Abstract
In this study, we have constructed and expressed inverted repeat chimeras from the first exons of the six known hydrophobins of the fungus Cladosporium fulvum, the causal agent of tomato leaf mold. We used quantitative PCR to measure specifically the expression levels of the hydrophobins. The targeted genes are silenced to different degrees, but we also detected clear changes in the expression levels of nontargeted genes. This work highlights the difficulties that are likely to be encountered when attempting to silence more than one gene in a multigene family.
Most filamentous fungi produce hydrophobins, which are small moderately hydrophobic proteins that are characterized by a strictly conserved array of eight cysteines (36). Two distinct types have been identified: class I and class II hydrophobins. These proteins have been extensively reviewed in recent years and play important roles in the formation of aerial hyphae, spore production and dispersal, the stabilization of fruiting body structures, and the virulence of some pathogenic fungi (3, 12, 26, 27, 28, 29, 31, 32, 35-38). In many cases, individual fungal species have more than one hydrophobin gene. For example, the tomato pathogen Cladosporium fulvum has at least six hydrophobins: HCf-1, HCf-2, HCf-3, and HCf-4 are typical class I hydrophobins, and HCf-5 and HCf-6 are class II hydrophobins.
Targeted gene deletion of HCf-1 demonstrated that HCf-1 is a major determinant of conidial hydrophobicity and, thus, contributes to the ability of conidia to be dispersed by water (34). While no evident phenotype was observed in mutants lacking HCf-2, HCf-3, HCf-4, and HCf-6, we were unable to isolate a mutant lacking HCf-5. In the presence of a multigene family, it is possible that different hydrophobins complement each other. On the other hand, the inability to isolate a knockout is often considered to be an indication that a gene is essential for viability.
As an alternative to allelic replacement, RNA-mediated interference (RNAi) has emerged as an effective method for silencing gene expression (2, 15, 19, 30). Specific inhibition of gene expression by RNAi has been demonstrated in a range of organisms from an initial report on Caenorhabditis elegans (5), trypanosomes (39), Drosophila melanogaster (10), mammalian cells (4), and the yeast Cryptococcus neoformans (14). In fungi, RNAi has been demonstrated in Neurospora crassa, where inverted repeat transgenes forming hairpin RNAs are efficient activators of RNA silencing (7). Since RNA-dependent silencing occurs in C. fulvum (8), we decided to use this tool to analyze the function of the six hydrophobins by simultaneously silencing up to six hydrophobin genes in C. fulvum. In this paper, we report success in obtaining targeted gene silencing of selected hydrophobins, including the simultaneous silencing of all hydrophobins in individual transformants. However, we also highlight the unexpected effects on targeted and nontargeted genes, including gene overexpression, and discuss the limits when applying this technique.
Silencing hydrophobins by expression of inverted repeat RNA.
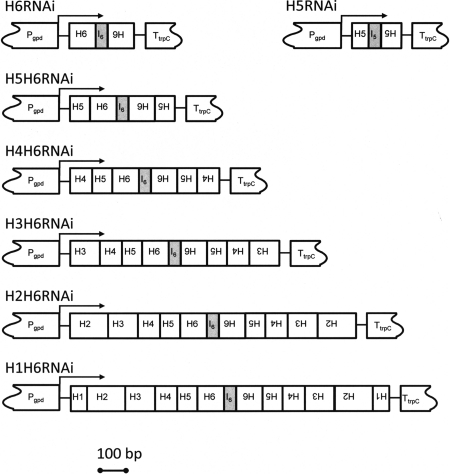
