded, and abasic site containing DNAs. Nucleic Acids Res. 19:4915-4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helloin, E., L. Jansch, and L. Phan-Thanh. 2003. Carbon starvation survival of Listeria monocytogenes in planktonic state and in biofilm: a proteomic study. Proteomics 3:2052-2064. [DOI] [PubMed] [Google Scholar]
- 25.Kathariou, S. 2002. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 65:1811-1829. [DOI] [PubMed] [Google Scholar]
- 26.Kazmierczak, M. J., S. C. Mithoe, K. J. Boor, and M. Wiedmann. 2003. Listeria monocytogenes σB regulates stress response and virulence functions. J. Bacteriol. 185:5722-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reference deleted.
- 28.Kusano, K., Y. Sunohara, N. Takahashi, H. Yoshikura, and I. Kobayashi. 1994. DNA double-strand break repair: genetic determinants of flanking crossing-over. Proc. Natl. Acad. Sci. USA 91:1173-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Latorre, L., A. Parisi, R. Fraccalvieri, G. Normanno, M. C. La Porta, E. Goffredo, L. Palazzo, G. Ciccarese, N. Addante, and G. Santagada. 2007. Low prevalence of Listeria monocytogenes in foods from Italy. J. Food Prot. 70:1507-1512. [DOI] [PubMed] [Google Scholar]
- 30.Loessner, M. J., R. B. Inman, P. Lauer, and R. Calendar. 2000. Complete nucleotide sequence, molecular analysis and genome structure of bacteriophage A118 of Listeria monocytogenes: implications for phage evolution. Mol. Microbiol. 35:324-340. [DOI] [PubMed] [Google Scholar]
- 31.Lynch, M., J. Painter, R. Woodruff, and C. Braden. 2006. Surveillance for foodborne-disease outbreaks—United States, 1998-2002. MMWR Surveill. Summ. 55(10):1-42. [PubMed] [Google Scholar]
- 32.MacCoss, M. J., C. C. Wu, and J. R. Yates III. 2002. Probability-based validation of protein identifications using a modified SEQUEST algorithm. Anal. Chem. 74:5593-5599. [DOI] [PubMed] [Google Scholar]
- 33.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meinersmann, R. J., R. W. Phillips, M. Wiedmann, and M. E. Berrang. 2004. Multilocus sequence typing of Listeria monocytogenes by use of hypervariable genes reveals clonal and recombination histories of three lineages. Appl. Environ. Microbiol. 70:2193-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mereghetti, L., P. Lanotte, V. Savoye-Marczuk, N. Marquet-Van Der Mee, A. Audurier, and R. Quentin. 2002. Combined ribotyping and random multiprimer DNA analysis to probe the population structure of Listeria monocytogenes. Appl. Environ. Microbiol. 68:2849-2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michaels, M. L., and J. H. Miller. 1992. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J. Bacteriol. 174:6321-6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nanduri, B., M. L. Lawrence, C. R. Boyle, M. Ramkumar, and S. C. Burgess. 2006. Effects of subminimum inhibitory concentrations of antibiotics on the Pasteurella multocida proteome. J. Proteome Res. 5:572-580. [DOI] [PubMed] [Google Scholar]
- 38.Nanduri, B., M. L. Lawrence, S. Vanguri, and S. C. Burgess. 2005. Proteomic analysis using an unfinished bacterial genome: the effects of subminimum inhibitory concentrations of antibiotics on Mannheimia haemolytica virulence factor expression. Proteomics 5:4852-4863. [DOI] [PubMed] [Google Scholar]
- 39.Nelson, K. E., D. E. Fouts, E. F. Mongodin, J. Ravel, R. T. DeBoy, J. F. Kolonay, D. A. Rasko, S. V. Angiuoli, S. R. Gill, I. T. Paulsen, J. Peterson, O. White, W. C. Nelson, W. Nierman, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, D. H. Haft, J. Selengut, S. Van Aken, H. Khouri, N. Fedorova, H. Forberger, B. Tran, S. Kathariou, L. D. Wonderling, G. A. Uhlich, D. O. Bayles, J. B. Luchansky, and C. M. Fraser. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 32:2386-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nesvizhskii, A. I., A. Keller, E. Kolker, and R. Aebersold. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75:4646-4658. [DOI] [PubMed] [Google Scholar]