The silencing constructs were generated by first creating an inverted repeat insert which contained the first exon of HCf-6 followed by its first intron and then with the exon of HCf-6 again but in the reverse orientation. This was carried out using PCR amplification of the appropriate fragments from previously described cDNA clones (18, 24). The resulting sense-intron-antisense sequence was amplified by PCR and then cloned between the Pgpd promoter and the TtrpC terminator of Aspergillus nidulans derived from pAN7-1 (21); the plasmid was called pH6RNAi. This procedure was repeated to create pH5RNAi using the equivalent exon and intron of HCf-5. The first exons of the other hydrophobins (in order, HCf-5, HCf-4, HCf-3, HCf-2, and HCf-1) were added in succession to create the silencing constructs named pH5H6RNAi, pH4-H6RNAi, pH3-H6RNAi, pH2-H6RNAi, and pH1-H6RNAi, respectively. A diagram of the silencing constructs used in the cotransformation of C. fulvum is shown in Fig. 1. These plasmids were introduced into C. fulvum protoplasts by cotransformation with pAN7-1, a plasmid conferring resistance to hygromycin (21). The success rate of the cotransformation was between 23 and 32%. The transformants resistant to hygromycin were selected at random and called H5RNAi1 to -21, H6RNAi1 to -21, H5-H6RNAi1 to -21, H4-H6RNAi1 to -15, H3-H6RNAi1 to -21, H2-H6RNAi1 to -19, and H1-H6RNAi1 to -42 (Table 1). We used quantitative PCR (qPCR) to measure the expression levels of the targeted hydrophobins relative to their wild-type levels. Sequences of TaqMan primers and probes specific to the six hydrophobins and the 28S rRNA gene of C. fulvum were designed. Duplicate samples were quantified for all six hydrophobins, and mean values were used to express mRNA levels relative to that of the 28S rRNA gene.
FIG. 1.
Diagram of the silencing constructs used in this study. Seven silencing plasmids were constructed. The silencing approach consisted at first of individually targeting the silencing of HCf-5 and HCf-6 with pH5RNAi and pH6RNAi constructs, respectively. For the “all six hydrophobins silenced” construct, the exons of five hydrophobins were added progressively to the pH6RNAi construct. H1 to H6 represent the first exon of each hydrophobin in either the forward or reverse orientation (as shown). I5 and I6 are the first introns of HCf-5 and HCf-6, respectively. The transcription of all chimeric genes was driven by the Aspergillus nidulans Pgpd promoter and by the terminator TtrpC.
TABLE 1.
Silencing constructs and strains used in this study
| Silencing constructa | Targeted hydrophobin gene(s) | Strains |
|---|---|---|
| pH5RNAi | HCf-5 | H5RNAi1 to -21 |
| pH6RNAi | HCf-6 | H6RNAi1 to -21 |
| pH5-H6RNAi | HCf-5, HCf-6 | H5-H6RNAi1 to -21 |
| pH4-H6RNAi | HCf-4, HCf-5, HCf-6 | H4-H6RNAi1 to -15 |
| pH3-H6RNAi | HCf-3, HCf-4, HCf-5, HCf-6 | H3-H6RNAi1 to -21 |
| pH2-H6RNAi | HCf-2, HCf-3, HCf-4, HCf-5, HCf-6 | H2-H6RNAi1 to -19 |
| pH1-H6RNAi | HCf-1, HCf-2, HCf-3, HCf-4, HCf-5, HCf-6 | H1-H6RNAi1 to -42 |
The seven silencing constructs were cotransformed in C. fulvum protoplasts with pAN7-1, conferring hygromycin resistance (21). The transformants growing on hygromycin media were selected and named: transformation with pH5RNAi resulted in strains H5RNAi1 to -21.
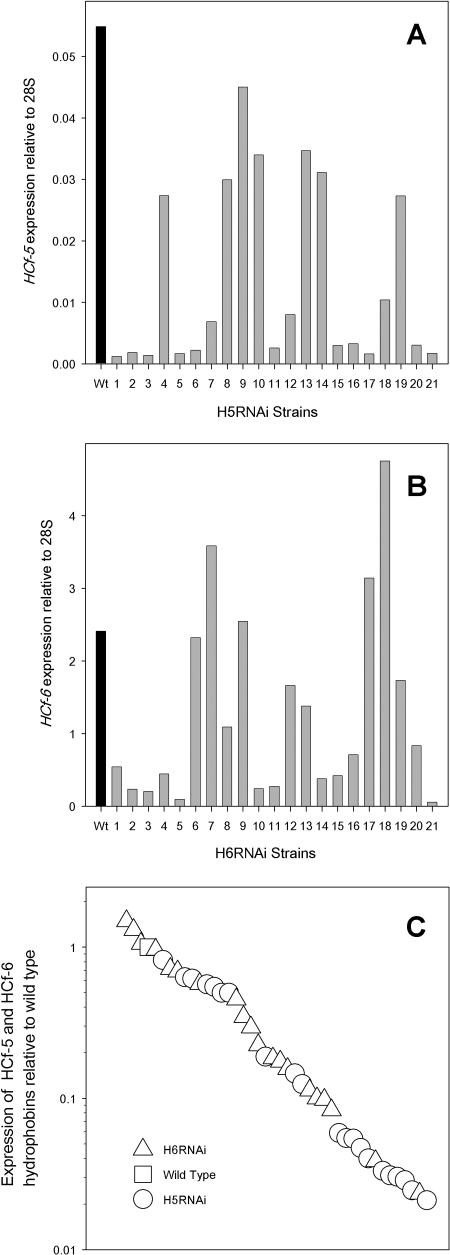
Initial experiments in which the individual class II hydrophobins HCf-5 and HCf-6 were targeted for silencing resulted in transgenic strains that displayed a wide range of levels for the respective RNAs (Fig. 2A and B). In the H5RNAi strains, HCf-5 was downregulated to some extent in all cases. In the H6RNAi strains, in addition to the silenced strains, some strains unexpectedly displayed increased levels of HCf-6 RNA. We plotted the expression levels of the targeted hydrophobins relative to those of the wild types of both the H5RNAi and the H6RNAi strains on the same graph (Fig. 2C). From this, it is evident that very similar ranges and distributions of silencing were obtained for both constructs. Overexpression of the targeted gene was also observed in the silencing genes of Venturia inaequalis (6). In our studies, the sequence used in silencing was not the same as the sequence used to measure RNA by qPCR; therefore, overexpression cannot be explained by the detection of the transgene RNA. We propose that some form of “compensation,” in which a feedback-regulatory mechanism enhances the expression of the targeted gene, is possible. In past experiments, we have tried repeatedly, and failed, to obtain C. fulvum strains lacking HCf-5 by gene knockout mediated by homologous recombination (J. R. Whiteford and P. D. Spanu, unpublished results), whereas we had no problems obtaining the gene knockout of the other hydrophobins (25, 33, 34). Failure to obtain mutants is sometimes equated to an indication that the genes are essential for survival. HCf-5 was expressed at low levels compared to those of the other six hydrophobins (unpublished data), and strong silencing (3% of the wild-type HCf-5 expression levels) was observed without affecting the growth of the fungus, which would suggest that HCf-5 is not essential for growth and that the failure to obtain a knockout may be due to, for example, the inaccessibility of the HCf-5 locus to homologous recombination. However, the silencing of Neurospora crassa pan-2 suggests that, in the case of genes expressed at low levels, the reduction of the mRNA steady-state level is not sufficient to recapitulate the phenotype of the pan-2 mutant strain (7). Therefore, because low levels of expression for the silenced strains might still be sufficient for gene activity and for gene function, the absence of phenotypes in silenced strains is actually of limited heuristic value and must be treated with caution.
FIG. 2.
RNA abundance of hydrophobins HCf-5 and HCf-6 in wild-type C. fulvum H5RNAi and H6RNAi transgenic strains, respectively. RNA was extracted from mycelium grown on agar plates. qPCR was used to determine the levels of hydrophobin RNA relative to that of the 28S rRNA gene (28S). (A) HCf-5 RNA in the wild type (Wt) and strains H5RNAi1 to -21; (B) HCf-6 RNA in the wild type and strains H6RNAi1 to -21; (C) the expression data for both HCf-5 and HCf-6 expression levels in the transgenic H5RNAi and H6RNAi strains, respectively, relative to the wild-type expression levels, plotted in rank order on a log axis.