- 41.Norton, D. M., J. M. Scarlett, K. Horton, D. Sue, J. Thimothe, K. J. Boor, and M. Wiedmann. 2001. Characterization and pathogenic potential of Listeria monocytogenes isolates from the smoked fish industry. Appl. Environ. Microbiol. 67:646-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petit, M. A., and D. Ehrlich. 2002. Essential bacterial helicases that counteract the toxicity of recombination proteins. EMBO J. 21:3137-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57:543-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramaswamy, V., V. M. Cresence, J. S. Rejitha, M. U. Lekshmi, K. S. Dharsana, S. P. Prasad, and H. M. Vijila. 2007. Listeria—review of epidemiology and pathogenesis. J. Microbiol. Immunol. Infect. 40:4-13. [PubMed] [Google Scholar]
- 45.Ramnath, M., K. B. Rechinger, L. Jansch, J. W. Hastings, S. Knochel, and A. Gravesen. 2003. Development of a Listeria monocytogenes EGDe partial proteome reference map and comparison with the protein profiles of food isolates. Appl. Environ. Microbiol. 69:3368-3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rangarajan, S., R. Woodgate, and M. F. Goodman. 2002. Replication restart in UV-irradiated Escherichia coli involving pols II, III, V, PriA, RecA and RecFOR proteins. Mol. Microbiol. 43:617-628. [DOI] [PubMed] [Google Scholar]
- 47.Rasmussen, O. F., P. Skouboe, L. Dons, L. Rossen, and J. E. Olsen. 1995. Listeria monocytogenes exists in at least three evolutionary lines: evidence from flagellin, invasive associated protein and listeriolysin O genes. Microbiology 141:2053-2061. [DOI] [PubMed] [Google Scholar]
- 48.Reeves, A., and W. G. Haldenwang. 2007. Isolation and characterization of dominant mutations in the Bacillus subtilis stressosome components RsbR and RsbS. J. Bacteriol. 189:1531-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Selby, C. P., and A. Sancar. 1990. Structure and function of the (A)BC excinuclease of Escherichia coli. Mutat. Res. 236:203-211. [DOI] [PubMed] [Google Scholar]
- 50.Severino, P., O. Dussurget, R. Z. Vencio, E. Dumas, P. Garrido, G. Padilla, P. Piveteau, J. P. Lemaitre, F. Kunst, P. Glaser, and C. Buchrieser. 2007. Comparative transcriptome analysis of Listeria monocytogenes strains of the two major lineages reveals differences in virulence, cell wall, and stress response. Appl. Environ. Microbiol. 73:6078-6088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sokolovic, Z., J. Riedel, M. Wuenscher, and W. Goebel. 1993. Surface-associated, PrfA-regulated proteins of Listeria monocytogenes synthesized under stress conditions. Mol. Microbiol. 8:219-227. [DOI] [PubMed] [Google Scholar]
- 52.Vazquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Dominguez-Bernal, W. Goebel, B. Gonzalez-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Voloshin, O. N., F. Vanevski, P. P. Khil, and R. D. Camerini-Otero. 2003. Characterization of the DNA damage-inducible helicase DinG from Escherichia coli. J. Biol. Chem. 278:28284-28293. [DOI] [PubMed] [Google Scholar]
- 54.Weeks, M. E., D. C. James, G. K. Robinson, and C. M. Smales. 2004. Global changes in gene expression observed at the transition from growth to stationary phase in Listeria monocytogenes ScottA batch culture. Proteomics 4:123-135. [DOI] [PubMed] [Google Scholar]
- 55.Wiedmann, M., J. L. Bruce, C. Keating, A. E. Johnson, P. L. McDonough, and C. A. Batt. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 65:2707-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolters, D. A., M. P. Washburn, and J. R. Yates III. 2001. An automated multidimensional protein identification technology for shotgun proteomics. Anal. Chem. 73:5683-5690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.