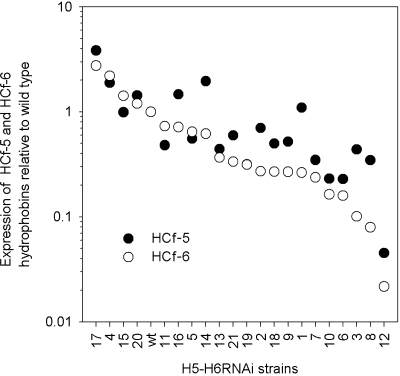
In addition to the single silencing of strains, we constructed multiple silenced H5-H6RNAi strains, H4-H6RNAi strains, H3-H6RNAi strains, H2-H6RNAi strains, and H1-H6RNAi strains. For H5-H6RNAi, C. fulvum strains transformed with pH5-H6RNAi showed a large range of HCf-5 and HCf-6 mRNA abundances which varied from two- to threefold overexpression of both hydrophobins in strain H5-H6RNAi17 to the silencing of HCf-6 down to 2% of the wild-type levels in strain H5-H6RNAi12 (Fig. 3). With regard to H4-H6RNAi, the transformants in which three hydrophobins (HCf-4, HCf-5, and HCf-6) were targeted also displayed a range of hydrophobin expression. This varied from the upregulation of individual hydrophobins to very strong silencing down to just over 0.1% of wild-type HCf-4 RNA. For H3-H6RNAi, H2-H6RNAi, and H1-H6RNAi, the transformants overall showed similar ranges of targeted hydrophobin RNA levels, which ranged from a maximum induction for HCf-3 (over 30-fold of the wild type) to a minimum induction of H1-H6RNAi for the same gene (just over 1% of the wild type). In experiments in which the number of target hydrophobins was larger, it was difficult to identify isolates in which all hydrophobins were silenced simultaneously. This was particularly evident in the H2-H6RNAi and H3-H6RNAi strains, where no silencing of HCf-5 and HCf-5/HCf6 was observed. In H1-H6RNAi, the hydrophobins HCf-2, HCf-3, HCf-4, HCf-5, and HCf-6 were silenced in about 50% of the cases, while HCf-1 was silenced in 24% of the isolates.
FIG. 3.
RNA abundance of HCf-5 and HCf-6 in C. fulvum H5-H6RNAi transgenic strains in which both hydrophobins are targeted simultaneously. The range of HCf-5 and HCf-6 expression levels in 21 strains relative to those of the wild type (wt) is plotted; strains are ranked in order of HCf-6 level.
The first evident conclusion from our observations is that there were wide variations (both up- and downregulation) in the hydrophobin levels in the multiply silenced transformants, as well as in the strains in which individual hydrophobins were targeted. This pattern has been found in other fungi, including Cryptococcus neoformans, Magnaporthe oryzae, Aspergillus fumigatus, and Histoplasma capsulatum (9, 14, 16, 22). The variability of silencing remains a problem in a number of eukaryotes, and it has been observed in other organisms submitted to RNAi like Drosophila melanogaster, Anopheles stephensi, Arabidopsis thaliana, and Chlamydomonas reinhardtii that the variability of the silencing level depends on the type of construct, transgene copy number, site of integration, and target gene (1, 11, 20, 23). We speculate that the level of silencing may be determined by a series of independent factors. For example, although the expression of the inverted repeat RNAs are all driven by the same A. nidulans GPD promoter, there are likely to be effects specific to the site of transgene integration which affect the expression levels of the silencing hairpin RNA.
Silencing of targeted hydrophobins affects the expression levels of nontargeted hydrophobins.
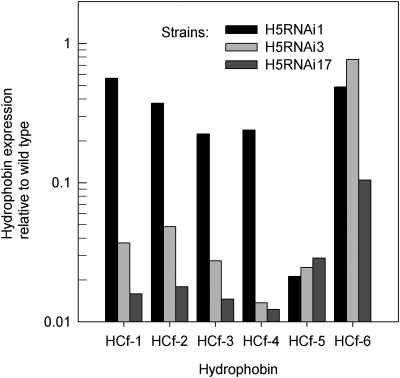
The effect of silencing specific hydrophobins on the expression levels of nontargeted hydrophobins was measured (Fig. 4). We selected three strains from each of the H5RNAi, H6RNAi, H5-H6RNAi, and H4-H6RNAi constructs. We show here detailed results of the analysis made of H5RNAi1, H5RNAi3, and H5RNAi17. The targeted HCf-5 was silenced down to about 3% of the wild-type levels in all three strains. Surprisingly, the expression of the nontargeted hydrophobins was also lower than that of the wild type, and the degree of cross-silencing varied widely across the strains.
FIG. 4.
RNA abundance of all six hydrophobins in three strains in which only HCf-5 was targeted by RNAi. The expression levels of HCf-1 to HCf-6 were measured in H5RNAi1, H5RNAi3, and H5RNAi17—all strains that showed strong silencing of the targeted HCf-5 hydrophobin. The levels of hydrophobin RNA are given relative to the respective levels of the wild-type C. fulvum strain.
Silencing hydrophobins during the life cycle of C. fulvum.
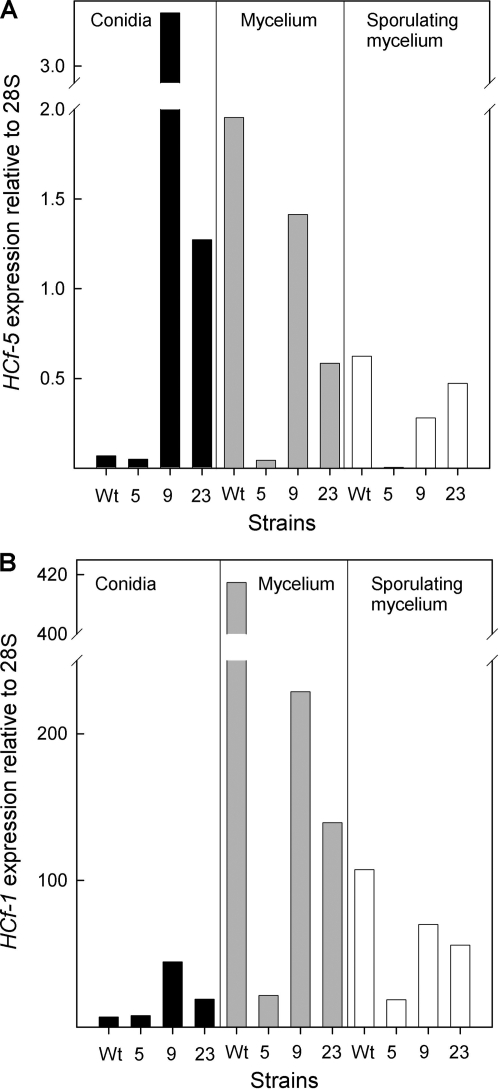
In the experiments described above, the hydrophobin levels were all determined in cultures grown in static liquid medium. In order to assess the effectiveness of the RNAi-induced silencing throughout the life cycle of C. fulvum, we monitored hydrophobin levels at different stages of the C. fulvum life cycle. Three stages were analyzed: conidium, mycelium (3 days after inoculation, i.e., prior to onset of conidiation), and conidiating mycelium (6 days after inoculation). We measured the relative amounts of HCf-1 and HCf-5 RNAs in the three strains H2-H5RNAi5, H2-H5RNAi9, and H2-H5RNAi23 and compared them to those of the wild type (Fig. 5). Interestingly, the expression levels of the gene targeted for silencing HCf-5 and the nontargeted gene HCf-1 showed the same profile: the expression level was lower than that of the wild type in all three strains in both mycelial stages, whereas in the conidium stage either the transformants showed marked overexpression (in H2-H5RNAi9 and -23) or expression was practically the same as in the wild type (in H2-H5RNAi5). This variation may be due to changes in the activity of the GPD promoter during development, in the level of expression of the endogenous hydrophobins, and in the efficiency of the proposed feedback-regulatory mechanisms mentioned above.
FIG. 5.
RNA abundance of HCf-5 (targeted) and HCf-1 (nontargeted) in a selection of H2-H6RNAi strains at different stages of development. The expression of HCf-5 RNA (A) and HCf-1 RNA (B) was determined by qPCR in isolated conidia, mycelium, and sporulated mycelium growing on solid media. The wild type (Wt) and three H2-H6RNAi strains, -5, -9, and -23, were used in this analysis, as they showed strong, weak, and moderate levels of silencing of the targeted HCf-5 hydrophobin, respectively, when measured during growth on agar medium.
Strains H2-H6RNAi5 and H1-H6RNAi23 in C. fulvum, the only strains presenting an “easily wettable” phenotype, were highly silenced for three of the class I hydrophobins. This phenotype was maintained throughout various rounds of subculturing. The silencing of the class I hydrophobins seems to be the cause of the “easily wettable” phenotype. This phenotype has already been observed in C. fulvum when cosuppressing HCf-1, in the knockout of HCf-1, and in the double knockouts of HCf-1 and HCf-2 (8, 34). The need for a high degree of silencing to observe a phenotype has already been reported by several authors, by whom it has been suggested that mRNA levels must drop below a threshold, which may be different for each protein depending on its mechanism of action and regulation (14, 22). In some cases, the reduction of the mRNA level for the targeted gene is not sufficient to induce the phenotype of the targeted gene mutant strain (7). Often, it is reported that there is a phenotypic difference between knockout and knockdown mutants. The hydrophobin mpg1 knockout mutant is known to be reduced in pathogenicity. However, most of the mpg1 knockdown mutants retained full pathogenicity (17). These observations indicate that although RNAi is successful in downregulating targeted genes, this method cannot be considered similar to knockout approaches employing complete gene replacement. This work highlights the difficulties that are likely to be encountered when attempting to silence more than one gene in a multigene family. We have demonstrated that there are serious issues related to variations of the levels of silencing for both targeted genes and related nontargeted genes whose primary sequences are not necessarily similar to those of the targeted ones (e.g., class I and class II hydrophobins). These variations may even differ at different stages of the life cycle. Therefore, studies in which gene function is investigated by RNAi-induced silencing need to be conducted in which such effects are monitored and taken into account when interpreting the results.
Footnotes
Published ahead of print on 14 November 2008.
REFERENCES
- 1.Brown, A. E., L. Bugeon, A. Crisanti, and F. Catteruccia. 2003. Stable and heritable gene silencing in the malaria vector Anopheles stephensi. Nucleic Acids Res. 31:E85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dykxhoorn, D., C. D. Novina, and P. A. Sharp. 2003. Killing the messenger: short RNAs that silence gene expression. Nat. Rev. Mol. Cell Biol. 4:457-467. [DOI] [PubMed] [Google Scholar]
- 3.Ebbole, D. J. 1997. Hydrophobins and fungal infection of plants and animals. Trends Microbiol. 5:405-408. [DOI] [PubMed] [Google Scholar]
- 4.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 5.Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, and S. E. Driver. 1998. Potent and specific genetic interference by double stranded RNA in Caenorhabditis elegans. Nature 391:806-811. [DOI] [PubMed] [Google Scholar]
- 6.Fitzgerarld, A., J. A. Van Kan, and K. M. Plummer. 2004. Simultaneous silencing of multiple genes in the apple scab fungus, Venturia inaequalis, by expression of RNA with chimeric inverted repeats. Fungal Genet. Biol. 41:963-971. [DOI] [PubMed] [Google Scholar]
- 7.Goldoni, M., G. Azzalin, G. Macino, and C. Cogoni. 2004. Efficient gene silencing by expression of double stranded RNA in Neurospora crassa. Fungal Genet. Biol. 41:1016-1024. [DOI] [PubMed] [Google Scholar]
- 8.Hamada, W., and P. Spanu. 1998. Cosuppression of the hydrophobin gene HCf-1 is correlated with antisense RNA biosynthesis in Cladosporium fulvum. Mol. Gen. Genet. 259:630-638. [DOI] [PubMed] [Google Scholar]
- 9.Kadotani, N., H. Nakayashiki, Y. Tosa, and S. Mayama. 2003. RNA silencing in the phytopathogenic fungus Magnaporthe oryzae. Mol. Plant-Microbe Interact. 16:769-776. [DOI] [PubMed] [Google Scholar]
- 10.Kennerdell, J. R., and R. W. Carthew. 1998. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell 95:1017-1026. [DOI] [PubMed] [Google Scholar]
- 11.Kerschen, A., C. A. Napoli, R. A. Jorgensen, and A. E. Muller. 2004. Effectiveness of RNA interference in transgenic plants. FEBS Lett. 566:223-228. [DOI] [PubMed] [Google Scholar]
- 12.Kershaw, M. J., and N. J. Talbot. 1998. Hydrophobins and repellents: proteins with fundamental roles in fungal morphogenesis. Fungal Genet. Biol. 23:18-33. [DOI] [PubMed] [Google Scholar]
- 13.Reference deleted.
- 14.Liu, H., T. R. Cottrell, L. M. Pierini, W. E. Goldman, and T. L. Doering. 2002. RNA interference in the pathogenic fungus Cryptococcus neoformans. Genetics 160:463-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahmood-ur-Rahman, I. Ali, T. Husnain, and S. Riazuddin. 2008. RNA interference: the story of gene silencing in plants and humans. Biotechnol. Adv. 26:202-209. [DOI] [PubMed] [Google Scholar]
- 16.Mouyna, I., C. Henry, T. L. Doering, and J. P. Latgé. 2004. Gene silencing with RNA interference in the human pathogenic fungus Aspergillus fumigatus. FEMS Microbiol. Lett. 237:317-324. [DOI] [PubMed] [Google Scholar]
- 17.Nakayashiki, H. 2005. RNA silencing in fungi: mechanisms and applications. FEBS Lett. 579:5950-5957. [DOI] [PubMed] [Google Scholar]
- 18.Nielsen, P. S., A. J. Clark, R. P. Oliver, M. Huber, and P. D. Spanu. 2001. HCf-6, a novel class II hydrophobin from Cladosporium fulvum. Microbiol. Res. 156:59-63. [DOI] [PubMed] [Google Scholar]
- 19.Paddison, P. J., J. M. Silva, D. S. Conklin, M. Schlabach, M. Li, S. Aruleba, V. Balija, A. O'Shaughnessy, L. Gnoj, K. Scobie, K. Chang, T. Westbrook, M. Clearly, R. Sachidanandam, W. R. McCombie, S. J. Elledge, and G. J. Hannon. 2004. A resource for large-scale RNA-interference-based screens in mammals. Nature 428:427-431. [DOI] [PubMed] [Google Scholar]
- 20.Piccin, A., A. Salameh, C. Benna, F. Sandrelli, G. Mazzotta, M. Zordan, E. Rosato, C. P. Kyriacou, and R. Costa. 2001. Efficient and heritable functional knock-out of an adult phenotype in Drosophila using a GAL4-driven hairpin RNA incorporating a heterologous spacer. Nucleic Acids Res. 29:E55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Punt, P. J., R. P. Oliver, M. A. Dingemanse, P. H. Pouwels, and C. A. M. J. J. van den Hondel. 1987. Transformation of Aspergillus based on the hygromycin-b resistance marker from Escherichia coli. Gene 56:117-124. [DOI] [PubMed] [Google Scholar]
- 22.Rappleye, C. A., J. T. Engle, and W. E. Goldman. 2004. RNA interference in Histoplasma capsulatum demonstrates a role for α-(1,3)-glucan in virulence. Mol. Microbiol. 53:153-165. [DOI] [PubMed] [Google Scholar]
- 23.Rohr, J., N. Sarkar, S. Balenger, B. R. Jeong, and H. Cerutti. 2004. Tandem inverted repeat system for selection of effective transgenic RNAi strains in Chlamydomonas. Plant J. 40:611-621. [DOI] [PubMed] [Google Scholar]
- 24.Segers, G. C., W. Hamada, R. P. Oliver, and P. D. Spanu. 1999. Isolation and characterisation of five different hydrophobin-encoding cDNAs from the fungal tomato pathogen Cladosporium fulvum. Mol. Gen. Genet. 261:644-652. [DOI] [PubMed] [Google Scholar]
- 25.Spanu, P. 1998. Deletion of HCf-1, a hydrophobin gene of Cladosporium fulvum, does not affect pathogenicity in tomato. Physiol. Mol. Plant Pathol. 52:323-334. [Google Scholar]
- 26.Stringer, M. A., and W. E. Timberlake. 1995. dewA encodes a fungal hydrophobin component of the Aspergillus spore wall. Mol. Microbiol. 16:33-44. [DOI] [PubMed] [Google Scholar]
- 27.Talbot, N. J. 1997. Fungal biology: growing into the air. Curr. Biol. 7:R78-R81. [DOI] [PubMed] [Google Scholar]
- 28.Talbot, N. J., D. J. Ebbole, and J. E. Hamer. 1993. Identification and characterization of Mpg1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell 5:1575-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tucker, S. L., and N. J. Talbot. 2001. Surface attachment and pre-penetration stage development by plant pathogenic fungi. Annu. Rev. Phytopathol. 39:385-417. [DOI] [PubMed] [Google Scholar]
- 30.Watson, J. M., A. F. Fusaro, M. Wang, and P. M. Waterhouse. 2005. RNA silencing platforms in plants. FEBS Lett. 579:5982-5987. [DOI] [PubMed] [Google Scholar]
- 31.Wessels, J. G. H. 1997. Hydrophobins: proteins that change the nature of the fungal surface. Adv. Microb. Physiol. 38:10-45. [DOI] [PubMed] [Google Scholar]
- 32.Wessels, J. G. H. 1999. Fungi in their own right. Fungal Genet. Biol. 27:134-145. [DOI] [PubMed] [Google Scholar]
- 33.Whiteford, J. R., H. Lacroix, N. J. Talbot, and P. D. Spanu. 2004. Stage-specific cellular localisation of two hydrophobins during plant infection by the pathogenic fungus Cladosporium fulvum. Fungal Genet. Biol. 41:624-634. [DOI] [PubMed] [Google Scholar]
- 34.Whiteford, J. R., and P. D. Spanu. 2001. The hydrophobin HCf-1 of Cladosporium fulvum is required for efficient water-mediated dispersal of conidia. Fungal Genet. Biol. 32:159-168. [DOI] [PubMed] [Google Scholar]
- 35.Whiteford, J. R., and P. D. Spanu. 2002. Hydrophobins and the interactions between fungi and plants. Mol. Plant Pathol. 3:391-400. [DOI] [PubMed] [Google Scholar]
- 36.Wösten, H. A. B. 2001. Hydrophobins: multipurpose proteins. Annu. Rev. Microbiol. 55:625-646. [DOI] [PubMed] [Google Scholar]
- 37.Wösten, H. A. B., and M. L. de Vocht. 2000. Hydrophobins, the fungal coat unravelled. Biochim. Biophys. Acta 1469:79-86. [DOI] [PubMed] [Google Scholar]
- 38.Wösten, H. A. B., M.-A. van Wetter, L. G. Lugones, H. C. van der Mei, H. J. Busscher, and J. G. H. Wessels. 1999. How a fungus escapes the water to grow into the air. Curr. Biol. 9:85-88. [DOI] [PubMed] [Google Scholar]
- 39.Zoraghi, R., and T. Seebeck. 2002. The cAMP-specific phosphodiesterase TbPDE2C is an essential enzyme in bloodstream form Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 99:4343-4348. [DOI] [PMC free article] [PubMed] [Google Scholar]